Therapeutic Potential of CRISPR/Cas9 Nuclear Scissors in the Modulation of MicroRNAs in Various Cancers: A Review
Article Information
Asra Iftakhara*
Institute of Pharmacy, Physiology and Pharmacology, University of Agriculture Faisalabad, Pakistan
*Corresponding Author: Dr. Asra Iftakhara, Institute of Pharmacy, Physiology and Pharmacology, University of Agriculture Faisalabad, Pakistan
Received: 28 February 2018; Accepted: 16 March 2018; Published: 22 March 2018
Citation: Asra Iftakhara. Therapeutic Potential of CRISPR/Cas9 Nuclear Scissors in the Modulation of MicroRNAs in Various Cancers: A Review. Journal of Cancer Research 2 (2018): 38-53.
View / Download Pdf Share at FacebookAbstract
Cancer is leading cause of death around the world. There are many strategies have been planned to tackle the cancer progression but each has its own merits and demerits. The microRNA`s are controlling 60 percent human genome, have role in progressive timing, organogenesis, hematopoiesis, cell proliferation, apoptosis and possibly tumorgenesis by amplification or deletion of genes. There are many drugs like 5-fluorouracil (5-FU), Celecoxib and genistein were used to modulate the miRNA`s expression but their success rate remained least. The clustered regularly interspaced short palindromic repeat (CRISPR/ Cas9) is emerging technology used to modulate the expression of miR-17, miR-200c, and miR-141 with 96 percent success rate. This review will provide us information about the potential therapeutic use of CRISPR/Cas9 against defaulted microRNA`s in the long-standing repression and beginning or inhibition of cancer.
Keywords
Cancer, MicroRNA`s, Therapeutic, CRISPR/Cas9
Cancer articles Cancer Research articles Cancer review articles Cancer PubMed articles Cancer PubMed Central articles Cancer 2023 articles Cancer 2024 articles Cancer Scopus articles Cancer impact factor journals Cancer Scopus journals Cancer PubMed journals Cancer medical journals Cancer free journals Cancer best journals Cancer top journals Cancer free medical journals Cancer famous journals Cancer Google Scholar indexed journals MicroRNA`s articles MicroRNA`s Research articles MicroRNA`s review articles MicroRNA`s PubMed articles MicroRNA`s PubMed Central articles MicroRNA`s 2023 articles MicroRNA`s 2024 articles MicroRNA`s Scopus articles MicroRNA`s impact factor journals MicroRNA`s Scopus journals MicroRNA`s PubMed journals MicroRNA`s medical journals MicroRNA`s free journals MicroRNA`s best journals MicroRNA`s top journals MicroRNA`s free medical journals MicroRNA`s famous journals MicroRNA`s Google Scholar indexed journals Therapeutic articles Therapeutic Research articles Therapeutic review articles Therapeutic PubMed articles Therapeutic PubMed Central articles Therapeutic 2023 articles Therapeutic 2024 articles Therapeutic Scopus articles Therapeutic impact factor journals Therapeutic Scopus journals Therapeutic PubMed journals Therapeutic medical journals Therapeutic free journals Therapeutic best journals Therapeutic top journals Therapeutic free medical journals Therapeutic famous journals Therapeutic Google Scholar indexed journals CRISPR/Cas9 articles CRISPR/Cas9 Research articles CRISPR/Cas9 review articles CRISPR/Cas9 PubMed articles CRISPR/Cas9 PubMed Central articles CRISPR/Cas9 2023 articles CRISPR/Cas9 2024 articles CRISPR/Cas9 Scopus articles CRISPR/Cas9 impact factor journals CRISPR/Cas9 Scopus journals CRISPR/Cas9 PubMed journals CRISPR/Cas9 medical journals CRISPR/Cas9 free journals CRISPR/Cas9 best journals CRISPR/Cas9 top journals CRISPR/Cas9 free medical journals CRISPR/Cas9 famous journals CRISPR/Cas9 Google Scholar indexed journals organogenesis articles organogenesis Research articles organogenesis review articles organogenesis PubMed articles organogenesis PubMed Central articles organogenesis 2023 articles organogenesis 2024 articles organogenesis Scopus articles organogenesis impact factor journals organogenesis Scopus journals organogenesis PubMed journals organogenesis medical journals organogenesis free journals organogenesis best journals organogenesis top journals organogenesis free medical journals organogenesis famous journals organogenesis Google Scholar indexed journals hematopoiesis articles hematopoiesis Research articles hematopoiesis review articles hematopoiesis PubMed articles hematopoiesis PubMed Central articles hematopoiesis 2023 articles hematopoiesis 2024 articles hematopoiesis Scopus articles hematopoiesis impact factor journals hematopoiesis Scopus journals hematopoiesis PubMed journals hematopoiesis medical journals hematopoiesis free journals hematopoiesis best journals hematopoiesis top journals hematopoiesis free medical journals hematopoiesis famous journals hematopoiesis Google Scholar indexed journals cell proliferation articles cell proliferation Research articles cell proliferation review articles cell proliferation PubMed articles cell proliferation PubMed Central articles cell proliferation 2023 articles cell proliferation 2024 articles cell proliferation Scopus articles cell proliferation impact factor journals cell proliferation Scopus journals cell proliferation PubMed journals cell proliferation medical journals cell proliferation free journals cell proliferation best journals cell proliferation top journals cell proliferation free medical journals cell proliferation famous journals cell proliferation Google Scholar indexed journals tumorgenesis articles tumorgenesis Research articles tumorgenesis review articles tumorgenesis PubMed articles tumorgenesis PubMed Central articles tumorgenesis 2023 articles tumorgenesis 2024 articles tumorgenesis Scopus articles tumorgenesis impact factor journals tumorgenesis Scopus journals tumorgenesis PubMed journals tumorgenesis medical journals tumorgenesis free journals tumorgenesis best journals tumorgenesis top journals tumorgenesis free medical journals tumorgenesis famous journals tumorgenesis Google Scholar indexed journals apoptosis articles apoptosis Research articles apoptosis review articles apoptosis PubMed articles apoptosis PubMed Central articles apoptosis 2023 articles apoptosis 2024 articles apoptosis Scopus articles apoptosis impact factor journals apoptosis Scopus journals apoptosis PubMed journals apoptosis medical journals apoptosis free journals apoptosis best journals apoptosis top journals apoptosis free medical journals apoptosis famous journals apoptosis Google Scholar indexed journals articles Research articles review articles PubMed articles PubMed Central articles 2023 articles 2024 articles Scopus articles impact factor journals Scopus journals PubMed journals medical journals free journals best journals top journals free medical journals famous journals Google Scholar indexed journals
Article Details
Abbrevation:
5-fluorouracil - (5-FU); Clustered regularly interspaced short palindromic repeat - (CRISPR/ Cas9); MicroRNAs - (miRNAs); Untranslated regions - (UTRs); Antisense oligonucleotides - (ASO); Single-stranded RNA - (ssRNA); Adenosine deaminases acting on RNA - (ADARs); Argonaute - (Ago); RNA-induced silencing complex - (RISC); Small interfering RNA - (siRNA); Argonaute 2 - (Ago2); CRISPR RNA - (crRNA); Trans-activating crRNA - (tracr RNA); Protospacer adjacent motif - (PAM); Homologous directed repair - (HDR); Non-homologous end joining - (NHEJ); Acute myeloid leukemia - (AML); Zinc finger Ebox binding homeobox 1 - (ZEB1); Karyopherin alpha 2 - (KPNA2)/c-Jun; Colony stimulating factor 1 - (CSF1); Repressor-element-1-silencing transcription factor - (REST); Triacylglycerol - (TG); Hepatocellular carcinoma - (HCC); Yes-associated protein 1 - (YAP1)
1. Introduction
The microRNAs (miRNAs) are considered as emerging potential therapeutic targets against cancer. The miRNAs are small non-coding molecules having the sequence of 22 nucleotide base pairs that play a key role in the modulation of various biological processes by regulating genes expression at post-transcriptional level either through translational suppression or mRNA degradation [1, 2, 3, 4].The miRNAs have been involved in various essential pathways like progressive timing, organogenesis, hematopoiesis, cell proliferation, apoptosis, and possibly tumorgenesis [1, 3, 4], estimated up to 60% of the total human protein-coding genes are forecasted to hold miRNA obligatory location inside their 3' untranslated regions (UTRs) [2]. The induced expression of miRNAs is introduced into the host via intra-tumoral injections or viral vector such as lentivirus. Likewise, expression of viral vectors is to demonstrate the identical faintness encountered in gene therapy, like restricted infectivity and essential intended for nuclear translocation of a comparatively large DNA vector, transcription and ultimate maturation of the gene product [5, 6, 7].
There are many drugs such as doxorubicin, cisplatin, genistein, and paclitaxel which are used in the miRNA expression up-surging [8, 9]. As genistein is used to target 574-3p and miR-151 in prostate cancer [8] unique neutral lipid emulsion was especially encountered to of let-7 and microRNA-34a in lung cancer [10] 5-fluorouracil (5-FU) was to target let-7b and let-7c renal cell carcinoma [11] and Celecoxib used to increase expression of miRNA-29c in gastric cancer cells [12]. Cancer is still a major problem in medical sciences therefore, in order to treat it due to its complexity in a mutant genome; there is a need to develop effective therapeutic approach [13]. To overcome the mutant expression via silencing of miRNA, antisense oligonucleotides (ASO) are being used. Usually, this technique generates off-target effects [14, 15]. In such circumstances, a precise and better technology is needed for the silencing of the miRNA to proceed the therapeutic approach for the treatment of cancer.
The clustered regularly interspaced short palindromic repeat (CRISPR) is an emerging technology for the protein stability [16] gene editing [17] and miRNA [18, 19]. It is a scheme of adaptive immunity of prokaryotes from which this technology is developed [20]. The CRISPR/Cas9 scheme originated about 90% and 50% of archaea and bacteria to act as a defensive barrier against viruses [20]. The objective of this review is possible use of CRISPR/Cas9 technique in the way of miRNA`s biogenesis (shown in diagram ) to edit mutant nucleotide sequence to treat various cancers.
2. Biogenesis of microRNA
The biogenesis of microRNA (miRNA) is a comprehensive process. RNA polymerase II acts on the DNA and transcribes to miRNA in the form of pri-miRNA. There are mainly two RNAase enzymes involved in the biogenesis of miRNA i.e. Drosha and Dicer. The Drosha is a member of RNAase III enzyme family that play an indispensable role in the generation of Pre-miRNA from Pri-miRNA in the nucleus, the pre-miRNA transported to the cytoplasm with help of an export protein while Dicer acts on Pri-miRNA in the cytoplasm and leads to its maturation as a miRNA that has 22 nucleotide sequences [1, 3, 21, 22]. The 70 to 80-nt long RNA hairpin sequence of Pri-miRNA and ssRNA extension situated outside the Pri-miRNA are required for the action of Drosha [1, 23] while any abnormality in the extension of Pri-miRNA hairpin leads to blockage of Drosha action [24]. It was thought, the DGCR8-Drosha complex anchors the cleavage pattern of Pri-miRNA into pri-miRNA a fixed space from either the terminal loop or basal single-stranded RNA (ssRNA) but later on it was concluded that the flanking sequence/structure directs the binding locus DGCR8-Drosha for the cleavage [25]. The Drosha can dichotomy on the other hand of exon 5 of the eIF4H gene into a precursor-miRNA. It was the evidence of the Drosha structure dependent splicing of the RNA strand and Drosha can function like a splicing enhancer and promotes exon inclusion [23]. The miRNA amendment can be done by RNA editing and decay like; adenylation and uridylation; RNA methylation and Argonaute loading [3]. The p68 and p72 DEAD-box RNA helicase are crucial subunits necessary for processing of Pri-miRNA and rRNA via Drosha-mediated pathway. Any dysfunctioning in the subunits leads to early lethality [22]. The Msi and Lin28 regulates miRNA biogenesis in the cropping step in the nucleus [26]. The cyclin D1 protein induces the abundant expression of Dicer, and cooperates in the biogenesis of micro RNA. Cyclic D1based control of cell migration and proliferation depends on Dicer. It is found that the dicer has less expression level in cyclin D1 deficient cancer cells [27]. It was concluded that Adenosine deaminases acting on RNA (ADARs) play role in RNA editing by forming a complex with dicer via direct protein to protein interface. It increases the rate of pre-miRNA cleave through increasing the efficacy of Dicer and facilitates in the loading of miRNA onto the RISC and subsequently, silencing of aimed RNA. The reduced expression of the ADAR1 might contribute in the formation of lethal results [28]. The TAp63 and p53 have a role in suppression of tumorgenesis and metastasis by regulating the Dicer. TAp63 binds and transactivates the Dicer promoter, demonstrating direct transcriptional regulation of Dicer [29]. The level of dicer varies amid normal and disease state cells. The production of dicer inhibited in response to multiple stress embrace reactive oxygen species (ROS), Ras oncogenes and phorbol ester ultimately leading to cancer. Furthermore, dsRNA and Type 1 interferons inhibit the production of Dicer while in contrast; IFN-? induces the Dicer expression [30]. Argonaute (Ago) protein plays the vital role associated with miRNA and responsible for RNA cleavage. AGO1 and AGO2 have a role in RNA silencing through the complementary sequence with the help of miRNA. In the absence of AGO2, the siRNA was unable to form RNA-induced silencing complex (RISC) and possibly lead to nuclease-resistant modification [31]. The Hsc70/Hsp90 chaperone complex was detected involved in the loading of small duplexes RNA into the RICS and small interfering RNA (siRNA) mediated Fkbp4/5 reduce expression that lead to a low expression level of AGO2 and vice versa. Thus it is also considered that the Fkbp4/5 have a role in RISC assembly [32]. The PABPC1 intermingled with AGO2 in the cytoplasm and increased the recruitment of mRNA to RISC [33]. As the catalytic engine of RISC, argonaute 2 (AGO2) is crucial for miRNA-induced silencing. It comprises of two conserved domains: a PAZ domain that binds to the 3? end of the mature miRNA and a PIWI domain that is structurally related to ribonuclease-H and functionally interrelates with the 5? end of the guide strand [34]. Biochemical analysis of TRBP-containing complexes exposed the association of Dicer?TRBP with Argonaute 2 (Ago2) that is a catalytic engine of RISC [35, 36].
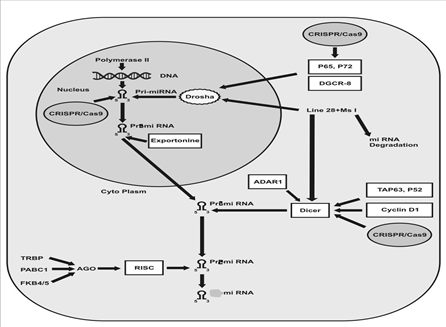
Figure 1: Suspected Target of CRISPR/Cas9 during miRNA biogenesis. This picture is demonstrating that we can edit the micro RNA to prevent the cancer. We can apply CRISPR/Cas9 tool in the a) seed sequence of miRNA at pri-miRNA level b) P65, P72 that the genes who are regulating the activity of drosha. It was concluded that the P65,P72 are tumor suppressor genes, their aberration causes the abnormality of miRNA c) Dicer is one of the most important enzyme in the biogenesis of miRNA that was found with aberrant expression, can be normalize.
3. CRISPR/ Cas9 Nuclear Scissors or Cutting Enzymes and Relevance with miRANs/Cancer
The transfusion of the CRISPR was taken through viral vectors such as adeno and lenti viral vectors into the cells in-vitro and in-vivo. The CRISPR contains gRNA and Cas9 protein. The gRNA expressed with polymerase III promoter H1 or U6, Cas9 expressed with polymerase II promoter, such as CMV, UBC or EF1?. The Cas9 is responsible to target cleavage of sequence that is a major concern in gene editing [37]. Now the off-target activity has been overcomed through genetic modification of Cas9 [38]. The Cas9 has two strands RuvC and HNH catalytic domains, RuvC is present close to the N-terminal region of Cas9 and flanking the domain of HNH domain is adjacent to the middle of the protein. The Ruvc and HNH domains cleave the targeted DNA complementary and non-complementary strands. The sgRNA, derived from the CRISPR RNA (crRNA) and trans-activating crRNA (tracr RNA) binds to the Cas9 and express it to the locus of interest by a 20-nt guide sequence through base pairing at mark position in the genome [39-42]. The target DNA sequence joins with the sgRNAnt sequence by protospacer adjacent motif (PAM). The cas9 cleave DNA targeted sequence of 3-4 nucleotides through up-streaming PAM and trigger to the DSBs. Thus, the DNA error free repairing occur through homologous directed repair (HDR) or non-homologous end joining (NHEJ)-mediated error-prone DNA repairing [39-42]. The studies have shown that the CRISPR/ Cas9 scheme significantly help out in the treatment of cancer as [17] perform an experiment on DU145 prostate cancer cell line to check out the validity of CRISPR/ Cas9 technology to knock out the NANOG and NANOGP8 oncogenes. The knockout of the NANOG and NANOGP8 significantly reduce the viability of Prostate cancerous cells. As the several genes like NF1, MED12, XPO1, CUL3, TADA2B, NF2, TADA1 NANOG and NANOGP8 have been identified, having a role in chemotherapy resistance, could mimic the stubbornness through gene editing tool CRISPR/ Cas9 [17, 43, 44]. The epigenetic alterations involved are DNA methylation and as well as histone modification that lead to the up or down regulation of genes that are controlling the cellular development, differentiation and proliferation [45]. There were frequently combined and missensed mutations found in TP53, PIK3CA, TTN, ATM, GATA3 FOXA1and KMT2D, PTEN, BRAF, IDH1, GNAS, KRAS [46], RTKs, ATK, c-KIT, mTOR, EGFR, IGF-1R [47, 48] DNMT3A, RUNX1, TET2, EZH2, APC, P53, SMAD4, and NF1 genes in myeloid and intestinal malignancy [49, 50] which could be normalized with the use of CRISPR/ Cas9 gene editing tool. It has been proved that about 60% of the human genome is being controlled by miRNA. Now, there is a need to apply the CRISPR/ Cas9 technology to control the expression of miRNA in order to control the expression of genes [51] so that the expression of miRNA with 96% success rate could be normalized. There 23 miRNAs identified with a high probability of targeting PHF6, while miR-128b and miR-574 have a top candidate for a possible oncogenic role in the genesis of T-ALL [52]. It is demonstrated that the miR-17, miR-200c, and miR-141 expression could be modulated by using the CRISPR/ Cas9 technology [51]. The Pri-miRNA directs miRNA gene transcription and afterwards Drosha and Dicer act on it leading in its maturation. The studies confirmed that structure of pri-miRNA decides the working effectiveness of Drosha in the biogenesis of miRNA. So, it needs to apply the CRISPR/ Cas9 at Pri-miRNA level [51, 41]. It is demonstrated that not only single clones with large pieces of deletion (6-18 bp) can lead to the failure of mature miR-17 biogenesis, but also the clone with only 2 deletions and 1 insertion can impede the exogenous expression of miR-17 as well [51]. A cross talk was observed when the CRISPR/ Cas9 scheme was applied to detect the expression level of miR-200 and miR-141. The CRISPR/ Cas9 induces the down-regulation of miR-200 that elevate the expression of ZEB1, ZEB1 down-regulating the expression of miR-141 automatically [52]. When the nt sequences of miR-17-92 cluster were ranged, they were roughly divided such as miR-20a, miR-17 and miR-18a; miR-19a and miR-19b-1; miR-92-1, and found silenced upon CRISPR/ Cas9 tool [51, 25]. Other study showed miR-93 expression knock out by CRISPR/ Cas9, only one single nucleotide deletion governs to non-functionality of miRNA. The CRISPR/ Cas9 inactivates the miRNA at the stage of Pri-miRNA by targeting the 5’ region of a microRNA and inhibits the action of Drosha [53]. Both miR-21 and miR-30a found onco-miR in studies, were efficiently silenced through CRISPR/ Cas9 tool via targeting the special loop region adjacent to the palindromic sequence of pri-miRNA [54].
4. Involvement of MicroRNAs in Different Cancers and their Associated Molecular Mechanisms
The studies have shown that the miRNA reside in the extracellular fluid and perform a crucial role in cross-talk between the cancerous and normal cells [55, 56]. The presence of miRNA in the plasma, serum and saliva helps as a marker in the diagnosis of cancer. As miR-121, miR-122 and miR-141 help in the diagnosis of colorectal and liver cancer [57], miR-181, let-7, miR-130b, miR-150, miR-145 and miR-199a-3p of liver stem cell carcinoma [58], miR-506, miR-101, miR-25, miR-29c, miR-182, miR-128 of ovary cancer [59]. Different types of cancers associated with microRNAs and specific target genes are elaborated in Table 1 and described below in detail .
Type of Cancer and MicroRNAs involved |
Specific Target Genes |
References |
|
Leukemia |
||
|
miR-146 |
FADD |
[60] |
|
miR-223 |
E2F1 |
[61] |
|
miR-150 |
Myb, Notch, Akt and DKC1 |
[62-65] |
|
miR-495 |
MEIS1 and PBX3 |
[66] |
|
miR-34b |
HSF1 |
[67] |
|
miR-17/92 |
PTEN and Bim |
[68] |
|
miR-34a |
p53, B-Myb and E2F1 |
[69] |
|
Breast Cancer |
||
|
miR-148b |
PIK3CA/p110?, ROCK1, NRAS, ITGA5 and CSF1 |
[70] |
|
miR-17~92 |
HIF1, VEGFA and TGFBR2 |
[71] |
|
miR-340 |
ZEB1 |
[72] |
|
MiR-34 Family |
p53, Wnt/b-catenin, Hedgehog and Notch. |
[73] |
|
miR-22 |
TIP60, NuA4 |
[74] |
|
Gastric Cancer |
||
|
miR-29c |
ADAM12-L |
[75, 76] |
|
miR-26b |
KPNA2/c-Jun |
[77] |
|
miR-214 |
CSF1 |
[78] |
|
miR-375 |
PDK1, |
[79] |
|
miR-21 |
RECK,PTEN, and PDCD4 |
[80,81] |
|
miR-106b-25 |
E2F1, TGF-beta, BCL2L11 and CDKN1A |
[82] |
|
Brain cancer |
||
|
miR-124a |
C/EBP, Atgl and Cgi-58 |
[83-85] |
|
miR-17 |
p21, Bim |
[86] |
|
miR-27a |
FOXO3a |
[87] |
|
miR-381 |
LRRC4 |
[88] |
|
miR-1908 |
PTEN |
[89] |
|
Colorectal and Prostate cancer |
||
|
miR-27a |
SGPP1 and Smad2 |
[90] |
|
miR-574 |
EGFR, RAC1 and EP300 |
[91] |
|
miR-135b |
TGFBR2 |
[92] |
|
miR-181a |
WIF-1, ?-catenin and E-cadherin |
[93] |
|
Liver Cancer |
||
|
miR-155-3p |
FBXW7 |
[94] |
|
miR-122 |
p53, Mdm2 |
[95] |
|
miR-1258 |
CKS1B |
[96] |
|
miR-182 |
NKG2A and NKG2D |
[97] |
|
miR-185 |
EMT |
[98] |
|
miR-186 |
YAP1 |
[99] |
|
Cervical Cancer |
||
|
miR-21 |
PDCD4, PTEN, CCL20, HIF-1? and p-Akt |
[100-102] |
|
miR-10a/b |
HOXA1 |
[103] |
|
miR-328 |
TCF7L2 |
[104] |
|
miR-138 |
c-Met |
[105] |
Table 1: miRNA and regulatory genes.
5. Leukemia
The studies have shown that the abnormality in dicer is a major constitute of miRNA biogenesis resulting in the aberrant development of T-cells and ultimately defective immune system [106]. The miR-146 is major constitute of innate and adaptive immunity as well as by direct targeting the FADD [60] and involved in the T-lymphocytes differentiation. The miR-146 generates the immune response via releasing the pro-inflammatory cytokines upon the pathogens exposure by targeting the NF-kB [107-109]. There are some transcriptional factors like NFI/CEBP and GATA1 found associated with the proper development of miR-223 at pri-miRNA stage [110]. The miR-223 negatively controls the expression of E2F1 protein that plays a role in cell cycle progression [61]. The miR-150 has a critical role in the development of B and T lymphocytes by controlling the transcriptional factor Myb and Notch, miR-150 down-regulation leads to lymphatic tissue tumor-genesis [62-64]. The secondary targets of miR-150 are Akt and DKC1 that are the regulator of Bim and P53 pro-apoptotic protein and finally lead to tumor suppression [65]. The Leukemia is actually the abnormality in the immune system. The miRNA play important role in the functioning and development of immune system [111]. There are sub-categories in the Leukemia. Acute myeloid leukemia (AML) is sub-category of leukemia bearing different types of mutation status of miRNAs. The miR-495 found down-regulated in most of the leukemic patients and has targeted genes including MEIS1 and PBX3 [66]. The miR-34b found up-regulated in healthy persons while significantly down-regulated in AML patients, miR-34b targets to HSF1, in AML HSF1 significantly up-regulated and raise the expression of Wnt-?-catenin pathway [67]. Among the studies miR-17/92 is found to be an important gene regulatory microRNA cluster, up-regulated in lymphatic leukemia and inhibits the PTEN and Bim pro-apoptotic genes expression [72]. The miR-34a is found to be associated with the p53 genes expression and detected as down-regulated in leukemia cells and p53 become also down-regulated. The miR-34a also restricts the expression of B-Myb and E2F1 [69, 112].
6. Breast Cancer
The miR-148b was noticed down-regulated in aggressive breast cancer. The miR-148b inhibit the malignancy by controlling the 130 genes of various signaling pathways, predominantly PIK3CA/p110?, ROCK1, NRAS, ITGA5 and CSF1 [70]. The miR-340 targets to Zinc finger E-box binding homeobox 1 (ZEB1) gene via directly binding to the 3?-UTR. The ZEB1 transcriptionally induces the suppression of miR-340 and also increase the expression of TGF-? mediated cancer progression and detected as a potential marker in breast cancer [72]. Aberrantly express miRNAs resulting into oncomiRNAs. The miRNA have a significant role in membrane-particles, tumorigenesis, exosomes, apoptotic bodies and provides cell-to-cell communication system. The MiR-34 family is comprised of miR-34a, miR-34b, and miR-34c and has been linked to the regulation of p53, Wnt/b-catenin, Hedgehog and Notch signaling pathways [73]. The miR-17~92 microRNA (miRNA) genes were found up-regulated and major contributor in cancer angiogenesis. The normal expression of miR-17~92 targets the angiogenesis genes HIF1?, VEGFA, and TGFBR2. The TGFBR2 3? UTR covers numerous well-preserved binding sites for miR-19a and miR-17/20a. [71]. MiR-22 was found up-regulated in breast cancer cells and promotes metastasis by targeting the lysine acetyl-transferase (TIP60). TIP60 was known as a portion of a well-preserved multi-subunit complex, NuA4, that is employed by numerous transcription factors toward their mark promoters, where it acetylates histones and involved in transcriptional regulation, DNA repairing, proliferation and apoptosis [113]. The miR-124 found down-regulated in breast cancer while normally it inhibits the metastasis by regulating the CTGF, RhoG, ROCK1 and ITGB1 [113].
7. Gastric Cancer
The miR-29c was found crucially down-regulated in gastric cancer and help in the progression of cancer [75]. While normally down-regulate ADAM12-L at mRNA levels [76]. MiR-26b is specifically linked with gastric cancer and noted down-regulated. Normally, miR-26b targets karyopherin alpha 2 (KPNA2)/c-Jun, induces suppression via direct binding to its 3'UTR region [77]. MicroRNA-214 inhibits the gastric cancerous cells proliferation by targeting the colony stimulating factor 1 (CSF1) it expresses the negative correlation with lymph node metastasis [78]. The miR-375 is also found down-regulated; it induces apoptotic effects via down-regulation of PDK1, which ultimately suppress the PI3K/Akt pathway [79]. The MiR-21 negatively regulates the TS genes including RECK, PTEN, and PDCD4 and leads to the gastric cancer invasion [80, 81]. The miR-106b-25 cluster noted up-regulated, it’s up-regulation leads to a functional abnormality of various genes mainly in E2F1 that is a check of DNA replication and ultimately aberrant expression of TGF-beta(major regulatory unit of cytokines) and as well as chemotherapy resistance by targeting the BCL2L11 and CDKN1A [82].
8. Brain Cancer
The loss of miR-124a was frequently detected in glioblastoma that attributes in the malignancy progression. The miR-124a to be regulated by the repressor-element-1-silencing transcription factor (REST, also known as a neuron-restrictive silencing factor) and miR-124a could be regulated via the regulation of REST [83], inhibits the C/EBP-? [84] and regulatory unit of Atgl and Cgi-58 and cellular triacylglycerol (TG) catabolism [85]. The micro RNA in the CNS like miR-17 effect to the p21, BIM genes and predominantly increase the tumor genesis both in-vivo and in-vitro [86] miR-27a modulate the expression of FOXO3a at mRNA level and leads the U87 growth suppression [87]. The miR-381 abnormal expression leads to effect on LRRC4 and ultimately promotes tumor growth in glioma cells [88] whereas miR-1908 directly hit to the PTEN promote cancer cell proliferation in Glioblastoma cells [89].
9. Colorectal and Prostate Cancer
The miR-27a down-regulation correlates with SGPP1 and Smad2 up-regulation and ultimately link to STAT3 regulatory pathway, mainly contribute in colorectal cancer. Increased miR-27a significantly repressed SGPP1 and Smad2 at both transcriptional and translational levels and inhibit the mutant cell proliferation by incorporating apoptosis [90]. It was found that the expression of miR-574-3p was significantly lowered in PCa cells. The lower stance of the miR-574-3p was associated with the advanced stage of the tumor. While its re-expression consequentially inhibited the migration, proliferation, and invasion of cancerous cells and induce apoptosis by regulation of Bcl-xL and triggering the caspace-3. It is also concluded that the miR-574-3p also regulates many important pathways such as Wnt and Jak-STAT signaling pathway by regulating the genes EGFR, RAC1, and EP300 via directly binding to 39 UTR [91]. The TGF-? signaling pathway known as tumor-suppressor pathway found usually deactivated in cancer and the indolent TGFBR2 is a cause of deviant TGF-? signaling pathway. The miRNA are major contributory units of post-transcriptional mechanism and detected that the miR-135b has a specific target for the 3’-untranslated region (3’-UTR) of TGFBR2. There was an inverse relation in TGFBR2 and miR-135b, the over-expression of miR-135b inhibits the expression of TGFBR2 and demonstrates itself as an oncogene in cancer by inhibiting the apoptosis and inducing the cell proliferation in colorectal cancer [92]. The over-expression of miR-181a contributes in colorectal cancer by targeting the WIF-1 gene and it is noted at an advance stage of the tumor. The miR-181 on ectopic expression suppressed the epithelial markers ?-catenin and E-cadherin, while enhanced the mesenchymal marker vimentin [93].
10. Liver Cancer
The miR-155-3p was unusually upregulated in hepatocellular carcinoma (HCC), its over-expression up-surge the abnormal cell proliferation and tumor genesis. The up-regulatory expression of miR-155-3p is linked with reduced levels FBXW7 mainly done constraining the expression of FBXW7 [94]. The miR-122 is an important regulatory unit to carry out normal cell cycle in hepatocytes by targeting the p53 and down-regulating the Mdm2 [95]. A study showed, miR-1258 significantly reduced in the HCC with its over-expression by targeting to CKS1B that contribute to liver cancer progression [96]. The miR-182 over-expression induces cytotoxicity against cancer by modulating the expression of NKG2A and NKG2D by mean of down and up regulation, respectively [97]. The miR-185 expression was significantly reduced in the HCC while its targeted gene six2 leads to metastasis. It also confirms that ectopic expression of miR-185 reversed EMT through the up-regulation of E-cadherin and down-regulation of vimentin in epithelial and mesenchymal HCC cells [98]. The miR-186 was found dominantly up-regulated in HCC and targets to Yes-associated protein 1 (YAP1) at mRNA level while normally, YAP1 inhibits the initiation of carcinogenesis [99].
11. Cervical Cancer
Cervical cancer is one of the most frequently occurring cancers among female. The miR-21 found over-expressed in cervical cancer and trigger the negative regulation of PDCD4 and PTEN [100, 101] and up-regulation of CCL20, HIF-1?, and p-Akt [102, 103] miR-376c significantly found down-regulated in clinical tissues of cervical cancer that directly regulates the BMI1 [104]. The studies evidenced that the over-activation of BMI1 leads to chemotherapy resistance in many cancer. BMI1 plays a crucial role in cell cycle, immortalization and senescence [105]. The miR-10a/b previously reported that it is down-regulated in gastric cancer, but Zou et al. [106] reported, miR-10a/b is a negative regulator of HOXA1, found significantly down-regulated in cervical cancer including the sample of mild, moderate and severe dysplasia and leads to more aggressiveness. The miR-328 and miR-138 significantly found reduced in cervical cancer [107, 108]. Both miRNAs were negative regulator of TCF7L2 (major functional unit of Wnt signaling) and c-Met, respectively [107, 108].
12. Conclusion
Although, various drugs and therapies have been established to treat cancer. The major problematic thing is the development of resistant in cancer cells against therapeutics. Therefore, it is need to develop some therapies against cancer with minimal side effect and great success rate. CRISPR/ Cas9 used for genome and epigenome editing scheme in the direction of therapeutic applications and opening the gate to treat numerous human and animal disease. CRISPR/ Cas9 is best technology to treat the cancer by targeting the miRNA`s rather than genes because majority of genes are controlled by miRNA`s.
References
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6 (2005): 376-85.
- Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455 (2008): 58-63.
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15 (2014): 509-524.
- Yin Y, Shen C, Xie P, et al. Construction of an initial microRNA regulation network in breast invasive carcinoma by bioinformatics analysis. Breast (2016): 1-10.
- McCormick F. Cancer gene therapy: fringe or cutting edge? Nat Rev Cancer 1(2001): 130-141.
- Roth JA. Adenovirus p53 gene therapy. Expert Opin Biol Ther 6 (2006): 55-61.
- Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 70 (2010): 5923-5930.
- Chiyomaru T, Yamamura S, Fukuhara S, et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One 8 (2013): e70372.
- Liu M, Zhang X, Hu CF, et al. MicroRNA-mRNA functional pairs for cisplatin resistance in ovarian cancer cells. Chin J Cancer 33 (2014): 285-294.
- Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 19 (2011): 1116-1122.
- Peng J, Mo R, Ma J, et al. let-7b and let-7c are determinants of intrinsic chemoresistance in renal cell carcinoma. World J Surg Oncol 13 (2015): 175.
- Saito Y, Suzuki H, Imaeda H, et al. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. Int J Cancer 132 (2013): 1751-1760.
- Maresso KC, Tsai KY, Brown PH, et al. Molecular cancer prevention: Current status and future directions. CA Cancer J Clin 65 (2015): 345-383.
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327 (2010): 198-201.
- Rupaimoole R, Han HD, Lopez-Berestein G, et al. MicroRNA therapeutics: principles, expectations, and challenges. Chin J Cancer 30 (2011): 368-370.
- Wu Y, Kang T. Protein stability regulators screening assay (Pro-SRSA): protein degradation meets the CRISPR?Cas9 library. Chin J Cancer 35 (2016): 60.
- Kawamura N, Nimura K, Nagano H, et al. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget 6 (2015): 22361-22374.
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31 (2013): 397-405.
- Chang H, Yi B, Ma R, et al. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 6 (2016): 22312.
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45 (2011): 273-297.
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436 (2005): 740-744.
- Fukuda T, Yamagata K, Fujiyama S, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9(2007): 604-611.
- Havens MA, Reich AA, Hastings ML. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet 10 (2014): e1004312.
- Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem 280 (2005): 27595-27603.
- Burke JM, Kelenis DP, Kincaid RP, et al. A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA. RNA 20 (2014): 1068-1077.
- Kawahara H, Okada Y, Imai T, et al. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem 286 (2011): 16121-16130.
- Yu Z, Wang L, Wang C, et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun 4 (2013): 2812.
- Ota H, Sakurai M, Gupta R, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153 (2013): 575-589.
- Xiaohua S, Deepavali C; Elsa FR. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer 13 (2013): 136-143.
- Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol 46 (2009): 1222-1228.
- Miyoshi K, Okada TN, Siomi H, et al. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA 15 (2009): 1282-1291.
- Martinez NJ, Chang HM, Borrajo Jde R, et al. The co-chaperones Fkbp4/5 control Argonaute2 expression and facilitate RISC assembly. RNA 19 (2013): 1583-1593.
- Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13 (2015): 17-24.
- Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284 (2009): 17897-17901.
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436 (2005): 740-744.
- Wang HW, Noland C, Siridechadilok B, et al. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol 16 (2009): 1148-1153.
- Fu Y, Sander JD, Reyon D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32 (2014): 279-284.
- Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351(2016): 84-88.
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339 (2013): 819-823.
- Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell 54 (2014): 234-244.
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23 (2014): R40-R46.
- Yue J, Du Z, Zhou FM, et al. Applications of CRSIPR/Cas9 in Cancer Research. Cancer Med Anticancer Drug 1(2016): 103.
- Smurnyy Y, Cai M, Wu H, et al. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat. Chem. Biol 10 (2014): 623-625.
- Neggers JE, Vercruysse T, Jacquemyn M, et al. Identifying drug-target selectivity of small-molecule CRM1/XPO1 inhibitors by CRISPR/Cas9 genome editing. Chem Biol 22 (2015): 107-116.
- Hendel A, Kildebeck EJ, Fine EJ, et al. Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Rep 7 (2014): 293-305.
- Mangone FR, Miracca EC, Feilotter HE, et al. ATM gene mutations in sporadic breast cancer patients from Brazil. Springer plus 4 (2015): 23.
- Demicco EG, Park MS, Araujo DM, et al. Solitary fibrous tumor: a clinicopathological study of 110 cases and proposed risk assessment model. Mod Pathol 25 (2012): 1298-1306.
- Zhang P, Pollock RE. Epigenetic regulators: New therapeutic targets for soft tissue sarcoma. cancer and microenvironment 1 (2014): e191.
- Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 32 (2014): 941-946.
- Drost J, van Jaarsveld RH, Ponsioen B, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521 (2015): 43-47.
- Chang H, Yi B, Ma R, et al. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 6 (2016): 22312.
- Mets E, Van Peer G, Van der Meulen J, et al. MicroRNA-128-3p is a novel oncomiR targeting PHF6 in T-cell acute lymphoblastic leukemia. Haematologica 99 (2014): 1326-1333.
- Zhao Y, Dai Z, Liang Y, et al. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci Rep 4 (2014): 3943.
- Jiang Q, Meng X, Meng L, et al. Small indels induced by CRISPR/Cas9 in the 5' region of microRNA lead to its depletion and Drosha processing retardance. RNA Biol 11 (2014): 1243-1249.
- Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 56 (2010): 1733-17341.
- Neviani P, Fabbri M. Exosomic microRNAs in the Tumor Microenvironment. Front Med (Lausanne) 2 (2016): 47.
- Yin J, Bai Z, Song J, et al. Differential expression of serum miR-126, miR-141 and miR-21 as novel biomarkers for early detection of liver metastasis in colorectal cancer. Chin J Cancer Res 26 (2014): 95-103.
- Chai S, Ma S. Clinical implications of microRNAs in liver cancer stem cells. Chin J Cancer 32 (2013): 419-426.
- Sun Y, Guo F, Bagnoli M, et al. Key nodes of a microRNA network associated with the integrated mesenchymal subtype of high-grade serous ovarian cancer. Chin J Cancer 34 (2015): 28-40.
- Curtale G, Citarella F, Carissimi C, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115 (2010): 265-273.
- Baek D, Villén J, Shin C, et al. The impact of microRNAs on protein output. Nature 455 (2008): 64-71.
- Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med 205 (2008): 1983-1991.
- Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131 (2007): 146-159.
- Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood 117 (2011): 7053-7062.
- Watanabe A, Tagawa H, Yamashita J, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia 25 (2011): 1324-1334.
- Jiang X, Huang H, Li Z, et al. MiR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc Natl Acad Sci USA 109 (2012): 19397-19402.
- Li G, Song Y, Zhang Y, et al. miR-34b Targets HSF1 to Suppress Cell Survival in Acute Myeloid Leukemia. Oncol Res 24 (2016): 109-116.
- Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9 (2008): 405-414.
- Zenz T, Mohr J, Eldering E, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 113 (2009): 3801-3808.
- Cimino D, De Pittà C, Orso F, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J 27 (2013): 1223-1235.
- Ma H, Pan JS, Jin LX, et al. MicroRNA-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett 376 (2016): 293-302.
- Hou LK, Yu Y, Xie YG, et al. miR-340 and ZEB1 negative feedback loop regulates TGF-?- mediated breast cancer progression. Oncotarget 7 (2016): 26016-26026.
- Prokopi M, Kousparou CA, Epenetos AA. The Secret Role of microRNAs in Cancer Stem Cell Development and Potential Therapy: A Notch-Pathway Approach. Front Oncol 4 (2015): 389.
- Pandey AK, Zhang Y, Zhang S, et al. TIP60-miR-22 axis as a prognostic marker of breast cancer progression. Oncotarget 6 (2015): 41290-41306.
- Saito Y, Suzuki H, Imaeda H, et al. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. Int J Cancer 132 (2013): 1751-1760.
- Duhachek-Muggy S, Zolkiewska A. ADAM12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer 15 (2015): 93.
- Tsai MM, Huang HW, Wang CS, et al. MicroRNA-26b inhibits tumor metastasis by targeting the KPNA2/c-jun pathway in human gastric cancer (2016): 8629.
- Wang M, Zhao C, Shi H, et al. Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: novel biomarkers and a mechanism for gastric cancer. Br J Cancer 110 (2014): 1199-1210.
- Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res 70 (2010): 2339-2349.
- Zhang Z, Li Z, Gao C, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest 88 (2008): 1358-1366.
- Zhang BG, Li JF, Yu BQ, et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep 27 (2012): 1019-1026.
- Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13 (2008): 272-286.
- Tivnan A, Zhao J, Johns TG, et al. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumour Biol 35 (2014): 1459-1465.
- Ponomarev ED, Veremeyko T, Barteneva N, et al. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-?-PU.1 pathway. Nat Med 17 (2011): 64-70.
- Das SK, Stadelmeyer E, Schauer S, et al. Micro RNA-124a regulates lipolysis via adipose triglyceride lipase and comparative gene identification 58. Int J Mol Sci 16 (2015): 8555-8568.
- Fontana L, Fiori ME, Albini S, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3 (2008): e2236.
- Ge YF, Sun J, Jin CJ, et al. AntagomiR-27a targets FOXO3a in glioblastoma and suppresses U87 cell growth in vitro and in vivo. Asian Pac J Cancer Prev 14 (2013): 963-968.
- Tang H, Liu X, Wang Z, et al. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res 1390 (2011): 21-32.
- Xia X, Li Y, Wang W, et al. MicroRNA-1908 functions as a glioblastoma oncogene by suppressing PTEN tumor suppressor pathway. Mol Cancer 14 (2015):154.
- Bao Y, Chen Z, Guo Y, et al. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS One 9 (2014): e105991.
- Chiyomaru T, Yamamura S, Fukuhara S, et al. Genistein up-regulates tumor suppressor microRNA-574-3p in prostate cancer. PLoS One 8 (2013): e58929.
- Li J, Liang H, Bai M, et al. Correction: miR-135b Promotes Cancer Progression by Targeting Transforming Growth Factor Beta Receptor II (TGFBR2) in Colorectal Cancer. PLoS One 10 (2015): e0145589.
- Ji D, Chen Z, Li M, et al. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer 13 (2014): 86.
- Tang B, Lei B, Qi G, et al. MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression. J Exp Clin Cancer Res 35 (2016): 93.
- Simerzin A, Zorde-Khvalevsky E, Rivkin M, et al. The liver-specific microRNA-122, the complementary strand of microRNA-122, acts as a tumor suppressor by modulating the p53/mouse double minute 2 homolog circuitry. Hepatology (2016): 28679.
- Hu M, Wang M, Lu H, et al. Loss of miR-1258 contributes to carcinogenesis and progression of liver cancer through targeting CDC28 protein kinase regulatory subunit 1B. Oncotarget (2016): 9728.
- Abdelrahman MM, Fawzy IO, Bassiouni AA, et al. Enhancing NK cell cytotoxicity by miR-182 in hepatocellular carcinoma. Hum Immunol 77 (2016): 667-673.
- Zhu SM, Chen CM, Jiang ZY, et al. MicroRNA-185 inhibits cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Six2. Eur Rev Med Pharmacol Sci 20 (2016): 1712-1719.
- Ruan T, He X, Yu J, et al. MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol Lett 11 (2016): 2941-2945.
- Yao Q, Xu H, Zhang Q-Q, et al. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells.Biochemical and Biophysical Research Communications 388 (2009): 539-542.
- Gomez-Gomez Y, Organista-Nava J, Ocadiz-Delgado R, et al. The expression of miR-21 and miR-143 is deregulated by the HPV16 E7 oncoprotein and 17?-estradiol. Int J Oncol 49 (2016): 549-558.
- Song N, Liang B, Wang D. The function of MiR-21 expression differences and pathogenesis on familial and triple negative breast Cancer serum. Pak. J. Pharm. Sci 29 (2016): 679-684.
- Yao T, Lin Z. MiR-21 is involved in cervical squamous cell tumorigenesis and regulates CCL20. Biochim Biophys Acta 1822 (2012): 248-260.
- Deng Y, Xiong Y, Liu Y. miR-376c inhibits cervical cancer cell proliferation and invasion by targeting BMI1. Int J Exp Pathol 97 (2016): 257-265.
- Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells 30 (2012): 372-378.
- Zou D, Zhou Q, Wang D, et al. The Downregulation of MicroRNA-10b and its Role in Cervical Cancer. Oncol Res 24 (2016) :99-108.
- Wang X, Xia Y. microRNA-328 inhibits cervical cancer cell proliferation and tumorigenesis by targeting TCF7L2. Biochem Biophys Res Commun 475 (2016): 169-175.
- Li J, Xia W, Su X, et al. Species-specific mutual regulation of p53 and miR-138 between human, rat and mouse. Sci Rep 6 (2016): 261-287.
- Muljo SA, Ansel KM, Kanellopoulou C, et al. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med 202 (2005): 261-269.
- Williams AE, Perry MM, Moschos SA, et al. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans 36 (2008): 1211-1215.
- Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103 (2006): 12481-12486.
- Zhao B, Zou J, Wang H, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA 108 (2011): 14902-14907.
- Fukao T, Fukuda Y, Kiga K, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell 129 (2007): 617-631.
