The Multiple Molecular Signatures in Gallbladder Carcinoma: from Basic Studies to Clinical Application
Article Information
Zibo Yin1, Guowei Yang1, Bowen Qian1, Tingming Liang2*, Li Guo1*
1Department of Bioinformatics, Smart Health Big Data Analysis and Location Services Engineering Lab of Jiangsu Province, School of Geographic and Biologic Information, Nanjing University of Posts and Telecommunications, Nanjing, China
2Jiangsu Key Laboratory for Molecular and Medical Biotechnology, College of Life Science, Nanjing Normal University, Nanjing, China
*Corresponding Authors: Tingming Liang, Jiangsu Key Laboratory for Molecular and Medical Biotechnology, College of Life Science, Nanjing Normal University, Nanjing, China
Li Guo, Department of Bioinformatics, School of Geographic and Biologic Information, Nanjing University of Posts and Telecommunications, Nanjing, China
Received: 03 June 2019; Accepted: 17 June 2019; Published: 05 July 2019
Citation: Zibo Yin, Guowei Yang, Bowen Qian, Tingming Liang, Li Guo. The Multiple Molecular Signatures in Gallbladder Carcinoma: from Basic Studies to Clinical Application. Journal of Bioinformatics and Systems Biology 2 (2019): 028-042.
View / Download Pdf Share at FacebookAbstract
Gallbladder carcinoma (GBC) is a malignant tumor in gastrointestinal system. In this review, we mainly focus on three molecular levels to illuminate the potential molecular mechanisms in gallbladder carcinoma. First, we review genes with mutation and methylation associated with occurrence and development of GBC. Second, we review non-coding RNA and key differential genes at the transcript level, especially for their interactions. Third, we review crucial proteins in GBC. Moreover, we also discuss the challenges of these molecular signatures in clinical applications. Finally, we discuss potential application of these crucial genes in prevention, diagnosis and treatment of GBC.
Keywords
Gallbladder carcinoma (GBC), Diagnosis, Treatment
Gallbladder carcinoma (GBC) articles, Diagnosis articles, Treatment articles
Gallbladder carcinoma (GBC) articles Gallbladder carcinoma (GBC) Research articles Gallbladder carcinoma (GBC) review articles Gallbladder carcinoma (GBC) PubMed articles Gallbladder carcinoma (GBC) PubMed Central articles Gallbladder carcinoma (GBC) 2023 articles Gallbladder carcinoma (GBC) 2024 articles Gallbladder carcinoma (GBC) Scopus articles Gallbladder carcinoma (GBC) impact factor journals Gallbladder carcinoma (GBC) Scopus journals Gallbladder carcinoma (GBC) PubMed journals Gallbladder carcinoma (GBC) medical journals Gallbladder carcinoma (GBC) free journals Gallbladder carcinoma (GBC) best journals Gallbladder carcinoma (GBC) top journals Gallbladder carcinoma (GBC) free medical journals Gallbladder carcinoma (GBC) famous journals Gallbladder carcinoma (GBC) Google Scholar indexed journals Diagnosis articles Diagnosis Research articles Diagnosis review articles Diagnosis PubMed articles Diagnosis PubMed Central articles Diagnosis 2023 articles Diagnosis 2024 articles Diagnosis Scopus articles Diagnosis impact factor journals Diagnosis Scopus journals Diagnosis PubMed journals Diagnosis medical journals Diagnosis free journals Diagnosis best journals Diagnosis top journals Diagnosis free medical journals Diagnosis famous journals Diagnosis Google Scholar indexed journals Treatment articles Treatment Research articles Treatment review articles Treatment PubMed articles Treatment PubMed Central articles Treatment 2023 articles Treatment 2024 articles Treatment Scopus articles Treatment impact factor journals Treatment Scopus journals Treatment PubMed journals Treatment medical journals Treatment free journals Treatment best journals Treatment top journals Treatment free medical journals Treatment famous journals Treatment Google Scholar indexed journals miRNA articles miRNA Research articles miRNA review articles miRNA PubMed articles miRNA PubMed Central articles miRNA 2023 articles miRNA 2024 articles miRNA Scopus articles miRNA impact factor journals miRNA Scopus journals miRNA PubMed journals miRNA medical journals miRNA free journals miRNA best journals miRNA top journals miRNA free medical journals miRNA famous journals miRNA Google Scholar indexed journals protein expression articles protein expression Research articles protein expression review articles protein expression PubMed articles protein expression PubMed Central articles protein expression 2023 articles protein expression 2024 articles protein expression Scopus articles protein expression impact factor journals protein expression Scopus journals protein expression PubMed journals protein expression medical journals protein expression free journals protein expression best journals protein expression top journals protein expression free medical journals protein expression famous journals protein expression Google Scholar indexed journals DNA articles DNA Research articles DNA review articles DNA PubMed articles DNA PubMed Central articles DNA 2023 articles DNA 2024 articles DNA Scopus articles DNA impact factor journals DNA Scopus journals DNA PubMed journals DNA medical journals DNA free journals DNA best journals DNA top journals DNA free medical journals DNA famous journals DNA Google Scholar indexed journals lncRNA-LET articles lncRNA-LET Research articles lncRNA-LET review articles lncRNA-LET PubMed articles lncRNA-LET PubMed Central articles lncRNA-LET 2023 articles lncRNA-LET 2024 articles lncRNA-LET Scopus articles lncRNA-LET impact factor journals lncRNA-LET Scopus journals lncRNA-LET PubMed journals lncRNA-LET medical journals lncRNA-LET free journals lncRNA-LET best journals lncRNA-LET top journals lncRNA-LET free medical journals lncRNA-LET famous journals lncRNA-LET Google Scholar indexed journals CRNDE articles CRNDE Research articles CRNDE review articles CRNDE PubMed articles CRNDE PubMed Central articles CRNDE 2023 articles CRNDE 2024 articles CRNDE Scopus articles CRNDE impact factor journals CRNDE Scopus journals CRNDE PubMed journals CRNDE medical journals CRNDE free journals CRNDE best journals CRNDE top journals CRNDE free medical journals CRNDE famous journals CRNDE Google Scholar indexed journals
Article Details
Abbreviations:
mRNA-messenger RNA; miRNA-microRNA; ncRNA-non-coding RNA; lncRNA-long non-coding RNA; GBC-gallbladder carcinoma1. Introduction
Gallbladder carcinoma (GBC) is the most common biliary tract malignancy and has a poor prognosis, frequently presenting at an advanced stage. Prevalence rates of up to 7.5 per 100,000 for men and 23 per 100,000 for women have been reported from Andean-area populations, North-American Indians, Mexican Americans and inhabitants of Northern India [1]. Fewer than 10% of GBC patients survive more than 5 years after treatment [2]. GBC is hard to detect in the early stage, and most patients cannot be cured by surgery after detection because of the invasion of cancerous cells. For dozens of years of clinical practice, surgery was the only effective way to treat GBC. However, surgery usually cannot cure patients because the pathogeny of GBC has not been completely disclosed and the invasive cancerous cells cannot be eliminated surgically. Therefore, novel methods may be important ways to cure GBC, such as targeted therapy, which has broad application [3].
In this review, we focus on the detection and treatment signatures of GBC with the goal of illustrating the crosstalk among DNA, RNA and protein levels (Figure 1), and we expect that this review will provide useful information for the clinical diagnosis and treatment of GBC.
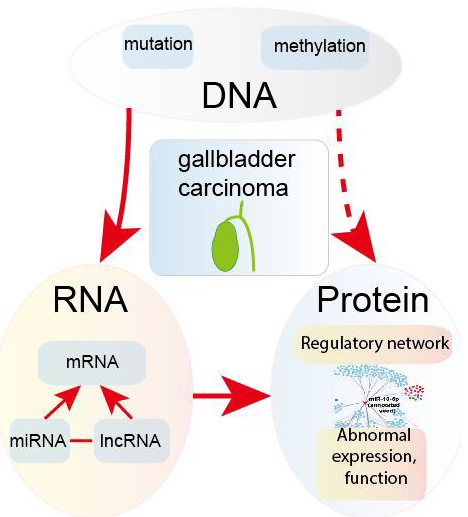
Figure 1: The three molecular signature levels are reviewed.
2. Genomic Signatures of GBC
Here, the molecular signatures at DNA level mainly include mutation and methylation. The mutation of coding genes provides great advantages in the diagnosis and guidance of treatment. Research in Chile showed that allele-specific mutations could affect the incidence of GBC [4]. Inherited rare germline mutations had been proved to be related to GBC [5] and epidermal growth factor receptor mutations may have advantages for the treatment of gallbladder cancer [6]. Furthermore, some key genes, like P53, K-ras, Keap1, PIK3CA, EGFR, P16 and B-raf, have been explored. For example, P53 mutation in exons 5-8 were found in GBC patients and K-ras in codon 12 was important in the early stage [7, 8]. Mutation of Keap1 was found in C249Y and S338L led to the loss of Nrf2 repression activity [9]. Moreover, PIK3CA mutation in exons 9 was found specifically and may be effective in targeted therapies [10]. EGFR mutation in exons 19-21 could sustain survival and proliferation of GBC cells [11] and high percentage of B-raf mutations in exon 15 was found [12]. Therefore, gene mutation in GBC is meaningful for the understanding and treatment of GBC (Table 1).
The methylation of DNA also greatly contributes to diagnosis and therapy. A recent study showed that the patterns of gene promoter methylation allowed them to be considered biomarkers for the early detection, diagnosis, prognosis and therapeutic selection [13]. Furthermore, gene-specific DNA methylation (such as APC, CDKN2A, ESR1, PGP9.5 and SSBP2) had the same function [14]. Another study in north-central India proved D4Z4 and DNF92 subtelomeric sequences to be hypermethylated and hypomethylated, respectively [15]. Aberrant hypermethylation of promoter regions was an early, progressive and cumulative event in GBC [16]. One study on the methylation profile of GBC showed that the methylation profile was different from that in a healthy individual, which proved that methylation was an early event [17]. MYC hypomethylation was only detected in tumoral samples and was associated with its protein expression (p=0.029) and MYC mutations were detected in 80% of GBC samples [18].
|
Genes |
Mutation position |
Ref. |
|
p53 |
exons 5,6,7,8 |
[7] |
|
k-ras |
codon 12 (GGT change to GAT) |
[8] |
|
Keap1 |
C249Y/S338L |
[9] |
|
PIK3CA |
exons 9 |
[10] |
|
EGFR |
exons19,20,21 |
[11] |
|
P16 |
exons 1, 2 |
[8] |
|
B-raf |
exon 15 |
[12] |
Table 1: The study of some key mutation genes in GBC.
3. Crucial mRNAs and ncRNAs in GBC
The most effective way to cure GBC is to diagnose the cancer in the early stage, but the disappointing reality is that GBC remains difficult to diagnose preoperatively. Furthermore, extension of the disease beyond the mucosa predicts a poor chance of long-term survival [19]. Increasing studies have shown that miRNAs, mRNAs and long non-coding RNAs are very effective in the diagnosis of GBC. mRNAs in GBC greatly affect physiological and pathophysiological conditions. Among these, human telomerase reverse transcriptase (hTERT) mRNA, a catalytic subunit of telomerase, had been determined to be effective for diagnosing the nature of the polypoid lesion in the gallbladder [20], which can help to diagnose GBC. Survivin, an inhibitor of apoptosis, played a possible role and was associated with poor prognosis [21]. SUMO-1 mRNA may be an interesting target in the diagnosis and treatment of GBC based on the differences expression in carcinoma of gallbladder, gallbladder tissues surrounding GBC, adenomatous polyp of gallbladder [22]. In addition, research on the positive expression of CD-146 [23], VEGF, Flt-1 and KDR [24] were also related to the incidence of GBC.
miRNA is involved in the initiation and progression of GBC (Table 2), and miRNA expression profiling can be used to identify signatures associated with diagnosis, staging, progression, prognosis and response to treatment [25]. miRNAs participate in a variety of regulatory pathways, and the expression level of miRNAs is closely connected to survival of patients with GBC. For example, patients with a high miRNA-155 expression level had distinctly shorter overall survival than patients with a low miRNA-155 expression level [26]. In addition, high miR-155 expression was related to aggressive GBC, and it may be a potential prognostic marker and therapeutic target [27]. miR-146b-5p inhibited growth of GBC by targeting epidermal growth factor receptor [28]. miR-335 may be associated with aggressive tumor behaviors and reduced expression of miR-335 may be a useful indicator for clinical outcome and could be a therapeutic target for primary GBC [29]. miR-29c-5p, a tumor-suppressive miRNA that may serve as a potential prognostic biomarker or therapeutic target for GBC, suppressed GBC progression by directly targeting CPEB4 and inhibiting the MAPK pathway [30]. Moreover, miR-133a-3p acted as a tumor suppressor by directly targeting the recombination signal-binding protein Jk (RBPJ) in GBC [31]. Another miRNA, miR-26a, may contribute to GBC proliferation by directly targeting HMGA2 and might be a prognostic factor and therapeutic target for GBC patients [32].
|
miRNA |
Expression(miRNA) |
mRNA |
Expression(mRNA) |
Ref. |
|
miR-146b-5p |
down |
EGFR |
up |
[28] |
|
miR-20a |
up |
Smad7 |
down |
[44] |
|
miR-29c-5p |
down |
CPEB4 |
up |
[30] |
|
miR-133a-3p |
down |
RBPJ |
up |
[31] |
|
miR-26a |
down |
HMGA2 |
up |
[32] |
|
miR-122 |
down |
PKM2 |
up |
[45] |
|
miR-143-3p |
down |
ITGA6 |
up |
[46] |
|
miR-101 |
down |
ZFX |
up |
[47] |
|
miR-30d-5p |
down |
LDHA |
up |
[48] |
|
miR-182 |
up |
CADM1 |
down |
[49] |
|
miR-30b |
down |
NT5E |
up |
[50] |
|
miR-340 |
down |
NT5E |
up |
[50] |
|
miR-125b-5p |
down |
Bcl2 |
up |
[51] |
|
miR-143-5p |
down |
HIF-1α |
up |
[52] |
|
miR-33a |
down |
IL-6 |
up |
[53] |
|
miR-223 |
down |
STMN1 |
up |
[54] |
|
miR-218-5p |
down |
PRKCE |
up |
[55] |
Table 2: Some abnormal miRNAs in GBC and their target mRNAs.
Through recent research, long non-coding RNAs also have been determined to play important roles in GBC pathogenesis [33]. Long non-coding RNA SPRY4-IT1 promoted GBC progression and may serve as a candidate target for new therapies in human GBC [34]. In addition, CRNDE was found an important contributor to GBC development as a scaffold to recruit the DMBT1 and c-IAP1 and affect PI3K-AKT pathway [35]. Furthermore, upregulation of HOXA-AS2 promoted proliferation and induced epithelial-mesenchymal transition in GBC, and it could be as a potential therapeutic target to inhibit GBC metastasis [36]. Another study showed that UCA1 promoted GBC cell proliferation and metastasis in vitro and suppressed the transcription of p21 and E-cadherin by recruiting an enhancer of the zeste homolog [37]. Moreover, lncRNA-H19 was a novel prognostic factor for GBC and might play important regulatory roles in the epithelial-mesenchymal transition (EMT) process [38]. Overexpression of lncRNA-LET conferred a proliferative advantage to tumor cells under hypoxic conditions. The ectopic expression of lncRNA-LET led to promotion of cell cycle arrest at G0/G1 phase and induction of apoptosis under hypoxic conditions. Abnormal expression of lncRNA-LET also suppressed gallbladder tumor growth, and lncRNA-LET was determined to a potential prognostic marker and therapeutic target for GBC [39].
Indeed, long non-coding RNAs have important interactions with mRNAs and miRNAs (Table 3 and Figure 2). For example, H19 regulated FOXM1 expression by competitively binding endogenous miR-342-3p [40] and GCASPC and miR-17-3p interaction regulated cells proliferation [41]. LINC00152 negatively regulated Bag-1 as a molecular sponge for miR-138 [42] and CCAT1 negatively regulated Bmil by spongeing miR-218-5p [43].
|
LncRNA |
Expression (LncRNA) |
miRNA |
Expression (miRNA) |
mRNA |
Expression (mRNA) |
Ref. |
|
LINC00152 |
up |
miR-138 |
down |
HIF-1a |
up |
[42] |
|
CCAT1 |
up |
miR-218-5p |
down |
Bmi1 |
up |
[43] |
|
HOTAIR |
up |
miR-130a |
down |
c-Myc |
up |
[56] |
|
H19 |
up |
miR-194-5p |
down |
AKT2 |
up |
[57] |
|
H19 |
up |
miR-342-3p |
down |
FOXM1 |
up |
[40] |
|
MALAT1 |
up |
miR-363-3p |
down |
MCL-1 |
up |
[58] |
|
MALAT1 |
up |
miR-206 |
down |
KRAS |
up |
[59] |
|
MALAT1 |
up |
miR-206 |
down |
ANXA2 |
up |
[59] |
|
PAGBC |
up |
miR-511 |
down |
PIK3R3 |
up |
[60] |
|
PAGBC |
up |
miR-133b |
down |
SOX4 |
up |
[60] |
|
TUG1 |
up |
miR-300 |
down |
TGF-β1 |
up |
[61] |
|
MINCR |
up |
miR-26a-5p |
down |
EZH2 |
up |
[62] |
|
GCASPC |
down |
miR-17-3p |
up |
pyruvate carboxylase |
up |
[41] |
Table 3: Some examples of the relationship between lncRNAs and micRNAs/mRNAs in GBC.
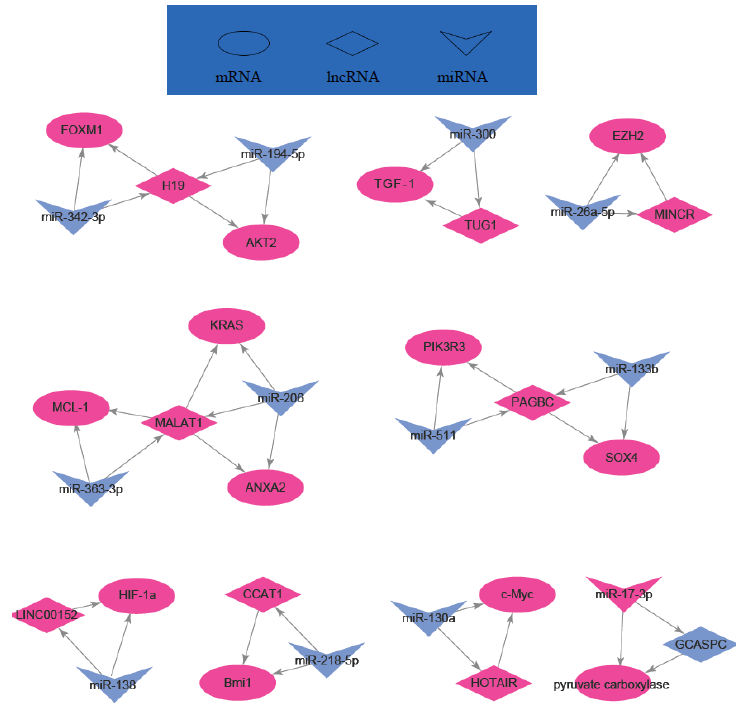
Figure 2: Regulation network of some lncRNAs, mRNAs and miRNAs in GBC.
4. Crucial proteins in GBC
Compared with molecules above mentioned, crucial proteins also contribute to diagnosis and treatment of GBC [65, 72]. The modification of proteins, mainly including protein phosphorylation and acetylation, may contribute to abnormal expression and/or function. Many proteins impacting on GBC have been reported (Table 4).
|
Protein |
Expression |
Tissue |
Ref. |
|
Bcl-2 |
up |
gallbladder tissue |
[63] |
|
c-erb-B2 |
up |
gallbladder tissue |
[64] |
|
MMP-2 |
up |
gallbladder tissue |
[65] |
|
TIMP-2 |
down |
gallbladder tissue |
[65] |
|
VEGF |
up |
gallbladder tissue specimens (formalin-fixed and paraffin-embedded) |
[66] |
|
FHIT |
down |
gallbladder tissue |
[67] |
|
MLH1 |
down |
gallbladder tissue |
[67] |
|
retinoblastoma |
up |
gallbladder tissue |
[68] |
|
p16INK4 |
down |
gallbladder tissue |
[68] |
|
S100A4 |
up |
gallbladder tissue |
[69] |
|
P53 |
up |
gallbladder tissue |
[69] |
|
p27 |
down |
gallbladder tissue |
[69] |
|
P16 |
down |
gallbladder tissue |
[69] |
|
RB |
down |
gallbladder tissue |
[69] |
|
Smad4 |
down |
gallbladder tissue |
[69] |
|
FHIT |
down |
gallbladder tissue |
[69] |
|
E-cadherin |
down |
gallbladder tissue |
[69] |
|
promyelocytic leukemia (PML) |
down |
gallbladder tissue |
[69] |
|
CDX2 |
up |
GBC cell lines |
[70] |
|
cyclin D1 |
up |
gallbladder tissue specimens (formalin-fixed and paraffin-embedded) |
[71] |
|
p16 |
down |
gallbladder tissue specimens (formalin-fixed and paraffin-embedded) |
[71] |
|
retinoblastoma |
up |
gallbladder tissue specimens (formalin-fixed and paraffin-embedded |
[71] |
|
MUC1 mucins |
up |
gallbladder tissue |
[72] |
|
MUC4 |
up |
gallbladder tissue |
[73] |
|
CD133 |
up |
gallbladder tissue |
[74] |
|
transcription factor specificity protein 1 (SP1) |
up |
gallbladder tissue/GBC-SD cell lines |
[75] |
Table 4: Some key proteins in GBC.
For example, the overexpression of Bcl-2 could promote tumor cell differentiation [63]. The overexpression of c-erb-B2 was related to the worse prognosis of GBC [64] and the ratio of MMP-2/TIMP-2 may be a new significant marker in early diagnosis [65]. Reduced Fhit expression might be involved in the development of GBC and be correlated with Mlh1 expression [67]. The high expression of retinoblastoma protein inhibited P16INK4 protein and related to the decreased survival in GBCs [68]. Furthermore, the overexpression of S100A4, P53 and the loss of p27, p16, RB, Smad4, FHIT, E-cadherin and PML expression led to poor survival. PML and P53 were found effective therapeutic targets for the disease [69]. The research on GBC cell lines showed that the overexpression of SP1 [75] and CDX2 appeared in most GBC cell lines, and the expression of CDX2 showed a relationship with the expression of MUC2 [70]. A study in gallbladder tissue specimens (formalin-fixed and paraffin-embedded) showed that cyclin D1 had a negative-correlation with P16 and affected the early stage of GBCs [71]. As mentioned above, the expression of protein may correlate with each other and affect disease diagnosis.
Phosphorylation also contributed to diagnosis and treatment of GBC [76]. For example, CD133 had a role in the migration of GBC cells through phosphorylation [74] and MUC4 interacted with ErbB2 to cause tumor growth, which was related to the hyperphosphorylation of ErbB2, MAPK and Akt [73]. Research on cirsimaritin showed that the pro-apoptotic effect of cirsimaritin could be reversed by down-regulating the phosphorylation of Akt [77] and affected the GBC-SD cell line. In addition, the protein phosphatase PHLPP was found to help treat GBC by inhibiting survivin phosphorylation [78]. Rise of CCK1 receptor expression is associated with the increase of protein lysine acetylation [79], showing that GBC can be treated by using histone deacetylase inhibitor [80].
5. Challenges and Conclusion
Although many factors are related to GBC, but GBC still cannot be diagnosed accurately in clinical settings. A clinical experiment in the Queen Mary Hospital, University of Hong Kong showed that 61% of patients had an inaccurate diagnosis [81], which showed that diagnosis is difficult in the early stage. Operative resection is still the only way to cure GBC, but unfortunately, only 38% of patients have been eligible for resection during the last 20 years. Furthermore, one report showed that none of the patients without surgery survived more than 5 years [82]. Therefore, the challenges of treating GBC are serious because only surgery in the early stage can cure GBC, but most patients are diagnosed at the intermediate or advanced stage.
Because of the current challenges and problems in the diagnosis and treatment of GBC, new methods for treating GBC must be developed. To this end, targeted therapy may be an effective way to cure GBC. Targeted therapy has been proved to affect GBC in clinical trials in the Clinical Center for Targeted Therapy at the MD Anderson Cancer Center [83]. GBC still cannot be readily cured by targeted therapy, especially in the advanced stage, and almost all patients who do not have surgery currently die within 1 year following clinical treatment [84]. Chemotherapy is another adjuvant therapy for GBC in clinical settings. During a randomized controlled study, patients who underwent chemotherapy after surgery had a higher 5-year survival rate than those with surgery alone. However, adverse drug reactions, such as anorexia and leukopenia, were usually associated with chemotherapy [85].
Research on the pathogenesis and prognosis of GBC has made great progress, and genes have been determined to be possible markers for prognosis. Lupeol in the EGFR/MMP-9 pathway was proved to induce apoptotic cell death in GBC, which can be used as a potential treatment method [86]. Long non-coding RNAs of LINC00152 and CRNDE in the PI3K-AKT pathway contribute to process of GBC carcinogenesis, and PI3K/AKT may be a potential therapeutic target for GBC [75, 85].
Taken together, in this review, the multiple molecular signatures in GBC were concluded. We hope that these potential signatures can provide some references for diagnosis and therapeutic of GBC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 61771251), the key project of social development in Jiangsu Province (No. BE2016773), the National Natural Science Foundation of Jiangsu (No. BK20171443), Sponsored by NUPTSF (Nos. NY215068 and NY217100), the Qinglan Project in Jiangsu Province, and the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD).
Conflict of Interest
None
References
- Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nature Reviews Cancer 4 (2004): 695-706.
- Adson MA. Carcinoma of the gallbladder. Surgical Clinics of North America 53 (1973): 1203-1216.
- Thomas MB. Targeted therapies for cancer of the gallbladder. Current Opinion in Gastroenterology 24 (2008): 372-376.
- Wistuba II, Sugio K, Hung J, et al. Allele-specific Mutations Involved in the Pathogenesis of Endemic Gallbladder Carcinoma in Chile. Cancer Research 55 (1995): 2511-2515.
- Yadav S, De SN, Kumari N, et al. Targeted Gene Sequencing of Gallbladder Carcinoma Identifies High-impact Somatic and Rare Germline Mutations. Cancer Genomics Proteomics 14 (2017): 495-506.
- Leone F, Cavalloni G, Pignochino Y, et al. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clinical Cancer Research An Official Journal of the American Association for Cancer Research 12 (2006): 1680.
- Hanada K, Itoh M, Fujii K, et al. K-ras and p53 mutations in stage I gallbladder carcinoma with an anomalous junction of the pancreaticobiliary duct. Cancer 77 (1996): 452-458.
- Kim YT, Jin K, Jang YH, et al. Genetic alterations in gallbladder adenoma, dysplasia and carcinoma. Cancer Letters 169 (2001): 59-68.
- Shibata T, Kokubu A, Gotoh M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135 (2008): 1358-1368.
- Deshpande V, Nduaguba A, Zimmerman SM, et al. Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma. BMC Cancer 11 (2011): 60.
- Francesco L, Giuliana C, Ymera P, et al. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clinical Cancer Research An Official Journal of the American Association for Cancer Research 12 (2006): 1680.
- Saetta AA, Papanastasiou P, Michalopoulos NV, et al. Mutational analysis of BRAF in gallbladder carcinomas in association with K-ras and p53 mutations and microsatellite instability. Virchows Archiv 455 (2004): 179-182.
- Letelier P, Brebi P, Tapia O, et al. DNA promoter methylation as a diagnostic and therapeutic biomarker in gallbladder cancer. Clinical Epigenetics 4 (2012): 1-11.
- Kagohara LT, Schussel JL, Subbannayya T, et al. Global and gene-specific DNA methylation pattern discriminates cholecystitis from gallbladder cancer patients in Chile. Future Oncology 11 (2015): 233-249.
- Poojary SS, Mishra G, Gupta S, et al. Dysfunction of subtelomeric methylation and telomere length in gallstone disease and gallbladder cancer patients of North Central India. Journal of hepato-biliary-pancreatic sciences 23 (2016): 276-282.
- Garcia P, Manterola C, Villaseca M, et al. Promoter methylation profile in preneoplastic and neoplastic gallbladder lesions. Molecular Carcinogenesis 48 (2009): 79-89.
- Takahashi T, Shivapurkar N, Riquelme E, et al. Aberrant promoter hypermethylation of multiple genes in gallbladder carcinoma and chronic cholecystitis. Clinical Cancer Research 10 (2004): 6126-6133.
- Ishak G, Leal MF, Santos NPCD, et al. Deregulation of MYC and TP53 through genetic and epigenetic alterations in gallbladder carcinomas. Clinical and Experimental Medicine 15 (2015): 421-426.
- Misra S, Chaturvedi A, Misra NC, et al. Carcinoma of the gallbladder. Surgical Clinics of North America 46 (2003): 1145-1151.
- Uchida N, Tsutsui K, Kobara H, et al. A case of gallbladder carcinoma diagnosed preoperatively by the detection of human telomerase reverse transcriptase mRNA in endoscopically obtained gallbladder bile. Endoscopy 35 (2003): 185-188.
- Nigam J, Chandra A, Kazmi HR, et al. Expression of survivin mRNA in gallbladder cancer: a diagnostic and prognostic marker?. Tumor Biology 35 (2014): 9241-9246.
- Zhang WX, Zhang YD, Peng J. The value researchof the SUMO-1 mRNA in the diagnosis of gallbladder carcinoma. China Journal of Modern Medicine 24 (2014): 43-45.
- Kapoor S. CD146 expression and its close relationship to tumor progression in systemic malignancies besides gall bladder carcinomas. Tumor Biology 34 (2013): 1273-1274.
- Fang HQ, Li HJ, Liu YB, et al. Effects of vascular endothelial growth factor (VEGF) antisense oligodeoxynucleotide on mRNA and expression of VEGF, flt-1, and kinase insert domain containing receptor and VEGF excretion in human gallbladder carcinoma cells. National Medical Journal of China 87 (2007): 3329-3334.
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America 103 (2006): 2257-2261.
- Zhang XL, Chen JH, QinCK. MicroRNA-155 expression as a prognostic factor in patients with gallbladder carcinoma after surgical resection. International Journal of Clinical and Experimental Medicine 8 (2015): 21241-21246.
- Kono H, Nakamura M, Ohtsuka T, et al. High expression of microRNA-155 is associated with the aggressive malignant behavior of gallbladder carcinoma. Oncology Reports 30 (2013): 17-24.
- Cai J, Xu L, Cai Z, et al. MicroRNA-146b-5p inhibits the growth of gallbladder carcinoma by targeting epidermal growth factor receptor. Molecular Medicine Reports 12 (2015): 1549-1555.
- Peng HH, Zhang YD, Gong LS, et al. Increased expression of microRNA-335 predicts a favorable prognosis in primary gallbladder carcinoma. Oncotargets and Therapy 6 (2013): 1625-1630.
- Shu YJ, Bao RF, Jiang L, et al. MicroRNA-29c-5p suppresses gallbladder carcinoma progression by directly targeting CPEB4 and inhibiting the MAPK pathway. Cell Death and Differentiation 24 (2017): 445-457.
- Huang Y, Wu Y, Dong J, et al. MicroRNA-133a-3p exerts inhibitory effects on gallbladder carcinoma via targeting RBPJ. American Journal of Cancer Research 6 (2016): 2448-2462.
- Zhou H, Guo W, Zhao Y, et al. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. International Journal of Oncology 44 (2014): 2050-2058.
- Manel E. Non-coding RNAs in human disease. Nature Reviews Genetics 12 (2011): 861-874.
- Yang L, Cheng X, Ge N, et al. Long non-coding RNA SPRY4-IT1 promotes gallbladder carcinoma progression. Oncotarget 8 (2016): 3104-3110.
- Shen S, Liu H, Wang Y, et al. Long non-coding RNA CRNDE promotes gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget 7 (2016): 72833-72844.
- Peng Z, Cao P, Zhu X, et al. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget 8 (2017): 33137-33143.
- Qiang C, Jin L, Wang S, et al. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget 8 (2017): 47957-47968.
- Wang SH, Wu XC, Zhang MD, et al. Upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition. American Journal of Cancer Research 6 (2016): 15-26.
- Ma MZ, Kong X, Weng MZ, et al. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Molecular Carcinogenesis 54 (2015): 1397-1406.
- Wang SH, Ma F, Tang ZH, et al. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. Journal of Experimental and Clinical Cancer Research 35 (2016): 160.
- Ma MZ, Zhang Y, Weng MZ, et al. Long Noncoding RNA GCASPC, a Target of miR-17-3p, Negatively Regulates Pyruvate Carboxylase-Dependent Cell Proliferation in Gallbladder Cancer. Cancer Research 76 (2016): 5361-5371.
- Cai Q, Wang Z, Wang S, et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biology 7 (2017): 160-247.
- Ma MZ, Chu BF, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death and Disease 6 (2015): e1583.
- Chang Y, Liu C, Yang J, et al. MiR-20a triggers metastasis of gallbladder carcinoma. Journal of Hepatology 59 (2013): 518-527.
- Lu W, Zhang Y, Zhou L, et al. miR-122 inhibits cancer cell malignancy by targeting PKM2 in gallbladder carcinoma. Tumor Biology (2015): 1-11.
- Jin YP, Hu YP, Wu XS, et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death and Disease 9 (2018): 182.
- Bao RF, Shu YJ, Hu YP, et al. miR-101 targeting ZFX suppresses tumor proliferation and metastasis by regulating the MAPK/Erk and Smad pathways in gallbladder carcinoma. Oncotarget 7 (2016): 22339-22354.
- He Y, Chen X, Yu Y, et al. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Molecular Carcinogenesis 57 (2018): 772-783.
- Qiu Y, Luo X, Kan T, et al. TGF-β upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting CADM1. Molecular Biosystems 10 (2014): 679-685.
- Wang N, Xiang X, Chen K, et al. Targeting of NT5E by miR-30b and miR-340 attenuates proliferation, invasion and migration of gallbladder carcinoma. Biochimie 146 (2017): 56-57.
- Yang D, Zhan M, Chen T, et al. miR-125b-5p enhances chemotherapy sensitivity to cisplatin by down-regulating Bcl2 in gallbladder cancer. Scientific Reports 7 (2017): 43109.
- He M, Zhan M, Chen W, et al. MiR-143-5p Deficiency Triggers EMT and Metastasis by Targeting HIF-1α in Gallbladder Cancer. Cellular Physiology and Biochemistry International Journal of Experimental Cellular Physiology Biochemistry and Pharmacology 42 (2017): 2078-2092.
- Zhang M, Gong W, Zuo B, et al. The microRNA miR-33a suppresses IL-6-induced tumor progression by binding Twist in gallbladder cancer. Oncotarget 7 (2016): 78640-78652.
- Lu W, Hu Y, Ma Q, et al. miR-223 increases gallbladder cancer cell sensitivity to docetaxel by downregulating STMN1. Oncotarget 7 (2016): 62364-62376.
- Wang H, Zhan M, Xu SW, et al. miR-218-5p restores sensitivity to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer. Cell Death and Disease 8 (2017): e2770.
- Ma MZ, Li CX, Zhang Y, et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Molecular Cancer 13 (2014): 156.
- Wang SH, Wu XC, Zhang MD, et al. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumor Biology 37 (2016): 9721-9730.
- Wang SH, Zhang WJ, Wu XC, et al. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. Journal of Cellular and Molecular Medicine 20 (2016): 2299-2308.
- Wang SH, Zhang WJ, Wu XC, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 7 (2016): 37857-37867.
- Wu XS, Wang F, Li HF, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. Embo Reports 18 (2017): 1837-1853.
- Ma F, Wang SH, Cai Q, et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomedicine and Pharmacotherapy 88 (2017): 863-869.
- Wang SH, Yang Y, Wu XC, et al. Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Letters 380 (2016): 122-133.
- Mikami T, Yanagisawa N, Baba H, et al. Association of Bcl-2 protein expression with gallbladder carcinoma differentiation and progression and its relation to apoptosis. Cancer 85 (2015): 318-325.
- Kim YW, Huh SH, Park YK, et al. Expression of the c-erb-B2 and p53 protein in gallbladder carcinomas. Oncology Reports 8 (2001): 1127-1132.
- Fan YZ, Zhang JT, Yang HC, et al. Expression of MMP-2, TIMP-2 protein and the ratio of MMP-2/TIMP-2 in gallbladder carcinoma and their significance. World Journal of Gastroenterology 8 (2002): 1138-1143.
- Zhi WQ, Wu K, Wang J, et al. Association of p53, p16, and vascular endothelial growth factor protein expressions with the prognosis and metastasis of gallbladder cancer. Journal of the American College of Surgeons 193 (2001): 380-383.
- Koda M, Yashima K, Kawaguchi K, et al. Expression of Fhit, Mlh1, and P53 protein in human gallbladder carcinoma. Cancer Letters 199 (2003): 131-138.
- Shi YZ, Hui AM, Li X, et al. Overexpression of retinoblastoma protein predicts decreased survival and correlates with loss of p16INK4 protein in gallbladder carcinomas. Clinical Cancer Research An Official Journal of the American Association for Cancer Research 6 (2000): 4096-4100.
- Chang HJ, Yoo BC, Sun WK, et al. Significance of PML and p53 protein as molecular prognostic markers of gallbladder carcinomas. Pathology and Oncology Research 13 (2007): 326-335.
- Xiang-Song W, Yoshimitsu A, Toru I, et al. Expression of homeodomain protein CDX2 in gallbladder carcinomas. Journal of Cancer Research and Clinical Oncology 131 (2005): 271-278.
- Ma HB, Hu HZ, Wang ZR, et al. Association of cyclin D1, p16 and retinoblastoma protein expressions with prognosis and metastasis of gallbladder carcinoma. World Journal of Gastroenterology 11 (2005): 744-747.
- Kawamoto T, Shoda J, Irimura T, et al. Expression of MUC1 mucins in the subserosal layer correlates with postsurgical prognosis of pathological tumor stage 2 carcinoma of the gallbladder. Clinical Cancer Research 7 (2001): 1333-1342.
- Miyahara N, Shoda J, Ishige K, et al. MUC4 interacts with ErbB2 in human gallbladder carcinoma: potential pathobiological implications. European Journal of Cancer 44 (2008): 1048-1056.
- Li C, Wang C, Xing Y, et al. CD133 promotes gallbladder carcinoma cell migration through activating Akt phosphorylation. Oncotarget 7 (2016): 17751-17759.
- Cai Q, Wang ZQ, Wang SH, et al. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. American Journal of Translational Research 8 (2016): 4068-4081.
- Ubersax JA, Ferrell JE, Jr. Mechanisms of specificity in protein phosphorylation. Nature Reviews Molecular Cell Biology 8 (2007): 530-541.
- Quan Z, Gu J, Dong P, et al. Reactive oxygen species-mediated endoplasmic reticulum stress and mitochondrial dysfunction contribute to cirsimaritin-induced apoptosis in human gallbladder carcinoma GBC-SD cells. Cancer Letters 295 (2010): 252-259.
- Qiu Y, Li X, Yi B, et al. Protein phosphatase PHLPP induces cell apoptosis and exerts anticancer activity by inhibiting Survivin phosphorylation and nuclear export in gallbladder cancer. Oncotarget 6 (2015): 19148-19162.
- Wu W, Ouyang B, Lu Z, et al. CCK1 receptor is involved in the regulation of protein lysine acetylation in GBC-SD cells and gallbladder carcinoma. Irish Journal of Medical Science 186 (2017): 1-6.
- Schulte J, Lim S, Friedrichs AN, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Research 69 (2009): 2065-2071.
- Al-Hadeedi SY, Moorehead RJ, Leaper DJ, et al. Carcinoma of the gallbladder: a diagnostic challenge. Journal of the Royal College of Surgeons of Edinburgh 36 (1991): 174-177.
- Reid KM, Ramos-De la Medina A, Donohue JH. Diagnosis and surgical management of gallbladder cancer: a review. Journal of Gastrointestinal Surgery 11 (2007): 671-681.
- Subbiah IM, Tsimberidou AM, Naing A, et al. Abstract 2669: Early-phase trials in patients with advanced gallbladder cancer and cholangiocarcinoma: The MD Anderson Clinical Center for Targeted Therapy experience. Cancer Research 72 (2012): 2669-2669.
- Bizama C, Garcia P, Espinoza JA, et al. Targeting specific molecular pathways holds promise for advanced gallbladder cancer therapy. Cancer Treatment Reviews 41 (2015): 222-234.
- Takada T, Amano H, Yasuda H, et al. Is postoperative adjuvant chemotherapy useful for gallbladder carcinoma? A phase III multicenter prospective randomized controlled trial in patients with resected pancreaticobiliary carcinoma. Cancer 95 (2002): 1685-1695.
- Liu Y, Bi T, Shen G, et al. Lupeol induces apoptosis and inhibits invasion in gallbladder carcinoma GBC-SD cells by suppression of EGFR/MMP-9 signaling pathway. Cytotechnology 68 (2016): 123-133.
