The Isolation, Cloning, Characterization and Conservation among the Psittacidae of a Sex-Specific, Tandemly Repeated DNA Component from the African Grey Parrot (Psittacus erithacus)
Article Information
Dineke H de Kloet, Josephine M Davis, Siwo R. de Kloet*
Animal Genetics Inc., 3382 Capital Circle NE, Tallahassee FL 32308, USA
*Corresponding Author: Dr. Siwo R. de Kloet, Animal Genetics Inc., 3382 Capital Circle NE, Tallahassee FL 32308, USA
Received: 12 April 2022; Accepted: 18 April 2022; Published: 25 May 2022
Citation: Dineke H de Kloet, Josephine M Davis, Siwo R. de Kloet. The Isolation, Cloning, Characterization and Conservation among the Psittacidae of a Sex-Specific, Tandemly Repeated DNA Component from the African Grey Parrot (Psittacus erithacus). Archives of Veterinary Science and Medicine 5 (2022): 004-023.
View / Download Pdf Share at FacebookKeywords
Curved; Repetitive; Sex-specific DNA; W chromosome; Psittaciformes: Sex determination
Curved articles; Repetitive articles; Sex-specific DNA articles; W chromosome articles; Psittaciformes articles: Sex determination articles
Article Details
Abbreviations
AMPPD=adamantine-methoxy-phosphoryloxy-phenyl-dioxetane; cpm=counts per minute; PVDF=polyvinylidene difluoride; SSC=0.15 M NaCl; 0.015 M sodium citrate pH 7.2; TE=0.01 M tris-HCl buffer pH 8.0; 0.001 M EDTA; TPE=0.08M Tris-phosphate; 0.008M EDTA pH 8
1. Introduction
Sex-specific, repeated DNA components have been identified in many higher animals including snakes, mammals and birds. Sex-specific repetitive DNA components can amount to between 0.3 and 3% of the total genomic DNA and can comprise a substantial part of the chicken female specific W chromosomal DNA [1]. The function, if any, of such sequences is unknown, as is their evolutionary origin.
In mammals, sex-specific, repeated DNA components are usually conserved as sex-specific elements only between relatively closely related species [2]. Related sequences may occur sex-nonspecifically elsewhere in the genome. The female-specific W-chromosome of virtually all colubrid species of snakes [3] contains long tandemly repeated GATA or GACA sequences but GATA or GACA repeats of shorter length occur again non-sex-specifically elsewhere in the snake genome. In birds, some repeated sex-specific sequence elements similar to a highly female-specific repeated DNA component of the chicken (Gallus gallus) [1] are also found in DNA of the female Japanese pheasant (Phasianus colchicus) and turkeys (Meleagris gallopavo), but males of these latter species contain also a fairly large amount of material with a similar sequence [4]. Other repeated sequences resembling those found in gallinaceous birds have been found in more distantly related avian families such as owls (Strigiformes), cranes (Gruiformes) and Falconiformes [5]. In contrast again, a female specific repeated DNA component of the lesser black-backed gull occurs as a sex-specific component in the closely related herring gull (Larus argentatus) but in other birds outside the order Charadriiformes related sequences (identified by cross hybridization) do not show any sex-specificity [6]. A repeated sex-specific DNA sequence in the zebrafinch (Taeniopygia guttata) [7] is not found in the chicken and therefore supposedly specific for the former species.
Parrots and parakeets (Psittaciformes) form a well-defined, old avian family [8-14] of considerable interest to programs in avian conservation, since many species have become rare or endangered and are hence of considerable interest to captive propagation projects. Most psittacine species are sexually monomorphic, which means that there are no obvious external signals which differentiate males from females. The sex of these birds has therefore to be determined cytologically by detection of the female-specific W chromosome in blood cells or feathers, surgically by observing the internal reproductive organs by laparoscopy or by quantitative determination of steroid hormone levels in fecal matter, blood or serum. These methods can require considerable experimental skill and can also be of considerable risk to the survival of the bird or can only be applied only to mature birds. More recently DNA-based methods have been developed for sex determination in birds. In an earlier paper [14] we have described a general method for the identification of sex-specific repeated DNA in parrots. In the present paper we describe this method, which relies of the identification, cloning and characterization of a highly sex-specific, tandemly repeated component in DNA of the African grey parrot (Psittacus erithacus) in more detail and report that it occurs widespread with only a few exceptions as a highly female specific element in the DNA of many psittacine species and is suitable for the practical application in gender determination of parrots and parakeets.
2. Materials and Methods
2.1 Blood samples were obtained from the following species (common English and scientific names) used in this investigation
African grey parrot (Psittacus erithacus), Alexander ringneck parakeet (Psittacula eutrapia*), Amboina king parrot (Allisterus amboinensis*), aymara parakeet (Amoropsittaca aymara), black-masked love bird (Agapornis personata ), blue and gold macaw (Ara ararauna), blue-crowned conure (Aratinga acuticauda), blue-fronted amazon (Amazona aestiva), blue-headed pionus (Pionus menstruus), Bourke’s parrot (Neopsephotis bourkii), brown-eared conure (Aratinga pertinax), budgerigar (Melopsittacus undulatus), chattering lory (Lorius garrulus), cherry-headed conure (Aratinga erythrogenys), cockatiel (Nymphicus hollandicus*), crimson-winged parrot (Apromiscus erythropterus*) .eastern rosella (Platycercus eximus*), eclectus parrot (Eclectus roratus*), gang gang cockatoo (Callocephalon fimbriatum*), Goffins cockatoo (Cacatua goffini), greater sulphur-crested cockatoo (C. galerita), Indian ringneck parakeet (Psittacula krameri*), kea (Nestor notabilis), Mexican double yellowhead amazon (Amazona ochrocephala), moluccan cockatoo (C. moluccensis), palm cockatoo (Probosciger aterrimus), pesquets parrot (Psittrichas fulgidus), Princess of Wales parakeet (Polytelis alexandrae*), red-tailed black cockatoo (Calyptorhynchus magnificus*), rose-breasted cockatoo (Eolophus roseicapilla), scarlet macaw (Ara macao), severe macaw (Ara severa), sun conure (Aratinga solstitalis) and white-tailed black cockatoo (C. funereus baudinii*).
2.2 Sex determination
The sex of the individual specimens was determined surgically, with the exception of the dimorphic species marked *), of which the sex was determined by appearance.
2.3 Isolation of DNA
DNA was isolated from blood, obtained by brachial vein puncture [15], Tris-base was used instead of Tris-buffer during phenol deproteinization. Final precipitation with two volumes of ethanol from the aqueous phase was done after the addition of ammonium acetate (final concentration 2.5 M). Alternately, DNA was also isolated with cationic detergents [16] using kits provided by Washington Biotechnology Inc. (Bethesda, MD USA) or InVitrogen (San Diego Ca USA), and a modification of the protocol recommended by the manufacturers for the isolation of mammalian DNA. In this modification, 0.1 ml avian blood, diluted with 0.1 ml 0.15 M NaCl, 0.02 M EDTA pH. 8.0 was lysed in 2 ml of a solution containing 10% dodecyltrimethyl-ammonium bromide, 1.5 M NaCl, 100 mM Tris-HCl buffer pH 8.8 and 60 mM EDTA, heated for 2 min at 68oC, cooled and shaken with 2 ml of chloroform. After centrifugation for 5 min at 3000xg, the aqueous layer was collected, and the DNA precipitated by the addition of 4 ml of 1.0% cetyl trimethyl- ammonium bromide and further purified by dissolving in 1.2 M NaCl and precipitation with ethanol as described in the protocols. DNA was dissolved in TE (0.01 M tris-HCl buffer pH 8.0, 0.001 M EDTA).
2.4 Restriction enzyme digestion, gel electrophoresis, Southern and slot blot hybridization and radioactive labelling of DNA fragments
Restriction endonucleases were obtained from Promega (Madison WI USA) or New England Biolabs (Ipswich MA USA) and used with the buffers and protocols provided by the manufacturers. Agarose (BRL-Life Technologies Gaithersburg MD USA) gel electrophoresis [1.0 or 1.2% gels, in TPE buffer [17] containing 0.5 μg/ml ethidium bromide] was carried out in a horizontal slab gel apparatus. DNA fragments were visualized with long wave UV light. For preparative isolation of sex-specific DNA fragments a 3% Nusieve agarose gel was used according to the recommendations of the vendor (FMC Bioproducts Rockland ME USA).
Polyacrylamide electrophoresis using 5% polyacrylamide gels for the detection of curved DNA was carried out as described by Kodama et al. [18] and Saitoh et al. [4]. Southern blotting, using nylon membranes (Nytran, Schleicher and Schuell Keene NH) was carried out as described by Maniatis et al. [17]. Slot blots were prepared, using the same nylon membranes in an apparatus manufactured also by Schleicher and Schuell. For slot blotting, 10 μg of genomic DNA in 200 μl of TE was denatured at room temperature with 200 μl of 1 M NaOH for 10 min followed by neutralization with 400 μl 1 M NaH2PO4 and addition of 200 μl of 10 x SSC (SSC = 0.15 M NaCl, 0.015 M sodium citrate pH 7.2), prior to application to the membrane. 1 μg DNA was applied per slot. Filters were baked for 60 min at 80° and UV irradiated for further immobilization of the DNA. Filter prehybridization and hybridization with 32P-labelled radioactive probes was carried out overnight at 60°C in the presence of heparin as a blocking agent [19]. Filters were washed 6 times with 1 x SSC (0.15 M NaCl, 0.015 M sodium citrate pH 7.0) each time for 5 min at room temperature and for 2 min with 1 x SSC containing 0.1% sodium dodecyl sulphate at 55°C. In this study DNA was labelled with 32Pα-dCTP by random primer extension [20], using a kit (BRL). Autoradiograms were made on Kodak-X-omat X ray film (Thermofisher, Atlanta, Ga, USA) with or without an intensifying screen (Cronex Dupont Boston MA). Blots were also analyzed with the aid of a Betascope 603 Blot Analyzer (Betagen Waltham MA).
2.5 Detection of sex-specific DNA fragments in DNA of a female African grey parrot (P. erithacus)
Southern blots of restriction digests of genomic DNA were screened for the presence of sex-specific DNA fragments with a genomic sex-specific probe prepared as described before [14], consisting of 50 ng heat denatured, 32P-labelled female African grey parrot DNA, preannealed to 200 μg unlabelled male African grey parrot Cot 2 DNA [21] in 0.4 ml 5 x SSC for 20 min at 65°C [14].
2.6 Cloning of sex-specific DNA fragments
For cloning, sex-specific DNA fragments were visually identified in an Msp1 digest fractionated by electrophoresis in 3% Nusieve (FMC) gels after staining with ethidium bromide. Gel slices containing the sex-specific monomers of approximately 600bp (identified as described above) were excised and the DNA fragment isolated by the powered glass procedure [22] using a gene clean kit (Bio 101 LaJolla CA). Fragments were ligated into Acc1 cut, dephosphorylated vector pGEM 3Z+, the resulting recombinant plasmid was used to transform E.coli 31/17 by the CaCl2 procedure [17] and the bacterial suspension plated on LB agar containing ampillicin. Clones containing sex-specific inserts were identified by colony hybridization [17] using the same sex-specific probe as described above for the identification of sex-specific DNA fragments in Southern blots and the positive bacteria grown on LB agar containing ampicillin for plasmid isolation and sequencing of the insert.
2.7 Nucleotide sequence determination
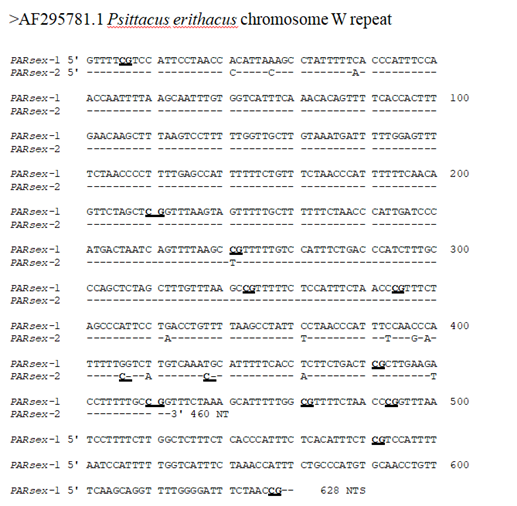
Nucleotide sequences were determined with an automatic sequencer (ABI Foster City CA USA), using the Sanger procedure [23] and forward and reverse primers. The sequence of one of the sex-specific monomers (PARsex-1) has been entered into the Genbank data base under Accession number AF295781.
2.8 Quantification of the sex-specific DNA component
Repeat copy number of the sex-specific component was determined as previously described for the centromeric DNA component [24, 25]. Briefly, genomic DNA was blotted onto a nylon membrane in graded amounts and probed with a 32P-labelled 37 bp fragment (268’-TTAAGCCGTTTTTGTCCATTTCTGACCCATCTTTGCC-305’) of AF295781, 5’-labelled with γ32P-ATP and polynucleotide kinase. Linearized plasmid DNA containing the 628 bp monomer insert served as a standard. The number of cpm bound to the membrane was determined with a Betascope (Betagen Waltham MA USA). The genomic content of the DNA repeat was computed by linear regression using a least square procedure from the radioactivity measured in the genomic DNA and the standard.
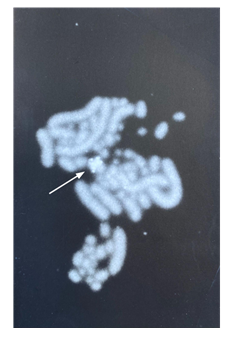
2.9 Cytological localization of the PARsex-1 repeat
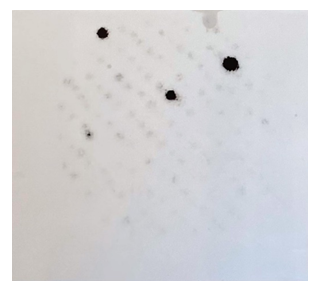
Chromosome spreads were prepared from lymphocytes of C. moluccensis as described by Belterman and de Boer [26]. This species was used because it produced better chromosome spreads than the female African grey parrots that were available. In situ hybridization of cloned, biotin labelled PARsex-1 monomer to metaphase chromosomes and visualization of the hybrids with streptavidine and mouse fluorescein labelled antistreptavidine was carried out as described by van der Ploeg et al. [27]. The probe was labelled with biotin-16-dUTP (BRL) by random primer extension [20]. Chromosomes were counterstained with propidium iodide. Photographs were made with a Nikon fluorescence microscope. "Slow-fade" (Molecular Probes, Eugene OR) was used to retard fading of the fluorescence.
2.10 Non-radioactive identification of sex-specific fragments in slotblots
For the non-radioactive identification and quantification of sex-specific DNA similar to PARsex-1 the 37 bp fragment described above was conjugated at it’s 5’-terminus [28, 29] with alkaline phosphatase by Keystone Laboratories, Camarillo, CA. Slot blots of DNA were prepared as decribed above. Hybridization of this probe was carried out in 50% formamide containing buffer as decribed by Maniatis et al. [17]. Detection of the hybridized probe was done with the chemiluminescent substrate AMPPD (adamantine-methoxy-phosphoryloxy-phenyl-dioxetane) [30, 31]. Photographs of the blots were made using Kodak-X-omat X ray film (Thermofisher, Atlanta, Ga, USA).
3. Results
3.1 Identification of sex-specific elements in DNA of the female African grey parrot
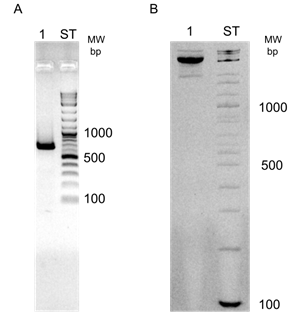
In an electrophoretic analysis of an Msp1 digest of male and female African grey parrot DNA staining with ethidium bromide reveals a very weak component of approximately 0.5 kB in female DNA which cannot be found in DNA of the male. In order to improve the detection a Southern blot of a Msp1 digest of DNA of the African grey parrot was probed with a small amount (50 ng) of 32P labelled female DNA, preannealed to a 4000 fold excess of unlabeled male African grey parrot Cot-2 DNA. Figure 1 shows clearly the presence of a component of approximately 600 bp in female DNA which cannot be detected in digests of DNA of males of this species.
Figure 1: Identification of female specific DNA components in a restriction digest (Msp1) of DNA of the African grey parrot. A Southern blot of 2.5 μgs DNA of male and female African grey parrot, digested with Msp1 was probed with a genomic female specific DNA probe of the African grey parrot prepared as described under "Materials and Methods".
Figure 2: Cloning of female specific DNA components of the African grey parrot. Material containing the female-specific component in the Msp1 digest of the African grey parrot (Figure 1) was ligated into Acc1 cut plasmid pGEM, the recombinant plasmid used to transform E. coli 31/17 and colonies carrying the female-specific PARsex-1 component identified by colony hybridization (dark colonies).
3.2 Cloning and nucleotide sequence of the sex-specific DNA component (PARsex-1) of the African grey parrot.
For further characterization, the sex-specific MspI component of approximately 600 bp was isolated from the gel, ligated into the Acc1 site of the vector pGEM 3Z+, cloned in E. coli 31/17, clones containing the sex-specific DNA component were identified by colony hybridization (Figure 2) using the radiolabeled probe described in the previous section [32] and the cloned fragments (PARsex-1) used for the determination of the nucleotide sequence (Figure 3, Genbank nr AF295781), copy number, genomic organization and chromosomal location of the component. Nucleotide sequence analysis of one of the cloned fragments (PARsex-1, Figure 3) shows that the actual length of the cloned repeat monomer is 629 nt (A=124, C=160, G=81, T=264). The fragment is AT rich (67% A + T) and is asymmetric with a pyrimidine-rich strand
Figure 3: The nucleotide sequences of a 628 NT female specific DNA fragment (PARsex-1) and an additional 460 NTS long subfragment of the African grey parrot (PARsex-2). CG dinucleotides are underlined. Non-conserved nucleotides in a second monomer sequence (PARsex-2) are indicated, (-) means the same base as in PARsex-1.
containing 67% (T + C). The corresponding sequence of a second monomer, generated by complete digestion with Msp1 (PARsex2, see Figure 6) differs in the first 460 nts by 16 nts or approximately 4% from the sequence of PARsex-1. Sequence analysis reveals also the presence of an internal repeat structure (consensus repeat sequence = (C,G,T)A1.00A1.00A0.75A0.40A0.05 NNNNN---) in which 60 oligo dA2-6 segments of PARsex-1 are repeated at average intervals of 628/60 or 10.5 nts (Figure 4). Such a sequence is typical of a curved DNA structure [4, 18, 33-35]. Polyacrylamide gel electrophoresis using a 5% gel in which curved DNA has an anomalous, slow rate of migration (Figure 5) [e.g. 18, 34, 36, 37], showed that this is indeed the case
Figure 4: Internal repeat structure of the PARsex-1 repeating unit. Position of the nucleotide preceeding an oligo dA2-6 segment of a fragment, fragment sequence, length and oligo dA2-6 segment length in the fragments are indicated. A consensus sequence was generated from the individual fragment sequences = (C,G,T)A1.00A1.00A0.75A0.40A0.16A0.05 N N N.
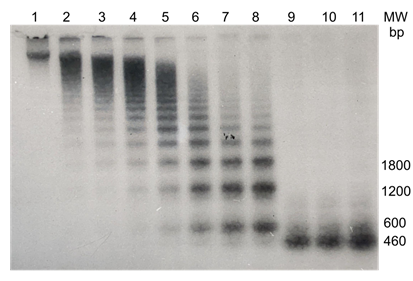
also for the sex-specific DNA component (PARsex-1) in the African grey parrot, in such gels, the cloned as well as the genomic sex-specific DNA fragments migrate, similar to the sex-specific DNA of chickens [18], as material of an apparent extremely high molecular weight. Figure 6 shows that in partial Msp1 digests of female African grey parrot DNA,
the sex-specific component forms a transient ladder type pattern upon electrophoresis in an agarose gel, with an initial monomer length of approximately 630 bp, indicative of a tandemly repeated organization. Exhaustive digestion results in the decrease of the amount this component, and the appearance of a component of approx. 460 nt, demonstrating that an
Figure 6: Genomic organization of PARsex-1-like material in DNA of the female African grey parrot. 15 μgs of female African grey parrot DNA in 300 μl restriction buffer B was digested at 37°C with 4 units of the restriction endonuclease Msp1. 10 μl samples were removed every five minutes and added to 10ul of 2X loading buffer, heated for 10 minutes in a boiling water bath and applied to the gel (lanes 1 - 8). After 40 min an additional 10 units of the enzyme were added to insure complete digestion and additional samples were taken every ten minutes (lanes 9 - 11). The samples were fractionated by electrophoresis through a 1% agarose gel. A Southern blot was made and probed with P32 labelled PARsex1.
additional Msp1 site (GCCGG) at nt 460 is digested more slowly by the enzyme, most likely as a result of a difference in the sequence flanking the MsP1 restriction site, as has been shown for other restriction enzymes [38]. Determination of the copy number shows that approximately 0.3% of the female African grey parrot DNA consists of elements with sequence similarity to the sex-specific repeat PARsex-1. Assuming a diploid DNA content in parrots of 3.6 pg [39], this value corresponds to a copynumber of approx. 1.3 x 104 repeats per genome. Such experiments reveal also that the genome of the male African grey parrot contains at most 0.0045% or 1.9 x 102 copies of material with some sequence similarity to the PARsex-1 repeat.
3.3 Chromosomal localization of the PARsex-1 repeat element
Figure 7 shows that PARsex-1 strongly hybridizes to one medium sized chromosome of a female Cacatua moluccensis. This species was chosen because it yielded superior chromosome preparations as compared with the African grey parrot females in our collection. Some hybridization occurs with other chromosomes or with chromosomes of a male of this species (data not shown).
|
Subfamily and species |
Female |
Male |
|
Ariini: |
||
|
Amazona aestiva |
7.8 (3) |
0.7 (3) |
|
Amazona ochracephala |
8.4 (2) |
0.7 (2) |
|
Amoropsittaca aymara |
15.8 (3) |
1.5 (2) |
|
Ara ararauna |
12.4 (4) |
1.2 (2) |
|
Ara macao |
14.7 (2) |
0.9 (2) |
|
Ara severa |
13.8 (1) |
2.4 (2) |
|
Aratinga acuticauda |
12.4 (3) |
1.6 (2) |
|
Aratinga erythrogenys |
15.8 (3) |
2.3 (3) |
|
Aratinga pertinax |
14.2 (2) |
2.1 (3) |
|
Aratinga solstitialis |
17.2 (3) |
2.7 (3) |
|
Pionus menstruus |
12.4 (1) |
0.9 (1) |
|
Cacatuini: |
||
|
Cacatua galerita |
33.7 (1) |
1.1 (3) |
|
Cacatua goffini |
30.3 (1) |
0.9 (3) |
|
Cacatua moluccensis |
35.0 (3) |
1.3 (3) |
|
Callocephalon fimbriatum |
30.2 (2) |
0.8 (1) |
|
Callyptorhynchus funereus |
38.3 (1) |
0.7 (1) |
|
Callyptorhynchus magnificus |
35.3 (1) |
0.9 (1) |
|
Eolophus roseicapella |
31.3 (3) |
1.1 (2) |
|
Nymphicus hollandicus |
34.5 (4) |
0.6 (2) |
|
Probosciger aterrimus |
12.7 (2) |
0.7 (1) |
|
Loriini: |
||
|
Lorius garrulus |
13.5 (2) |
1.3 (1) |
|
Nestorinii: |
||
|
Nestor notabilis |
0.5 (1) |
0.3 (1) |
|
Platycercini: |
||
|
Melopsittacus undulates |
23.5 (3) |
0.7 (2) |
|
Platycercus eximus |
25.6 (2) |
0.4 (1) |
|
Psittacini: |
||
|
Psittacus erithacus |
100 (3) |
1.5 (2) |
|
Psittaculini: |
||
|
Agapornis personata |
8.3 (2) |
0.3 (2) |
|
Alisterus amboinensis |
40.2 (2) |
2.2 (1) |
|
Apromiscus erythopterus |
16.4 (1) |
1.3 (2) |
|
Eclectus roratus |
7.0 (2) |
4.8 (4) |
|
Neospephotis bourkii |
15.0 (3) |
11.3 (4) |
|
Polytelis alexandrae |
24.3 (2) |
0.5 (2) |
|
Psittacula eutrapia |
20.6 (2) |
1.3 (3) |
|
Psittacula krameri |
30.3 (2) |
2.1 (2) |
|
Psittrichasini: |
||
|
Psittrichas fulgidus |
8.5 (1) |
0.3 (1) |
Table I: Conservation of sex-specificity of PARsex in selected Psittaciformes. 1 ug of alkali denatured DNA of the species indicated in the Table was applied to Nytran filter paper in a slot blot apparatus and probed with P32-labelled PARsex-1. Values represent the average hybridization of PARsex-1 with DNA of the species in the table, expressed as a percentage of the hybridization with DNA of the African grey parrot. Numbers in parenthesis indicate the number of individuals of each species used.
3.4 Conservation of PARsex-1 among other psittacine species
Table I shows that PARsex-1 hybridizes in a sex-specific manner with DNA of many psittacine species, belonging to different subfamilies. Strong hybridization is observed with DNA of females of Alisterus amboinensis, Psittacus erithacus, Polytelis alexandrae, Platycercus eximus and females of the Cacatuiini, with the exception of Probosciger aterrimus. A weaker but still highly sex-specific signal can be observed with the Ariini, Psittaculini and Loriini.
The data show also that PARsex-1 hybridizes rather well to DNA of female as well as male Eclectus roratus and Neopsephotis bourkii but that only very little hybridization can be detected with DNA of the single male and female of Nestor notabilis which were available for analysis.
3.5 Sex-specific pattern of PARsex-1 in restriction digests of Eclectus roratus and Neopsephotis bourkii
The data in Table I show that PARsex-1 hybridizes at most only 1.6 times as strongly to DNA of females as compared to DNA of males of males Eclectus roratus and Neopsephotis bourkii. The restriction pattern in Fig 8 shows that in the latter species (shown for N. bourkii only) distinct sex-specific as well as nonspecific PARsex-1-like components occur. Whereas a major component of approx. 600 bp is only observed in a Msp1 digest of female DNA, components of a high molecular occur in the male as well as the female DNA
Figure 8: Pattern of PARsex1-like restriction fragments in DNA of male and female Neopsephotis bourkii. 1.5 μg of DNA of male and female Neopsephotis bourkii was digested with the restriction endonuclease MspI, electrophoresed though an agarose gel and a Southern blot was made and probed with PARsex-1.
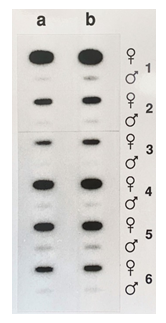
Figure 9: Non-radioactive detection of PARsex-1-like sex-specific DNA among the Psittacidae. 1 μg (ln a), 2μg (ln b) μg of alkali denatured DNA of males and females of Psittacus erithacus (1), Lorius garrulus (2), Agapornis personata (3), Cacatua galerita (4), Ara ararauna (5) or Amazona aestiva (6) was applied to a Nytran filter with a slot blot apparatus and probed with alkaline phosphatase labeled PARsex-1.
3.6 Nonradioactive sex identification in different Psittacidae
The data in Figure 9 show that the use of an alkaline phosphatase labeled oligonucleotide probe derived from PARsex-1 in combination with chemiluminescent detection allows the sensitive, non radioactive identification of the sex of several members of the Psittacidae. Again, where there is a strong signal obtained with DNA of females of these species, DNA of males is virtually devoid of this material.
4. Discussion
The results described in this paper demonstrate the presence of an abundant, tandemly repeated DNA component (PARsex-1) in the DNA of the female African grey parrot (Psittacus erithacus). The repeat monomer length of PARsex-1, cloned as an Msp1 fragment obtained after exhaustive digestion, is approximately 460 nts. Apparently this fragment is derived from another Msp1 fragment of approximately 628 bp as the result of a slow digestion of an additional Msp1 site in this latter fragment. The copynumber of the repeat is approximately 1.3 x 104. Assuming that in parrots, as in chickens, the W chromosome represents approx. 1.0% of the diploid genome [40], this result indicates that in the African grey parrot the sex-specific repeat accounts for a substantial fraction, about 30% (or about 40% if one considers the 628 bp repeat from which the 460 bp repeat has been derived) of the DNA of the W chromosome. The size and copynumber of the repeat are similar to the values found for female specific repeated DNA components in the chicken (Gallus gallus domesticus) where a 0.7 kb Xho1 fragment is repeated 1.4 x 104 fold and the turkey (Meleagris gallopavo) where a 0.4 kb Pst1 fragment is repeated approx. 104 fold [1, 4]. They differ however somewhat from the values found for a sex-specific component in the DNA of the female lesser black-backed gull (Larus fuscus) where a much larger, 8.6 kb repeat is present in 2.5 x 104 copies [6]. An analysis of the nucleotide sequence of PARsex-1 revealed an internal repeat structure in which clusters of 2 to 5 adenine residues occur with an average periodicity of 10.5 bp. Such a repeat structure is considered to be typical for "curved DNA" [33, 41], and was revealed by polyacrylamide gel electrophoresis [34] where the PARsex-1 monomer migrates as material of extremely high molecular weight (Figure 5). Why such curved DNA elements have a reduced electrophoretic mobility in polyacrylamide gels is not very well understood. Although it has long been thought that the curved nature itself and the associated altered dimensions of such DNA elements were directly responsible for the reduced electrophoretic mobility of such DNA segments upon polyacrylamide electrophoresis, currently direct interaction of the nucleotide sequence of these particular curved DNA segments with the polyacrylamide gel matrix is considered to be partly responsible [35, 42]. However, this explanation may be again only partly correct, since curved DNA elements migrate anomalousy upon electrophoresis in free solution [43]. Further, additional complications arise, because the curved nature of DNA elements with phased dA tracts disappears again in solutions with high concentrations of monovalent cations [44]. The curved nature of PARsex-1 however is not unique for a sex-specific DNA element, for the chicken, turkey and pheasant (Phasianus colchicus) sex-specific repeated DNA sequences have a similar bent structure [4]. The difference between the psittacine and the chicken sequence is that whereas in the chicken the oligo dT3-5 and oligo dA3-5 sequence elements each alternate at a distance of approx. 21 bp on the same DNA strand, only oligo dA clusters (and consequently dT clusters on the opposite strand) are found in psittacine sex-specific W-DNA. Yet oligo dA and/or alternating oligo dA and oligo dT is also not a general property of all avian sex-specific DNA sequences, since the sequence of a repeated female specific 0.4 kb BamH1 fragment of the bobwhite quail (Colinus virgianius) or a sex-specific DNA sequence in the zebra finch [7] lack this feature. Another difference between the chicken Xho1 repeat monomer and PARsex-1 is the relative scarcity of CG dinucleotides in the latter [approx 11 (in PARsex-1) vs 34 per chicken repeat monomer]. Sequences related to the PARsex-1 repeat element of the African grey parrot occur in the genome of females of many psittacines belonging to different subfamilies of the Psittacidae, like the Cacatuini, the Ariini, the Loriini etc. Again a limited amount of related sequence material can be detected in the genome of the males of these species under conditions of stringency applied in this investigation. Only in the Kea (Nestor notabilis) we could not detect a sex-specific hybridization signal with any certainty, because the hybridization values in males and females in this species were very low. Whether this observation reflects a distant phylogenetic relationship between this species and the other Psittacidae [8,-12] or whether it is the result of a loss of PARsex-1-like sequences from the W chromosome in this species, is unknown. More individuals of this species will have to be investigated to resolve this issue. Similar to what has been observed in some gallinaceous birds [4] small, but variable amounts of material cross-hybridizing with PARsex-1 occur also in the males of many psittacine species. In the African grey parrot this material amounts to at most 2.5% of that found in the female. Although minor amounts of PARsex-1-like material can be found as a non-sex-specific component in many psittacine species, only in Eclectus roratus and in the Bourke’s parakeet (Neopsephotus bourkii) significant hybridization of PARsex-1 occurs with male as well as female DNA. Southern blot experiments showed that Msp1 digests of DNA of the latter species contain in addition to a female specific PARsex-1 component of approx. 600 bp, a major amount of PARsex-1-like material occurs as material of very high molecular weight in DNA of males and females. A similar result was found in the case of the Eclectus parrot. Although it is probably coincidental, it may be of interest that the Eclectus parrot and the Bourke’s parakeet are both among the dimorphic psittacine species. The widespread occurrence of a female specific component with sequence similarity in DNA of virtually all members of the Psittacidae suggests that this component arose early in the evolution of this avian family, whose oldest known fossils date from the lower Eocene [11]. The observation that the sex-specific repeat monomers of a member of the Psittacidae and some members of the Gallidae (and owls(AF231331.1, AF231326.1, AF291661.1, and de Kloet unpubl. data), share a similar sequence organization (A2-6 clusters at approx. 10.5 nt intervals vs. resp. alternating A3-5 or T3-5 clusters repeated at 21 nt intervals) suggests that this characteristic may be a rather general and conserved property of avian sex-specific DNA components. A strong conservation of a sex-specific, tandemly repeated DNA component in a family of birds such as the Psittacidae) contrasts with male specific tandemly reiterated DNA components in mammals, where such sequences are often sex-specific only in a single species [45-48]. A conserved sex-specific repeated sequence has however been found among the Bovidae [49], but this sequence occurs interspersed. The unique nature of the Y chromosome, which limits recombinational exchange as a mechanism for the suppression of mutational change, has often been cited as the reason for the accumulation of species and chromosome specificity of Y linked, tandemly repeated sequences [49-51].
A similar reasoning may also explain the largely unique nature of the W-chromosome specific repeated sequences in parrots. Conservation of such sequences might only occur if such sequences are linked to positively selected genetic loci [52]. Whether the conservation of PARsex-1 means a close linkage of these repeats to conserved elements involved in sex determination or fertility [53] in the Psittacidae, remains to be investigated.
5. Conclusion
The present investigation describes the isolation and partial characterization of a sex-specific repeated DNA component in parrots. The sex-specificity is widespread among many, often phylogenetically distantly related psittacine species, suggesting an ancient origin of the nucleotide sequence.
The sequence is further recognized by the occurrence of oligo dA sequence elements at regular intervals of about 10.5 bp, a feature which is associated with a bent structure.
Acknowledgements
The authors are indebted to Mrs. Sharon Wolf, Avicultural Breeding and Research Center, Loxahatchee, FL, USA; Mr. Robert Prather, Tallahassee, FL, USA; Mr. Rory Brienen, Tallahassee, FL, USA and Mr. Harry de Koning, St. Oedenrode, The Netherlands for the generous gift of blood samples of the species used in this study. The authors are indebted to Dr. Steven Miller for his assistance with DNA sequencing and Dr. Jeannette E. Brooks for her help with the nonradioactive labeling and the hybridocytochemistry.
Conflict of Interest
The authors declare no conflicts of interest.
References
- Tone M, Nakano N, Takao E, et al. Demonstration of W chromosome-specific repetitive DNA sequences in the domestic fowl, Gallus g. domesticus. Chromosoma 86 (1982): 551-569.
- Cooke HJ, Schmidtke J, Gosden JR. Characterization of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma 87 (1982): 491-502.
- Singh L, Purdom IF, Jones KW. Satellite DNA and evolution of sex chromosomes. Chromosoma 59 (1976): 43-62.
- Saitoh H, Harata M, Mizuno S. Presence of female-specific bent-repetitive DNA sequences in the genomes of turkey and pheasant and their interactions with W-protein of chicken.Chromosoma 98 (1989): 250-258.
- Yamada K, Nishida-Umehara C, Ishijima J, et al. A novel family of repetitive DNA sequences amplified site-specifically on the W chromosomes in Neognathous birds. Chromosome Res 14 (2006): 613-627.
- Griffiths R, Holland PWH. A novel avian W chromosome specific DNA repeat sequence in the lesser black-backed gull (Larus fuscus). Chromosoma 99 (1990): 243- 250.
- Itoh Y, Kampf K, Arnold AP. Molecular cloning of zebra finch W chromosome repetitive sequences: evolution of the avian W chromosome . Chromosoma 117 (2008): 111-121.
- Smith GA. Systematics of Parrots. Ibis 117 (1975): 18-68.
- Cracraft J. Continental drift, paleoclimatology, and the evolution and biogeography of birds. J Zool Lond 169 (1973): 455-545.
- Feduccia A. The Age of Birds. Harvard University Press, Cambridge, USA (1980).
- Harrison CJO. The earliest parrot: a new species from the British eocene. Ibis 124 (1982): 203-210.
- de Kloet RS, de Kloet SR. The evolution of the spindlin gene in birds: sequence analysis of an intron of the spindlin W and Z gene reveals four major divisions of the Psittaciformes. Mol Phylogenet Evol 36 (2005):706-721.
- Wright TF, Schirtzinger EE, et al. A multilocus molecular phylogeny of the parrots (Psittaciformes): support for a Gondwanan origin during the cretaceous. Mol Biol Evol 10 (2008): 2141-2156.
- de Kloet DH, de Kloet SR. Molecular determination of the sex of parrots. Focus (BRL) 14 (1992): 106-108.
- Gros-Bellard M, Oudet P, Chambon P. Isolation of high molecular weight DNA from mammalian cells. Eur J Biochem 36 (1973): 31-38.
- Gustincich S, Manfioletti G, DelSal G, et al. A fast method for high-quality genomic DNA extraction from whole human blood. Biotechniques 11 (1991): 298-302.
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning; a laboratory manual. Cold Spring Harbour Laboratory. Cold Spring Harbour, New York (1982).
- Kodama H, Saitoh H, Tone M, et al. Nucleotide sequences and unusual electrophoretic behaviour of the W chromosome-specific repeating DNA units of the domestic fowl, Gallus gallus Chromosoma 96 (1987): 18-25.
- Singh L, Jones KW. The use of heparin as a simple cost effective means of controlling background in nucleic acid hybridization procedures. Nucleic Acids Res 12 (1984): 5627-5638.
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction fragments to high specific radioactivity. Anal Biochem 132 (1983): 6-13.
- Britten RJ, Kohne DE. Repeated segments of DNA. Sci Am 222 (1970): 24-31.
- Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA 76 (1979): 615-619.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA 74 (1977): 5463-5467.
- Madsen CS, McHugh KP, de Kloet SR. Characterization of a Major Tandemly Repeated DNA Sequence (RBMII) Prevalent Among Many Species of Waterfowl (Anatidae). Genome 35 (1992a): 1037-1044.
- Madsen CS, deKloet DH, Brooks JE, et al. Highly repeated DNA sequences in birds: the structure and evolution of an abundant, tandemly repeated 190 bp DNA fragment in parrots. Genomics 14 (1992b): 462-473.
- Belterman RHR, de Boer LEM. A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica 65 (1984): 39-82.
- Van der Ploeg M, Raap T, Tankes H. Practical Boerhaave Course: Hybridocytochemistry. University of Leiden Press, Leiden, The Netherlands (1990).
- Albretsen C, Haukanes BI, Aasland R, et al. Optimal conditions for hybridization with oligonucleotides: a study with myc-oncogene DNA probes. Anal Biochem 170 (1988): 193-202.
- Cate RL, Ehrenfels CW, Wysk M, et al. Genomic Southern analysis with alkaline-phosphatase-conjugated oligonucleotide probes and the chemiluminescent substrate AMPPD Genet Anal Tech Appl 8 (1991): 102-106.
- Dimo-Simonin N, Brandt-Casadevall C, Gujer HR. Chemiluminescent DNA probes: evaluation and usefulness in forensic cases. Forensic Sci Int 57 (1992): 119-127.
- Jablonski E, Moomaw EW, Tullis RH, et al. Preparation of oligodeoxynucleotide-alkaline phosphatase conjugates and their use as hybridization probes. Nucleic Acids Res 14 (1986): 6115-6128.
- Grunstein M, Wallis J. Colony hybridization. Methods in Enzymology 68 (1979): 379-389.
- Ulanovsky L, Bodner N, Trifonov EN, et al. Curved DNA: Design, synthesis and circularization. Proc Natl Acad Sci USA 83 (1986): 862-866.
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res 15 (1987): 247-265.
- Stellwagen NC, Stellwagen E. Effect of the matrix on DNA electrophoretic mobility. J Chromatography 1216 (2009): 1917-1929.
- Stellwagen E, Lu Y, Stellwagen NC. Curved DNA molecules migrate anomalously slowly in free solution. Nucleic Acids Res 33 (2005): 4425-4432.
- Fiorini A, Gouveia F de S, Fernandez MA. Scaffold/Matrix Attachment Regions and intrinsic DNA curvature. Biochemistry (Mosc) 71 (2006): 481-488.
- Wolfes H, Fliess A, Pingoud A. A comparison of the structural requirements for DNA cleavage by the isoschizomers HaeIII, BspRI and BsuRI. Eur J Biochem 150 (1985): 105-110.
- Shields GF. Organization of the avian genome. In: Perspectives in Ornithology (Brush AH, Clark GA Eds.) pp. 271-290, Cambridge Univ. Press, Cambridge, UK (1983).
- Bloom SE, Delaney ME, Muscarella DE. In: Manipulating the avian genome (Etches RJ, Verrinder Gibbins AM Eds), CRC Press, Boca Raton, USA (1993): 39-59 .
- Crothers DM, Haran TE, Nadeau JG. Intrinsically bent DNA. Journal of Biol Chemistry 265 (1990): 7093-7096.
- Fukudome K, Iwasaki K, Matsumoto S, et al. Undesirable contamination of DNA electrophoresed through polyacrylamide and agarose gels as revealed by reversing-pulse electric birefringence signals. Biopolymers 31 (1991): 1455-1458.
- Stellwagen NC. Electrophoresis of DNA in agarose gels, polyacrylamide gels and in free solution. Electrophoresis 30 (2009) (Suppl 1): S188-195.
- Stellwagen E, Peters JP, Maher LJ 3rd, et al. DNA A-tracts are not curved in solutions containing high concentrations of monovalent cations. Biochemistry 52 (2013): 4138-4148.
- Cooke HJ. Repeated sequences specific to human Nature 262 (1976): 182-186.
- Kunkel LM, Smith KD, Boyer SH. Human Y-chromosome-specific reiterated DNA. Science 191 (1976): 1189-1190.
- Phillips SJ, Birkenmeyer EH, Callahan R, et al. Male and female mouse DNA's can be discriminated using retroviral probes. Nature 297 (1982): 241-243.
- Lamar EE, Palmer E. Y-encoded, species specific DNA in mice: evidence that the Y chromosome exists in two polymorphic forms in inbred strains. Cell 37 (1984): 171-177.
- Matthews ME, Reed, KC. A DNA sequence that is present in both sexes of Artiodactyla is repeated on the Y chromosome of cattle, sheep and goats. Cytogenet Cell Genet 56 (1991): 40-44.
- Stephan W. Tandem-repetetive noncoding DNA: Forms and forces. Mol Biol Evol 6 (1989): 198-212.
- Charlesworth B. The evolution of sex chromosomes. Science 251 (1991): 1030-1033.
- Bouchard RA. Moderately repetitive DNA in evolution. Int Rev Cytol 76 (1982): 113-193.
- Lucchesi JC. Gene dosage compensation and the evolution of sex chromosomes. Science 202 (1978): 711-716. 711-716.