Virtual Docking Screening and QSAR Studies to Explore PI3K and mTOR Inhibitors Acting on AKT in Cancers
Article Information
Ilham kandoussi1*, Oussama Benherrif1, Wiame Lakhlili1, Jamal Taoufik2, Azeddine Ibrahimi1
1Laboratory of Biotechnology (MedBiotech), Faculty of Medicine and Pharmacy, Mohammed V University, Rabat, Morocco
2Laboratory of Therapeutic Chemistry, Faculty of Medicine and Pharmacy, Mohammed V University, Rabat, Morocco
*Corresponding Author: Ilham Kandoussi, Laboratory of Biotechnology (MedBiotech), Faculty of Medicine and Pharmacy, Mohammed V University, Rabat, Morocco
Received: 20 November 2020; Accepted: 11 December 2020; Published: 04 January 2021
Citation: Ilham kandoussi, Oussama Benherrif, Wiame Lakhlili, Jamal Taoufik, Azeddine Ibrahimi. Virtual Docking Screening and QSAR Studies to Explore PI3K and mTOR Inhibitors Acting on AKT in Cancers. Journal of Cancer Science and Clinical Therapeutics 5 (2021): 036-048.
View / Download Pdf Share at FacebookAbstract
The PI3K / AKT / mTOR pathway is an important regulator of a wide range of cellular processes including survival, proliferation, growth, metabolism, angiogenesis and metastasis. PI3K induces translocation of Ser / Thr Akt kinase, Akt is activated by PDK1 and mTOR2-dependent phosphorylations. Aberrant activation of the PI3K / AKT / mTOR pathway is frequently observed in many human malignancies and the combination of compounds simultaneously targeting different related molecules in the PI3K / AKT / mTOR pathway leads to synergistic activity. To explore the common AKT / mTOR and AKT / PI3K competitor ATP inhibitors we performed a 2D AKT-SAR model to predict the bioactivity of PI3K and mTOR inhibitors on AKT, the interaction of the best inhibitors was evaluated by docking analysis and compared to that of GSK690693 and Ipatasertib. An AKT-SAR model with a correlation coefficient (R2) of 0.85212 and an RMSE of 0.09266 was obtained, which was validated and evaluated by a cross-validation method LOO, the most predicted inhibitors of PI3K and mTOR respectively PIC50 activities between [9.30 - 8.54], and [9.79 - 8.91]. After docking and several comparisons, inhibitors with better predictions showed better affinity and interaction with AKT compared to GSK690693 and Ipatasertib. We therefore found that 8 PI3K inhibitors and 11 mTOR inhibitors met the Lipinski and Veber criteria and could be future drugs.
Keywords
QSAR; Virtual screening; PI3K/AKT/mTOR; Docking; Dual ATP inhibitors
QSAR articles; Virtual screening articles; PI3K/AKT/mTOR articles; Docking articles; Dual ATP inhibitors articles
QSAR articles QSAR Research articles QSAR review articles QSAR PubMed articles QSAR PubMed Central articles QSAR 2023 articles QSAR 2024 articles QSAR Scopus articles QSAR impact factor journals QSAR Scopus journals QSAR PubMed journals QSAR medical journals QSAR free journals QSAR best journals QSAR top journals QSAR free medical journals QSAR famous journals QSAR Google Scholar indexed journals Virtual screening articles Virtual screening Research articles Virtual screening review articles Virtual screening PubMed articles Virtual screening PubMed Central articles Virtual screening 2023 articles Virtual screening 2024 articles Virtual screening Scopus articles Virtual screening impact factor journals Virtual screening Scopus journals Virtual screening PubMed journals Virtual screening medical journals Virtual screening free journals Virtual screening best journals Virtual screening top journals Virtual screening free medical journals Virtual screening famous journals Virtual screening Google Scholar indexed journals PI3K/AKT/mTOR articles PI3K/AKT/mTOR Research articles PI3K/AKT/mTOR review articles PI3K/AKT/mTOR PubMed articles PI3K/AKT/mTOR PubMed Central articles PI3K/AKT/mTOR 2023 articles PI3K/AKT/mTOR 2024 articles PI3K/AKT/mTOR Scopus articles PI3K/AKT/mTOR impact factor journals PI3K/AKT/mTOR Scopus journals PI3K/AKT/mTOR PubMed journals PI3K/AKT/mTOR medical journals PI3K/AKT/mTOR free journals PI3K/AKT/mTOR best journals PI3K/AKT/mTOR top journals PI3K/AKT/mTOR free medical journals PI3K/AKT/mTOR famous journals PI3K/AKT/mTOR Google Scholar indexed journals Docking articles Docking Research articles Docking review articles Docking PubMed articles Docking PubMed Central articles Docking 2023 articles Docking 2024 articles Docking Scopus articles Docking impact factor journals Docking Scopus journals Docking PubMed journals Docking medical journals Docking free journals Docking best journals Docking top journals Docking free medical journals Docking famous journals Docking Google Scholar indexed journals Dual ATP inhibitors articles Dual ATP inhibitors Research articles Dual ATP inhibitors review articles Dual ATP inhibitors PubMed articles Dual ATP inhibitors PubMed Central articles Dual ATP inhibitors 2023 articles Dual ATP inhibitors 2024 articles Dual ATP inhibitors Scopus articles Dual ATP inhibitors impact factor journals Dual ATP inhibitors Scopus journals Dual ATP inhibitors PubMed journals Dual ATP inhibitors medical journals Dual ATP inhibitors free journals Dual ATP inhibitors best journals Dual ATP inhibitors top journals Dual ATP inhibitors free medical journals Dual ATP inhibitors famous journals Dual ATP inhibitors Google Scholar indexed journals angiogenesis articles angiogenesis Research articles angiogenesis review articles angiogenesis PubMed articles angiogenesis PubMed Central articles angiogenesis 2023 articles angiogenesis 2024 articles angiogenesis Scopus articles angiogenesis impact factor journals angiogenesis Scopus journals angiogenesis PubMed journals angiogenesis medical journals angiogenesis free journals angiogenesis best journals angiogenesis top journals angiogenesis free medical journals angiogenesis famous journals angiogenesis Google Scholar indexed journals metastasis articles metastasis Research articles metastasis review articles metastasis PubMed articles metastasis PubMed Central articles metastasis 2023 articles metastasis 2024 articles metastasis Scopus articles metastasis impact factor journals metastasis Scopus journals metastasis PubMed journals metastasis medical journals metastasis free journals metastasis best journals metastasis top journals metastasis free medical journals metastasis famous journals metastasis Google Scholar indexed journals human malignancies articles human malignancies Research articles human malignancies review articles human malignancies PubMed articles human malignancies PubMed Central articles human malignancies 2023 articles human malignancies 2024 articles human malignancies Scopus articles human malignancies impact factor journals human malignancies Scopus journals human malignancies PubMed journals human malignancies medical journals human malignancies free journals human malignancies best journals human malignancies top journals human malignancies free medical journals human malignancies famous journals human malignancies Google Scholar indexed journals threonine kinase articles threonine kinase Research articles threonine kinase review articles threonine kinase PubMed articles threonine kinase PubMed Central articles threonine kinase 2023 articles threonine kinase 2024 articles threonine kinase Scopus articles threonine kinase impact factor journals threonine kinase Scopus journals threonine kinase PubMed journals threonine kinase medical journals threonine kinase free journals threonine kinase best journals threonine kinase top journals threonine kinase free medical journals threonine kinase famous journals threonine kinase Google Scholar indexed journals solid tumors articles solid tumors Research articles solid tumors review articles solid tumors PubMed articles solid tumors PubMed Central articles solid tumors 2023 articles solid tumors 2024 articles solid tumors Scopus articles solid tumors impact factor journals solid tumors Scopus journals solid tumors PubMed journals solid tumors medical journals solid tumors free journals solid tumors best journals solid tumors top journals solid tumors free medical journals solid tumors famous journals solid tumors Google Scholar indexed journals
Article Details
Abbreviations:
PI3K- Phosphoinositide 3-Kinase; mTOR- Mammalian Target of Rapamycin; QSAR- Quantitative Structure-Activity Relationship; Ser- Serine; Thr- Threonine; LOO- Leave-one-out; PSA- Polar Surface Area; PLS- Partial Least Square
Background
The PI3K / AKT / mTOR pathway is hyperactivated or altered in many types of cancer and regulates a wide range of cellular processes, including survival, proliferation, growth, metabolism, angiogenesis, and metastasis. It is regulated by upstream signaling proteins and regulates many downstream effectors by collaborating with various compensatory signaling pathways [1]. There are three AKT isoforms (AKT1, AKT2 and AKT3), which, in general, are widely expressed. AKT is activated by a dual regulatory mechanism that requires both translocation to the plasma membrane and phosphorylation at Thr308 and Ser473 [2, 3]. The generation of PIP3 after PI3K activation recruits AKT by direct interaction with its PH domain. At the membrane level, another PH domain-containing serine / threonine kinase, called 3-phosphoinositide-dependent protein kinase 1 (PDK1), phosphorylates AKT on Thr308 [4]. Phosphorylation of Thr308 is necessary and sufficient for activation of AKT [5]. Studies have shown that overexpression of phosphorylated AKT (p-AKT) is associated with a poor prognosis for a number of solid tumors [6, 7] and some haematological malignancies [8, 9].
Various activating mutations in oncogenes, are found in various malignant tumors in almost every limb of the pathway. Substantial advances in the discovery of PI3K / AKT / mTOR alterations and their roles in tumorigenesis have led to the development of new targeted molecules with potential for the development of effective anticancer treatment [10]. Inhibition of the PI3K / AKT / mTOR pathway increases antitumor activity [5]. Preclinical data has shown that the combination of compounds simultaneously targeting different molecules of the PI3K / AKT / mTOR pathway leads to synergistic activity [10]. We provide in this study an in silico strategy for the exploration of PI3K AND mTOR inhibitors acting on AKT and therefore competitive dual inhibitors of ATP AKT / PI3K and AKT / mTOR. The marked interest in the development of new AKT / PI3K and AKT / mTOR inhibitors as potential agents for the treatment of cancer prompted us to explore the possibility of developing these inhibitors on the basis of a QSAR / AKT model in order to predict the bioactivity of PI3K and mTOR inhibitors on AKT. The interaction of the best will be evaluated by docking analysis.
2. Methodology
2.1 Dataset generation
Active inhibitors against AKT were extracted from BINDING DATABASE (https://www.bindingdb.org), their IC50 (molecule concentration leading to 50% inhibition) was transformed into logarithmic scale pIC50. 200 compounds with pIC50 greater than 8 were chosen for the present QSAR study. Also, mTOR and PI3K inhibitors with pIC50 greater than 8 have been selected in order to predict their activity against AKT using the AKT-QSAR model and explore their dual activity.
2.2 QSAR model generation
184 2D descriptors available on the MOE 2008.10 (obtained from Chemical Computing Group (CCP); Monreal, QC, Canada) [11] were calculated for the 200 compounds. Invariant and insignificant descriptors were initially eliminated; than the QSAR contingency descriptor selection and intercorrelation matrices between descriptor pairs were used to extract the 49 most relevant molecular descriptors which were employed for the distance calculation of each database entry. All 200 selected compounds were distributed randomly in training set with 140 compounds (70% of the data) and test set consisting of 60 compounds (30% of the data). Partial least squares (PLS) analysis based on the leave-one-out (LOO) method was used to correlate molecular descriptors with PIC50 values.
2.3 QSAR model validation
The internal validation procedure evaluates the relative predictive performance of the QSAR model, on the one hand by the correlation coefficient (R2) used to measure the correlation between the experimental pIC50 and predicted interest values in order to observe the variability between the variables in the set test, and secondly by the Root Mean Square Error (RMSE) a parameter used to evaluate the relative error of the QSAR model. The model is also tested by cross-validation using the LOO (leave-one-out) method and the computation of the correlation coefficient (R2) and mean squared error [RMSE], Z-Score $ Z-SCORE and $ XZ-SCORE are used to detect the outliers. External validation consists of evaluating the activities of the predictions and calculating the numerical parameters using the model.
2.4 Activity prediction
The QSAR-AKT model constructed and validated was used to predict the activity of two groups of PI3K and mTOR inhibitors against AKT, A first one is PI3K inhibitors and second one mTOR inhibitors. These inhibitors extracted from the Binding database (https://www.bindingdb.org/bind/index.jsp) have a pIC50 greater than 8 with 477 and 1008 inhibitors for PI3K and mTOR respectively. After calculating the predicted activity, the 40 inhibitors with the best predictions in each group were chosen for docking into AKT to explore their dual activity.
2.5 Molecular docking
The 3D coordinates of the mTOR inhibitors as well as the PI3K that showed the best predicted activity on QSAR-AKT model have been generate from 2D. 3MVH (PDB ID) is the AKT crystallized structure recovered from the PDB database for the docking analysis. MGL tools 1.5.6 with AutoGrid4 and AutoDock vina (Scripps) [12] were used for docking studies. The AKT structure was hydrogenated using MGL tools and PyMol was used to visualize the results [13]. In this work we adopted the same docking strategy used by the authors in previous studies [14]. The Grids boxes were generated around the active site of the two three-dimensional structures of the AKT kinase protein using MGL tools 1.5.6. The grids boxes were set to have between 16 and 20Å of edge with coordinates x=15.978, y=0.806, z=29.623the coordinates were determined using the potential substrate binding residues as centroids (in the hinge region and the activation loop) [15].
GSK690693 and Ipatasertib, known AKT inhibitors were also docked into AKT and were used as control to evaluate docking results. Their 3D structure was extracted from Pubchem. GSK690393 is a pan-AKT ATP selective inhibitor for AKT kinases [16]. Its potent pharmacodynamic and antitumor activity has been demonstrated in several human tumor cell lines and xenografts [17], growth inhibition and apoptosis in acute lymphoblastic leukemia cell lines [18]. Ipatasertib is a small molecule inhibitor of AKT, highly competitive orally administered. In cell line and xenograft models, Ipatasertib has been shown to be active in a wide range of cancer types, including breast, ovarian, colorectal and lung cancers [19].
3. Results
3.1 QSAR analysis
The AKT-QSAR model was built on the basis of 49 molecular descriptors. After the 200 molecules QSAR regression analysis, a PI3K-SAR model with a correlation coefficient (R2) of 0.85212 and a RMSE of 0.09266 was obtained, which was validated and evaluated by a cross-validation method LOO. The predictive performance of this model was represented by cross-validated RMSE with 0.53855 and R2 with cross-validation of 0.68474. Figure 1 shows a plot of experimental and predicted inhibitory potency in pIC50 values of the training and test set compounds showing a comparable and similar distribution between the two groups. No outliers were observed in the test set data, and all compounds were well-predicted with a
residual value less than one log unit.
3.2 Virtual screening
The AKT-QSAR model developed and validated was used to predict the activity of PI3K and mTOR inhibitors against PI3K. After calculating the predicted activity, the 40 inhibitors presenting the best predictions in each group are chosen to perform their docking into AKT. The best predicted PI3K and mTOR inhibitors present respectively pIC50 activities between [8.54-9.30], and [8.9-9.79].
3.3 Molecular docking studies
The docking scores (affinity) of the docked the PI3K and mTOR inhibitors in the catalytic site of AKT were set between -7.9 and -11.9 kcal/mol for the PI3K inhibitors group and between -7.8 and -11.9 kcal/mol for the mTOR inhibitors group; GSK690693 and Ipatasertib (AKT reference inhibitors) presented respective scores of -8.1 and -8.6 kcal/mol as reported on Table 1 and 2.
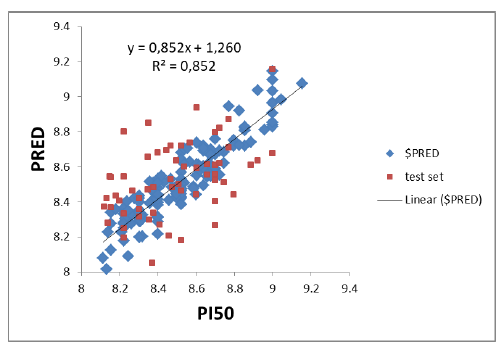
Figure 1: Relationship between observed and predicted data from QSAR-AKT model. The compounds of training set are in red and those of test set are in blue. Abbreviations: Pred, Predict; QSAR, quantitative structure-activity relationship.
|
BindingDB Reactant_set_id |
pIC50 |
Weight |
$PRED |
Number of H bonds |
Active site residues And Bonds length in Å |
Affinity- kcal/mol |
|
235143 |
8.669 |
428.423 |
8.620 |
5 |
3.2- 3.4 D292/3.1 L156 / 3.1 E228 / 2.8 A230 |
9.6 |
|
241313 |
8.316 |
484.950 |
8.651 |
2 |
3.3 L295 / 3.3 G292 |
9.5 |
|
50225680 |
9.031 |
502.578 |
8.817 |
4 |
3.2 K179 / 3.0 T195 / 3.2 - 3.5 E191 |
10.4 |
|
50225682 |
8.853 |
474.569 |
8.622 |
3 |
3.2 D292 / 3.1 G294 / 3.0 L295 |
9.1 |
|
50342300 |
9.638 |
515.951 |
8.654 |
2 |
3.3 D292 / 3.4 D438 |
10.8 |
|
50342301 |
8.769 |
495.534 |
8.549 |
2 |
3.3 D292 / 3.4 D438 |
11.5 |
|
50342304 |
8.677 |
511.532 |
8.583 |
2 |
3.1 K158 / 3.6 T195 |
11.3 |
|
50348643 |
8.187 |
632.740 |
8.796 |
4 |
3.2 - 3.4 E234 / 3.0 K179 / 3.2 G294 |
11.4 |
|
50409987 |
8.346 |
424.44 |
8.875 |
6 |
3.3 E234 / 3.0 D292 / 3.2 - 3.4 T291 / 3.2 E278 / 3.2 K158 |
8.9 |
|
50554599 |
8.522 |
515.578 |
8.562 |
3 |
3.4 - 3.0 K158 / 3.3 K179 |
9.0 |
|
50554601 |
8.221 |
543.632 |
8.542 |
3 |
2.9 T195 / 3.1 - 3.4 T291 |
9.9 |
|
50554602 |
9 |
558.646 |
8.599 |
3 |
3.4 - 3.1 T291 / 2.8 T195 |
9.8 |
|
50554603 |
8.167 |
592.664 |
8.877 |
4 |
3.4 K279 / 3.2 K180 / 3.4 - 3.2 T291 |
10.2 |
|
50554604 |
8.494 |
613.726 |
8.652 |
1 |
3.1 K179 |
9.1 |
|
50554605 |
8.619 |
572.674 |
8.587 |
3 |
3.5 - 3.2 T291 / 2.8 T195 |
10.3 |
|
50570130 |
9 |
400.389 |
8.677 |
4 |
3.4 K163 / 3.2 D292 / 3.3 K179 / 2.8 E191 |
10.1 |
|
50570911 |
8.698 |
457.493 |
8.953 |
4 |
3.5 K276 / 3.4 D274 / 3.2 E234 / 2.8 T195 |
8.3 |
|
50570918 |
8.154 |
527.633 |
8.798 |
3 |
3.1 K179 / 3.4 G159 / 3.2 E234 |
10.1 |
|
50570924 |
8.744 |
556.670 |
8.651 |
3 |
3.4 - 3.2 T291 / 2.9 T195 |
9.6 |
|
50570926 |
9.096 |
554.654 |
8.688 |
2 |
3.4 G294 / 3.3 K179 |
7.9 |
|
50581032 |
9.301 |
534.575 |
9.038 |
3 |
3.4 G294 / 3.3 E198 / 2.9 T195 |
10.5 |
|
50581034 |
9.397 |
489.506 |
8.929 |
4 |
3.2 E234 / 2.7 D292 / 3.0 K179 / 3.3 T195 |
9.9 |
|
50581036 |
9 |
552.565 |
9.275 |
3 |
3.3 E234 / 3.3 K276 / 2.9 K179 |
11.9 |
|
50581038 |
9.698 |
646.703 |
8.909 |
2 |
3.3 K179 / 2.9 E234 |
10.2 |
|
50581040 |
9.698 |
659.747 |
8.961 |
2 |
3.4 H194 3.4 D 439 |
8.9 |
|
50581042 |
9.522 |
661.763 |
8.995 |
1 |
3.4 H294 |
9.4 |
|
50581044 |
9.301 |
664.693 |
9.145 |
3 |
3.1 - 3.4 D292/ 3.3 E 234 |
10.1 |
|
50581046 |
9.698 |
679.752 |
9.233 |
2 |
3.1 K179 / 3.3 V164 |
9.3 |
|
50581053 |
9.397 |
471.517 |
8.691 |
3 |
3.4 K179 / 2.8 D292 / 3.3 E234 |
10.1 |
|
50602366 |
8.769 |
386.414 |
8.634 |
2 |
3.1 - 3.5 T291 |
9.5 |
|
50632672 |
9.221 |
349.354 |
8.609 |
4 |
3.2 E234 / 2.8 Y437 / 2.6 L156 / 3.3 D439 |
10.1 |
|
50703938 |
9.154 |
483.600 |
8.561 |
3 |
3.5 G311 / 3.2 T195 / 3.5 L156 |
9.1 |
|
50703939 |
8.677 |
497.627 |
8.626 |
2 |
3.2 E228 / 3.2 E234 |
9.8 |
|
50703940 |
9 |
511.655 |
8.641 |
3 |
2.9 E234 / 3.4 G311 / 2.9 T195 |
9.6 |
|
50881593 |
9.096 |
512.573 |
9.309 |
5 |
3.4 - 3.3 D292 / 3.3 N279 / 3.4 E278 / 2.8 L156 |
9.8 |
|
50927439 |
8.130 |
416.445 |
8.852 |
2 |
3.5 N279 / 3.5 L156 |
10.7 |
|
51043991 |
8.167 |
573.721 |
8.554 |
3 |
3.4 H194 / 3.3 E191 / 3.0 T195 |
11.0 |
|
51065310 |
8.397 |
460.541 |
8.955 |
2 |
3.3 K179 / 3.5 D439 |
9.1 |
|
51065317 |
8.346 |
419.488 |
8.600 |
3 |
3.4 - 3.1 K179 / 3.1 D439 |
9.8 |
|
51065318 |
9.148 |
389.462 |
8.583 |
1 |
3.4 - 3.1 K179 / 3.1 D439 |
9.7 |
|
GSK690693 |
8.698 |
425.5 |
***** |
5 |
1.8 G311 / 3.5 K179 / 3.0 T195 / 2.2 E191 / 2.9 G159 |
8.1 |
|
IPATASERTIB |
8.301 |
458 |
***** |
1 |
2.8 E234 |
8.5 |
The docking (affinity) scores of PI3K inhibitors in the catalytic site of AKT were set between -7.9 and -11.9 kcal / mol. GSK690693 and Ipatasertib (AKT reference inhibitors) showed scores of -8.1 and -8.6 kcal / mol
Table 1: Docked interaction analysis of PI3K inhibitors screened into AKT.
|
BindingDB Reactant_set_id |
PIC50 |
Weight |
$PRED |
Number of H bonds |
Active site residues And Bonds length in Å |
Affinity - kcal/mol |
|
240612 |
8.866 |
485.497 |
8.913 |
1 |
3.4 E234 |
10.1 |
|
240614 |
8.756 |
499.524 |
8.980 |
0 |
0 |
11.1 |
|
240621 |
8.653 |
515.979 |
8.934 |
2 |
3.5 G162 / 3.2 K276 |
10.3 |
|
240722 |
9.091 |
495.561 |
8.933 |
2 |
3.2 K179 / 3.4 N279 |
9.2 |
|
240723 |
8.835 |
496.549 |
8.924 |
1 |
3.2 D439 |
10.2 |
|
240724 |
8.787 |
509.588 |
9.000 |
1 |
3.2 K179 |
8.9 |
|
240755 |
9.677 |
543.578 |
8.921 |
3 |
3.3 K276 / 3.1- 3.1 T311 |
10.6 |
|
240758 |
9.309 |
557.604 |
8.983 |
1 |
3.0 K179 |
11.3 |
|
240765 |
9.346 |
574.06 |
8.938 |
1 |
3.2 K179 |
11.4 |
|
270328 |
8.193 |
462.468 |
8.916 |
4 |
3.3 - 3.0 K179 / 3.2 - 3.1 G294 |
10.1 |
|
270339 |
9.292 |
493.860 |
9.106 |
4 |
3.4 - 2.9 K179 / 3.4 - 3.3 G294 |
10.7 |
|
270346 |
9.060 |
477.405 |
9.124 |
4 |
3.3 - 2.9 K179/3.1 - 3.2 G294 |
9.9 |
|
270348 |
9.292 |
460.403 |
9.005 |
4 |
3.4- 2.9 K129/ 3.1 - 3.2 G294 |
9.7 |
|
270352 |
9.292 |
459.415 |
8.984 |
3 |
2.9 K179/ 3.2 - 3.2 G294 |
9.3 |
|
270356 |
8.886 |
459.415 |
8.957 |
1 |
3.2 K179 |
11.0 |
|
270358 |
8.698 |
476.858 |
8.973 |
3 |
3.6K276 / 3.2 K179 / 3.5 T291 |
10.6 |
|
270368 |
8.568 |
472.44 |
8.939 |
2 |
3.3 G294 /3.0 K179 |
10.7 |
|
270370 |
9.102 |
474.431 |
9.089 |
3 |
3.3 - 2.9 G294 / 3.1 K179 |
11.1 |
|
270372 |
8.602 |
474.431 |
9.006 |
3 |
3.4 K276 / 3.2 K158 / 2.9 T159 |
10.9 |
|
270379 |
8.187 |
474.431 |
9.066 |
4 |
3.3 - 3.2 G294 / 2.9 - 3.3 K179 |
11.5 |
|
270380 |
8.346 |
457.424 |
8.945 |
5 |
3.3 - 3.0 / 3.1 - 3.5 K179 / 3.4 E234 |
10.1 |
|
270383 |
8.455 |
460.403 |
9.005 |
4 |
3.4 - 2.9 K179 / 3.1 - 3.1 G294 |
9.8 |
|
270386 |
8.200 |
476.402 |
9.171 |
5 |
3.1 E234 / 2.9 - 3.5 K179 /3.2 - 3.2 G294 |
10.8 |
|
50570952 |
8.522 |
457.493 |
8.953 |
4 |
2.8 T195 / 3.4 D274 / 3.5 K276 / 3.2 E234 |
8.6 |
|
50581033 |
8.283 |
534.575 |
9.038 |
3 |
3.5 T195 / 3.5 K179 / 2.9 E234 |
10.3 |
|
50581035 |
8.619 |
489.506 |
8.929 |
4 |
3.3 T195 / 2.7 D192 / 3.1 K179 / 2.7 D291 / 3.1 E234 |
9.9 |
|
50581037 |
8.823 |
552.565 |
9.275 |
3 |
3.4 K276 / 2.9 K179 / 3.3 E234 |
11.9 |
|
50581041 |
8.420 |
659.747 |
8.961 |
1 |
3.4 H194 |
8.4 |
|
50581043 |
8.958 |
661.763 |
8.995 |
3 |
3.2 E234 / 3.2 G294 / 3.4 T195 |
8.1 |
|
50581045 |
8.602 |
664.693 |
9.145 |
3 |
3.1 - 3.5 D292 / 3.3 E234 |
10.2 |
|
50581047 |
9.522 |
679.752 |
9.233 |
2 |
2.8 D292 / 3.4 K158 |
7.8 |
|
50966358 |
8.522 |
388.394 |
9.023 |
4 |
3.3 - 3.4 - 3.3 T291 / 3.3 E278 |
8.0 |
|
50966360 |
8.301 |
387.410 |
9.092 |
3 |
3.3 D292 / 3.5 T291 / 3.3 E234 |
9.9 |
|
50966362 |
8.522 |
399.422 |
9.084 |
2 |
3.3 K158 / 3.0 T195 |
10.0 |
|
50966363 |
8.698 |
385.394 |
9.000 |
2 |
3.3 E234 / 3.4 D294 |
9.3 |
|
50966364 |
8.397 |
398.433 |
8.917 |
1 |
3.4 N279 |
10.1 |
|
50966368 |
8.522 |
398.433 |
8.917 |
3 |
3.0 D439 / 3.3 D292 / 3.5 T291 |
10.2 |
|
50966398 |
8.522 |
415.420 |
9.793 |
3 |
3.4 - 3.2 D292 / 3.3 T195 |
8.7 |
|
50966403 |
8.221 |
385.394 |
9.711 |
2 |
3.1 K179 / 3.3 D292 |
9.1 |
|
50966404 |
8.522 |
349.358 |
9.310 |
3 |
3.5 - 3.5 D292 / 3.3 E234 |
8.5 |
|
GSK 690693 |
8.698 |
425.5g |
***** |
5 |
1.8 G311 / 3.5 K179 / 3.0 T195 / 2.2 E191 / 2.9 G159 |
8.1 |
|
IPATASERTIB |
8.301 |
458 |
***** |
1 |
2.0 G159 |
8.5 |
The docking (affinity) scores of mTOR inhibitors in the catalytic site of AKT were set between-7.8 and -11.9 kcal / mol. GSK690693 and Ipatasertib (AKT reference inhibitors) showed scores of -8.1 and -8.6 kcal / mol
Table 2: Docked interaction analysis of mTOR inhibitors screened into AKT.
4. Discussion
Almost all intervenanants of the pathway may be affected by various mutations inducing malignant tumors. The discovery of PI3K / AKT / mTOR alterations and their role in tumorigenesis have allowed the development of new molecules with inhibitory activity and continuous development for an anticancer treatment. In order to explore new AKT/PI3K and AKT/mTOR common inhibitors, a 2D-QSAR model for AKT was constructed to predict the activity of one group of PI3K inhibitors and one of mTOR, then do the docking for those who presented the best prediction (the top 40 in each group) whose results were compared with that of GSK 690693 and Ipatasertib. The AKT inhibitors that were used in the construction of the QSAR model and those of PI3K and mTOR whose activity for AKT was predicted were extracted from Binding Database and selected according to their bioactivity IC50, the model was realized by the PLS method by the MOE software. AKT and mTOR inhibitors docking score compared to GSK 690693 and Ipatasertib score show that the PI3K inhibitors and those of mTOR respectively have an affinity between -7.9 and -11.9 kcal/mol and -7.8 and -11.9 kcal/mol while GSK 690693 and Ipatasertib only -8.1 and -8.5 kcal/mol, respectively, therefore, 39 PI3K inhibitors and 37 mTOR inhibitors show higher affinity than that of GSK 690693 and 38 PI3K inhibitors and 35 mTOR inhibitors show higher affinity than that of Ipatasertib. Thus, the affinity of Ipatasertib is more effective than that of GSK 690693 the opposite for the interactions. Visualization of these interactions showed that the different compounds adapt to the ATP binding site forming hydrogen bonds, the best PI3K-inhibitor establishes 6 bonds Hydrogens with AKT, that of mTOR establishes 5 bonds against the GSK 690693 and Ipatasertib have respectively 5 and only 1 connection. The important score of the affinity of some inhibitors despite the low number of hydrogen bonds can be explained by the hydrophobic interactions and that of Van Der Waal. Thus, the AKT affinity of most of the PI3K and mTOR inhibitors studied is more effective than that of GSK 690693 and Ipatasertib and the majority interaction is more effective than that of Ipatasertib. A phase 1 clinical study demonstrated robust inhibition of the AKT pathway by Ipatasertib at clinically feasible doses [20] and other studies of various types of tumors, including breast cancer, have shown an acceptable safety profile but characterized by gastrointestinal effects, asthenia or fatigue and rash [21].
A double comparison of the inhibitors studied with respect to GSK690693 which, given the affinity and the number of hydrogen bonds greater than 4 mediated by AKT, showed that 10 PI3K inhibitors (Table 3) and 11 mTOR inhibitors (Table 4) all have better results than GSK690693, but taking into account Lipinski's rules (no more than 5 hydrogen bond donors, no more than 10 hydrogen bond acceptors, an octanol-water partition coefficient no greater than 5,5) and those of Veber determining the good bioavailability (PSA (Polar surface) ≤ 140 Å2 (absolute polarity measurement), rotary links ≤ 10 (measure of flexibility, we found that 7 PI3K inhibitors (Table 5) of which 50409987 the most relevant (6 hydrogen bonds and an affinity -8,9/ mol) and 10 mTOR inhibitors (Table 6), of which the 270386 is the most relevant (5 hydrogen bonds and an affinity of -10.8 kcal / mol) corresponded to all criteria and could be future drugs (Figure 2), but knowing that GSK690693 has demonstrated broad in vitro activity, and that in vivo results against solid tumors demonstrate that GSK690693 has only modest antitumor activity and thus the use of GSK690693 as a unique agent in the treatment Malignant tumors in children will probably have limited value without further optimization [22], according to our study, inhibitors with an affinity and an interaction as important as GSK690693, which can act as inhibitors of AKT in addition to their action. On either the PI3K or the mTOR would be important.
PI3K inhibitors with better affinity and number of hydrogen bonds than GSK 690693
Table 3: Comparison of PI3K inhibitors studied to GSK 690693.
mTOR inhibitors with better affinity and number of hydrogen bonds than GSK 690693
Table 4: Comparison of mTOR inhibitors studied to GSK 690693.
|
BindingDB Reactant_set_id |
Name |
|
5E+07 |
2-[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-2-[(5-fluoro-6-methoxypyridin-3-yl)amino]imidazo[1,2-a]pyridin-6-yl]propan-2-ol |
|
5E+07 |
7-(2-(1-(2-(dimethylamino)acetyl)-5-methoxyindolin-6-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamino)isoindolin-1-one |
|
5E+07 |
(2S)-2-hydroxy-1-[3-[4-[2-(5-methyl-2-propan-2-yl-1,2,4-triazol-3-yl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl]pyrazol-1-yl]azetidin-1-yl]propan-1-one |
|
5E+07 |
4,6-dihydroxy-2-((5-methoxy-2-(pyridin-3-yl)-1H-indol-3-yl)methylene)benzofuran-3(2H)-one |
|
5E+07 |
1-[4-[4-morpholin-4-yl-7-(2-oxoethyl)pyrrolo[2,3-d]pyrimidin-2-yl]phenyl]-3-pyridin-4-ylurea |
|
5E+07 |
1-(2-((7-fluoro-5-methoxy-2-(1,3,5-trimethyl-1H-pyrazol-4-yl)-1H-indol-3-yl)methylene)-3-oxo-2,3-dihydrobenzofuran-5-yl)-3-methylurea |
|
5E+07 |
5-(1-(4-amino-6-methyl-1,3,5-triazin-2-yl)-1H-benzo[d]imidazol-2-ylamino)benzene-1,3-diol |
Table 5: PI3K inhibitors meeting Lipinski and Veber criteria.
|
BindingDB Reactant_set_id |
Name |
|
270380 |
(4S)-3-[3-[4-(5-amino-1,3,4-oxadiazol-2-yl)phenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-4-(4-fluorophenyl)-1,3-oxazolidin-2-one |
|
270386 |
(4S)-3-[3-[4-(5-amino-1,3,4-oxadiazol-2-yl)-3-fluorophenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-4-(5-fluoropyridin-2-yl)-1,3-oxazolidin-2-one |
|
270328 |
(4R)-3-[3-[3-fluoro-4-(1H-1,2,4-triazol-5-yl)phenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-4-(4-methyl-1,3-thiazol-2-yl)-1,3-oxazolidin-2-one |
|
270339 |
(4S)-4-(3-chloro-4-fluorophenyl)-3-[3-[3-fluoro-4-(1H-1,2,4-triazol-5-yl)phenyl]pyrazolo [1,5-a]pyrimidin-5-yl]-1,3-oxazolidin-2-one |
|
270346 |
(4S)-4-(3,4-difluorophenyl)-3-[3-[3-fluoro-4-(1H-1,2,4-triazol-5-yl)phenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-1,3-oxazolidin-2-one |
|
270348 |
(4S)-4-(5-fluoropyridin-2-yl)-3-[3-[3-fluoro-4-(1H-1,2,4-triazol-5-yl)phenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-1,3-oxazolidin-2-one |
|
270379 |
(4S)-4-(3-fluoro-6-methylpyridin-2-yl)-3-[3-[3-fluoro-4-(1H-1,2,4-triazol-5-yl)phenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-1,3-oxazolidin-2-one |
|
270383 |
(4S)-4-(5-fluoropyridin-2-yl)-3-[3-[3-fluoro-4-(1H-1,2,4-triazol-5-yl)phenyl]pyrazolo[1,5-a]pyrimidin-5-yl]-1,3-oxazolidin-2-one |
|
50570952 |
1-[4-[4-morpholin-4-yl-7-(2-oxoethyl)pyrrolo[2,3-d]pyrimidin-2-yl]phenyl]-3-pyridin-4-ylurea |
|
50581035 |
1-[(2E)-2-[[7-fluoro-5-methoxy-2-(1,3,5-trimethylpyrazol-4-yl)-1H-indol-3-yl]methylidene]-3-oxo-1-benzofuran-5-yl]-3-methylurea |
Table 6: mTOR inhibitors meeting Lipinski and Veber criteria.
GSK690693 (A) and Ipatasertib (B) respectively have 5 and 1 linkages with the catalytic site of AKT whereas 50409987 inhibitor of PI3K (C) and 270386 inhibitor of mTOR (D) respectively have 6 and 5 hydrogen bonds. Notes: red zones: oxygen atom, blue zones, nitrogen atom and green zones, other. The hydrogen bonds are represented by the dashed yellow. The numbers represent the size of the hydrogen bonds established between the ligand and the receptor.
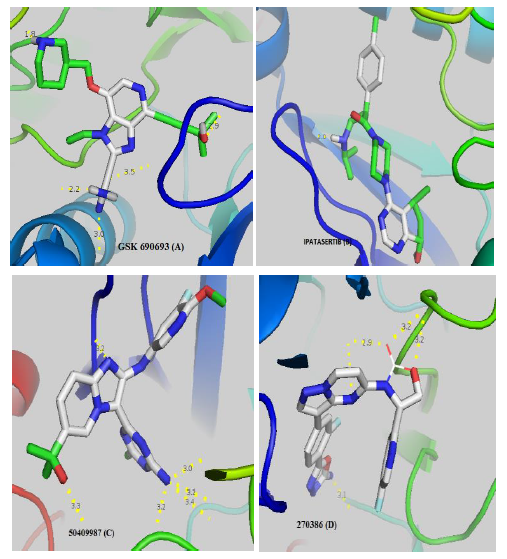
Figure 2: Visualization of the different interactions between the compounds and the active site of AKT via the hydrogen bonds.
5. Conclusion
The hyperactivation of PI3K / Akt / mTOR in cancer, associated with the crucial role of AKT signaling in tumorigenesis, has led to significant efforts to generate inhibitors to target this pathway. Through the construction of a QSAR-AKT model, we were able to predict potent PI3K and mTOR inhibitor activity on AKT select the best and reveal their interactions with AKT by docking while comparing them to AKT reference inhibitors. Promising results have been obtained. The dual inhibitors AKT / PI3K and AKT / mTOR have been predicted, pending the testing of these compounds directly on the cancer cell lines in our next study.
References
- Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol Biosyst 11 (2015): 1946-1954.
- Bellacosa A, Chan TO, Ahmed NN, et al. Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene 17 (1998): 313-325.
- Andjelkovic M, Alessi DR, Meier R, et al. Role of translocation in the activation and function of protein kinase B. J Biol Chem 272 (1997): 31515-31524.
- Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J 346 (2000): 561-576.
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nat Rev Cancer 2 (2002): 489.
- Schmitz KJ, Otterbach F, Callies R, et al. Prognostic relevance of activated Akt kinase in node-negative breast cancer: a clinicopathological study of 99 cases. Mod Pathol Off J U S Can Acad Pathol Inc 17 (2004): 15-21.
- Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J Clin Oncol Off J Am Soc Clin Oncol 23 (2005): 1473-1482.
- Rudelius M, Pittaluga S, Nishizuka S, et al. Constitutive activation of Akt contributes to the pathogenesis and survival of mantle cell lymphoma. Blood 108 (2006): 1668-1676.
- Min YH, Eom JI, Cheong JW, et al. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: its significance as a prognostic variable. Leukemia 17 (2003): 995-997.
- Polivka J, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther 142 (2014): 164-175.
- Vilar S, Moro GC S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery [Internet]. Current Topics in Medicinal Chemistry (2008).
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31 (2010): 455-461.
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24 (2010): 417-422.
- Lakhlili W, Chevé G, Yasri A, et al. Determination and validation of mTOR kinase-domain 3D structure by homology modeling. OncoTargets Ther 8 (2015): 1923-1930.
- Lakhlili W, Yasri A, Ibrahimi A. Structure–activity relationships study of mTOR kinase inhibition using QSAR and structure-based drug design approaches. OncoTargets Ther 9 (2016): 7345-7353.
- Heerding DA, Rhodes N, Leber JD, et al. Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H-imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase. J Med Chem 51 (2008): 5663-5679.
- Rhodes N, Heerding DA, Duckett DR, et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res 68 (2008): 2366-2374.
- Levy DS, Kahana JA, Kumar R. AKT inhibitor, GSK690693, induces growth inhibition and apoptosis in acute lymphoblastic leukemia cell lines. Blood 113 (2009): 1723-1729.
- Targeting Activated Akt with GDC-0068, a Novel Selective Akt Inhibitor That Is Efficacious in Multiple Tumor Models. Clinical Cancer Research (2019).
- Yan Y, Serra V, Prudkin L, et al. Evaluation and clinical analyses of downstream targets of the Akt inhibitor GDC-0068. Clin Cancer Res Off J Am Assoc Cancer Res 19 (2013): 6976-6986.
- Saura C, Roda D, Roselló S, et al. A First-in-Human Phase I Study of the ATP-Competitive AKT Inhibitor Ipatasertib Demonstrates Robust and Safe Targeting of AKT in Patients with Solid Tumors. Cancer Discov 7 (2017): 102-113.
- Carol H, Morton CL, Gorlick R, et al. Initial Testing (Stage 1) of the Akt Inhibitor GSK690693 by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer 55 (2010): 1329-1337.
