Structure Based Insights into the Association of Fluoroquinolones with Mycobacterial DNA-Gyrase Complexes
Article Information
Siva Kumar G, Harmanpreet singh, Elizabeth Sobhia M*
1Department of pharmacoinformatics, National Institute of Pharmaceutical Education and Research (NIPER), Mohali, Punjab, India
*Corresponding author: Elizabeth Sobhia M, Department of pharmacoinformatics, National Institute of Pharmaceutical Education and Research (NIPER), Mohali, Punjab, India
Received: 30 July 2021; Accepted: 06 August 2021; Published: 03 September 2021
Citation: Siva Kumar G, Harmanpreet singh, Elizabeth Sobhia M. Structure Based Insights into the Association of Fluoroquinolones with Mycobacterial DNA-Gyrase Complexes. Journal of Bioinformatics and Systems Biology 4 (2021): 103-116.
View / Download Pdf Share at FacebookAbstract
A type II topoisomerase enzyme, DNA gyrase is responsible for catalysing the cleavage of double stranded DNA. It has been already demonstrated through detailed biochemical studies that fluoroquinolones potentially inhibit DNA resealing process by binding to cleaved sites of the DNA and gyrase enzyme. The present study makes use of a series of MD simulations to understand the complex biological interplay of DNA and the gyrase enzyme along with the fluoroquinolone molecule by reconstructing the broken backbone of DNA of Mtb DNA gyrase crystal structure complex. The molecular insights from the two series of molecular dynamics on cleaved and uncleaved DNA gyrase revealed many turns and twists into the molecular mechanism of fluoroquinolone drugs, which are presumably useful for the exploration of binary drug-enzyme complex and ternary enzyme-DNA-drug complex.
Keywords
DNA gyrase, Fluoroquinolones, Molecular docking, Molecular dynamics, Free energy calculations
DNA gyrase articles; Fluoroquinolones articles; Molecular docking articles; Molecular dynamics articles; Free energy calculations articles
DNA gyrase articles DNA gyrase Research articles DNA gyrase review articles DNA gyrase PubMed articles DNA gyrase PubMed Central articles DNA gyrase 2023 articles DNA gyrase 2024 articles DNA gyrase Scopus articles DNA gyrase impact factor journals DNA gyrase Scopus journals DNA gyrase PubMed journals DNA gyrase medical journals DNA gyrase free journals DNA gyrase best journals DNA gyrase top journals DNA gyrase free medical journals DNA gyrase famous journals DNA gyrase Google Scholar indexed journals Fluoroquinolones articles Fluoroquinolones Research articles Fluoroquinolones review articles Fluoroquinolones PubMed articles Fluoroquinolones PubMed Central articles Fluoroquinolones 2023 articles Fluoroquinolones 2024 articles Fluoroquinolones Scopus articles Fluoroquinolones impact factor journals Fluoroquinolones Scopus journals Fluoroquinolones PubMed journals Fluoroquinolones medical journals Fluoroquinolones free journals Fluoroquinolones best journals Fluoroquinolones top journals Fluoroquinolones free medical journals Fluoroquinolones famous journals Fluoroquinolones Google Scholar indexed journals Molecular docking articles Molecular docking Research articles Molecular docking review articles Molecular docking PubMed articles Molecular docking PubMed Central articles Molecular docking 2023 articles Molecular docking 2024 articles Molecular docking Scopus articles Molecular docking impact factor journals Molecular docking Scopus journals Molecular docking PubMed journals Molecular docking medical journals Molecular docking free journals Molecular docking best journals Molecular docking top journals Molecular docking free medical journals Molecular docking famous journals Molecular docking Google Scholar indexed journals Molecular dynamics articles Molecular dynamics Research articles Molecular dynamics review articles Molecular dynamics PubMed articles Molecular dynamics PubMed Central articles Molecular dynamics 2023 articles Molecular dynamics 2024 articles Molecular dynamics Scopus articles Molecular dynamics impact factor journals Molecular dynamics Scopus journals Molecular dynamics PubMed journals Molecular dynamics medical journals Molecular dynamics free journals Molecular dynamics best journals Molecular dynamics top journals Molecular dynamics free medical journals Molecular dynamics famous journals Molecular dynamics Google Scholar indexed journals Free energy calculations articles Free energy calculations Research articles Free energy calculations review articles Free energy calculations PubMed articles Free energy calculations PubMed Central articles Free energy calculations 2023 articles Free energy calculations 2024 articles Free energy calculations Scopus articles Free energy calculations impact factor journals Free energy calculations Scopus journals Free energy calculations PubMed journals Free energy calculations medical journals Free energy calculations free journals Free energy calculations best journals Free energy calculations top journals Free energy calculations free medical journals Free energy calculations famous journals Free energy calculations Google Scholar indexed journals drug-enzyme complex articles drug-enzyme complex Research articles drug-enzyme complex review articles drug-enzyme complex PubMed articles drug-enzyme complex PubMed Central articles drug-enzyme complex 2023 articles drug-enzyme complex 2024 articles drug-enzyme complex Scopus articles drug-enzyme complex impact factor journals drug-enzyme complex Scopus journals drug-enzyme complex PubMed journals drug-enzyme complex medical journals drug-enzyme complex free journals drug-enzyme complex best journals drug-enzyme complex top journals drug-enzyme complex free medical journals drug-enzyme complex famous journals drug-enzyme complex Google Scholar indexed journals Moxifloxacin molecule articles Moxifloxacin molecule Research articles Moxifloxacin molecule review articles Moxifloxacin molecule PubMed articles Moxifloxacin molecule PubMed Central articles Moxifloxacin molecule 2023 articles Moxifloxacin molecule 2024 articles Moxifloxacin molecule Scopus articles Moxifloxacin molecule impact factor journals Moxifloxacin molecule Scopus journals Moxifloxacin molecule PubMed journals Moxifloxacin molecule medical journals Moxifloxacin molecule free journals Moxifloxacin molecule best journals Moxifloxacin molecule top journals Moxifloxacin molecule free medical journals Moxifloxacin molecule famous journals Moxifloxacin molecule Google Scholar indexed journals MMGBSA technique articles MMGBSA technique Research articles MMGBSA technique review articles MMGBSA technique PubMed articles MMGBSA technique PubMed Central articles MMGBSA technique 2023 articles MMGBSA technique 2024 articles MMGBSA technique Scopus articles MMGBSA technique impact factor journals MMGBSA technique Scopus journals MMGBSA technique PubMed journals MMGBSA technique medical journals MMGBSA technique free journals MMGBSA technique best journals MMGBSA technique top journals MMGBSA technique free medical journals MMGBSA technique famous journals MMGBSA technique Google Scholar indexed journals
Article Details
1. Introduction
Type II Topoisomerases are ubiquitous enzymes that catalyse DNA reorganisation, therefore are essential for many cellular events that happen during cell life [1]. The Type IIA enzymes are present in eukaryotes as well as in bacteria and share various structural and evolutionary features. Topoisomerase IV and DNA gyrase belong to Type IIA category enzymes that are expressed in bacteria. Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis encodes for only one Type II topoisomerase, i.e., DNA gyrase [2]. DNA gyrase is a heterotetrameric enzyme composed of two subunits of each GyrA and GyrB where GyrA subunit containscatalytically active DNA nicking and resealing region and GyrB subunit contains ATPase region. Gyrase enzyme executes its role by cleaving the double-stranded DNA after wrapping around it [3]. Gyrase was first identified as a cellular target for drugs of the fluoroquinolones class in 1971 [4, 5]. After this discovery, these drugs were widely employed to treat urinary tract infections (UTI), chronic bronchitis, skin and tissue infections, pneumonia and various other types of bacterial infections. But unfortunately, their clinical utility is being threatened by their widespread use leading to the steady growth of quinolone-resistant bacterial strains [6]. The most significant clinical form of resistance for quinolones is target mediated resistance as specific mutations in the enzyme weakens the interaction between enzyme and drug molecule [7-9]. The mechanism of action of DNA gyrase and fluoroquinolones association with binary DNA-gyrase complex is still under experimental investigations [10-12]. Recently in the year 2019, Mayer’s research group had added experimental evidence to the gyrase catalysed supercoiling mechanism which might be helpful for drug discovery [13]. It strongly prompted the scientific community to understand complex enzyme-drug-DNA mechanism and accelerate drug discovery as antibiotic resistance one of the most important public health threats of the 21st century.This complicated mechanism of DNA gyrase needs to be explored further, especially the process of drug enzyme binding and its intercalation into DNA. It motivated us to look back into the DNA-gyrase association with fluoroquinolones to understand molecular level complexity in ternary enzyme-DNA-drug complex by using MD simulation and post-MM-GBSA studies.The focus of our study is to investigate the molecular mechanism of drug enzyme association and further movement of drug molecule towards DNA cleavage site from the gyrase binding pocket.
2. Methodology
2.1 Steps involved in joining the cleaved DNA strands
The backbone of DNA was reconstructed by sketching broken bonds followingmethod analogous to a previous study [14]. Torsional angles of the new bonds were optimised manually in accordance with the adjacent nucleotide linkages using 3D-builder module of Maestro [15]. The sketched part of DNA was minimised to allow the optimisation of bond lengths and dihedral angles. Care was observed that original coordinates of DNA base pairs do not get disturbed and interchain hydrogen bonds between them stay intact.
2.2 Molecular dynamics simulations
The repaired double-stranded DNA with gyrase complex was submitted for 500 ns of MD simulations after removing the moxifloxacin molecule from the Quinolone Binding Pocket (QBP). The complex was prepared for simulation with Desmond’s system builder panel in the Maestro program by adding it to a cubic box filled with TIP3P water solvent system and applying periodic boundary conditions [16]. The prepared system was neutralized by adding counter ions and physiological salt concentration was limited to 0.15M. The binary DNA-gyrase, ternary DNA-gyrase-drug complex was designated with the OPLS3e force field [17]. The binary and ternary system was subjected for 500ns each using NPT ensemble at a temperature of 300 K and atmospheric pressure (1 bar) with the default settings of relaxation before simulations [18]. The system was equilibrated using standard Desmond protocol at constant pressure and temperature (NPT ensemble, 300 K, 1 bar) and the Berendsen coupling protocol with one temperature group. Hydrogen atoms bond length was constrained using the SHAKE algorithm [19]. Particle Mesh Ewald (PME) summation method was used to model long-range electrostatic interactions [20]. A cut off of 10Å was assigned for van der Waals and short-range electrostatic interactions. The binary uncut DNA-gyrase complex and ternary uncut DNA-gyrase-drug complex was subjected to 500ns of molecular dynamics at 300K temperature and 1 bar pressure without restraints after reaching the stability point.
The best pose of docked moxifloxacin with uncut DNA, gyrase was chosen for another 500ns molecular dynamics simulations to investigate the moxifloxacin association with gyrase and uncut DNA using Desmond software. As discussed in the primary 500ns simulation studies to obtain relative biological assembly of DNA by reducing the space between the broken parts of DNA, the same steps were repeated to prepare the system for 500ns production run after achieving the system equilibration.
2.3 Molecular docking studies
The 3D structure of the moxifloxacin considered in this study was prepared using the build module of Maestro 11 [15] after assigning appropriate charges. The LigPrep module was used to generate the low energy ionization, tautomeric and stereoisomeric states at physiological pH (7.0±2.0) [21, 22]. The repaired DNA gyrase structure was prepared using Protein Preparation Wizard by adding the missing hydrogen atoms, assigning the right bond order and optimising the orientations of hydroxyl group in Serine, Threonine and Tyrosine, amino group in Asparagine and Glutamate and ionisation state in Histidine [23]. The receptor grid was generated by defining Ser91, Asp94, DT10, DG11 in the quinolone binding pocket (QBP) of the crystal structure of Mtb-DNA gyrase, PDB ID: 5BS8. The grid box was extended up to 15Å for the inner box and 25Å for the outer box covering the entire binding site cavity. The prepared ligand was docked on the generated grid of the DNA gyrase structure using extra precision (XP) Glide docking program.Each of the poses wasanalysed for molecular interactions and Glide docking score [24, 25]. The moxifloxacin molecule with desired intermolecular interactions and best docking score was selected for the molecular dynamics simulation study [26].
2.4 Free energy calculations
Ligand binding free energies and ligand strain energies for moxifloxacin and DNA gyrase were calculated by Molecular Mechanics Generalized Born Surface Area (MMGBSA) method [27, 28] using Prime MMGBSA module of Schrödinger suite to determine binding free energy for moxifloxacin and unbroken DNA and gyrase complexes [29]. The calculation utilized VSGB 2.0 asimplicit solvent model [30]. The binding free energy was calculated using the following equation:
ΔG binding = ER: L - (ER + EL) + ΔGSA + ΔGSOLV
Where ER: L is the energy of the whole ligand-protein complex structure, ER+EL represents the sum of free energies of the receptor and ligand in the free form. ΔGSA is the difference in the surface area energy of the receptor-ligand complex and the sum of surface area energies of receptor and ligand separately. ΔGSOLV depicts the difference in the solvation energy of the docked structure and the sum of total of the solvation energies of receptor and ligand in an unbound state.
3. Results and Discussion
3.1 MD studies for uncleaved DNA gyrase-moxifloxacin complex
Molecular dynamics simulations of nucleic acid in its uncleaved state along with gyrase enzyme was carried out for 500ns to explore its physics-based dynamic behaviour [31]. It is known that the distance between adjacent DNA base pairs is 3.7Å, whereas the distance between the resealed ends of both strands of DNA was 8.7Å, as shown in Figure 1.
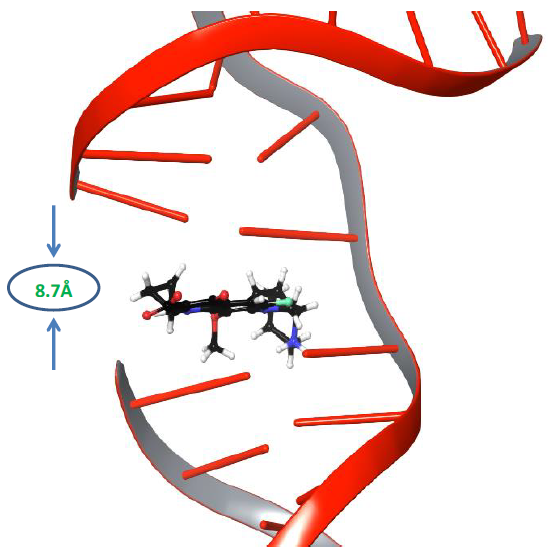
Figure 1: The distance between the cleaved ends of the broken strand of DNA.
Moxifloxacin molecule,which rests between both cleaved strands of DNA, was removed from the complexbefore the simulation. The Phospho Tyrosine Residue (PTR) was also converted to tyrosine by removing the phosphate group attached covalently to Tyr129, which is responsible for cleavage of both strands of DNA. An improperly repaired DNA and gyrase were subjected to a 500ns molecular dynamics simulation study to investigate the behaviour of DNA under dynamic conditions. We anticipated that significant changes would occur within the cleaved site of DNA under dynamic conditions. After the simulation, trajectory analysis was carried out to investigate the changes at the DNA repair site. Interestingly the distance between the two adjacent base pairs (DT10-DG11) of cleaved strandsof DNA was reduced from 8.7Å to 4.7Å after 364ns of the simulation study, as shown in Figure 2. It denotes that the molecular dynamics simulations reduced almost 4Å of distance gap between the adjacent base pairs of both the cleaved ends of the DNA strand.
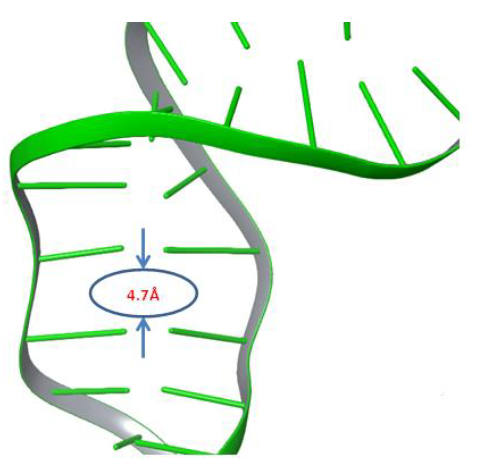
Figure 2: The gyrase enzyme created space between broken strands of DNA filled by 4Å.
Nevertheless, the maximum distance between the two adjacent base pairs of DNA did not match with the ideal value of distanceinthe biological assembly of DNA. Further molecular dynamics simulations mayadequately repair and get the relative biological assembly of DNA by reducing the adjacent base pair distance at the cleaved sites. The present healed double-stranded DNA-gyrase complex was further subjected to molecular docking studies to revealthe binding course of moxifloxacin to gyrase in the absence of cleaved DNA strands.
3.2 Moxifloxacin binding with DNA gyrase complex
The simulation frame 3645 was extracted as a PDB structure at 364ns as the distance between resealed adjacent base pairs reduced drastically, which was akin to the biological assembly of DNA. Our primary aim was to examine the binding site of moxifloxacin in the gyrase and DNA complex in the absence of DNA cleavage. The PDB structure was prepared and subjected toreceptor grid generation module to prepare a grid based on the DNA nucleotide bases and gyrA active site residues of QBP. The basesinteracting with moxifloxacin (DT10-Echain, DG11, DC14, DA15-Fchain) and active site residues of gyrA Arg128, Ser91, Asp94 information were used to prepare grid box for ligand docking. The ligand moxifloxacin was prepared using LigPrep module to get possible ionisation or protonation states at physiological pH(7.0±2.0). The Glide XP ligand docking protocol was followed to dock moxifloxacin and examine its bindingpatterntothe DNA-gyrase complex in the absence of DNA cleavage.
Interestingly, the moxifloxacin binds well to gyrase and uncleaved DNA complex, and one salt bridge interaction was observed with nucleotide baseof major groove. The quinone part of moxifloxacin showed interaction with Arg128 and the protonated NH2+ group of piperidine ring interacted with DNA nucleotide base to form a salt bridge. The docking pose analysis revealed that the drug binds in between the gyrase and the DNA, which is in close proximity to the enzyme recognitionsite of DNA (prone to be cleavage through catalytic Tyr129 of gyrA enzyme, which attacks at both DNA strands). Figure 3 shows moxifloxacin binding comfortably with Met127, Arg128, Asp94 via H-bond, salt bridge network. The binding pose of moxifloxacin revealed new insights into the binding of binary gyrase and DNA complex with drug molecule.
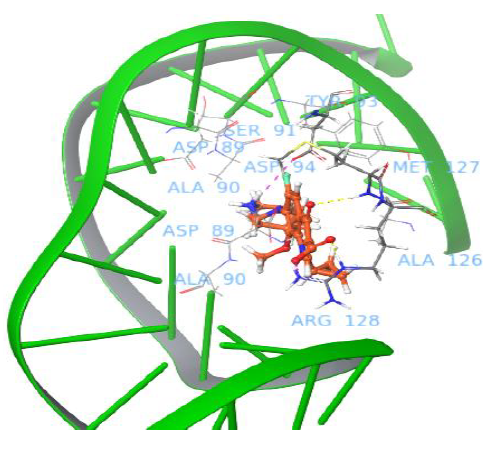
Figure 3: Binding pose of moxifloxacin with uncleaved DNA and gyrase complex.
Our computational molecular mechanistic studies offered molecular insights to possible routes to emphasise that DNA intercalating agents will slowly slip into broken DNA strands when it is cleaved with the help of gyrase enzyme. To the best of our knowledge this is the first study that statesmoxifloxacin passage from gyrase binding pocket to the cleaved site of DNA. These findings would help the scientific community tobetter understand the fluoroquinolone translocation pathway and design novel inhibitors blocking two host-guest mechanism of DNA gyrase[32].
3.3 MD studies to investigate the binding of moxifloxacin around uncleaved DNAand gyrase complex
The prepared uncleaved state of DNA and gyrase enzyme complex showed a little different binding location to accommodate the moxifloxacin molecule. Although it has more than four intermolecular interactions with surrounding residues, it is confirmed that binding strength with gyrase enzyme is less, which raises a question that how it will move into the cleaved region of DNA as it cannot persist with gyrase for long. To explore the doubts on quinolone molecule passage pathway from gyrase to uncut DNA, we carried out another 500ns molecular dynamics simulation study.
From the trajectory analysis, the following important points shed light on the movement of fluoroquinolone onto cleaved strands of DNA. Moxifloxacin molecule showed H-bond network with Arg128, Met127 and salt bridge networks with Asp94, Arg128 through the rigid docking studies.These intermolecular interactions were maintained for a while under dynamic conditions,as shown in Figure 4.
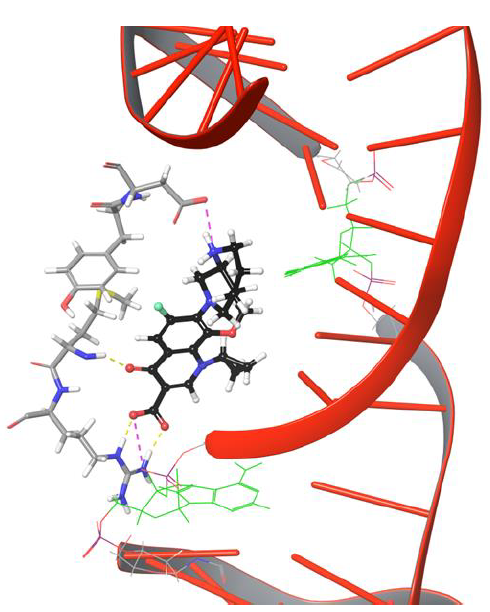
Figure 4: Moxifloxacin interaction networks with uncut DNA and gyrase complex.
As the moxifloxacin started to move towards DNA cleavage site (see Figure 5, DG 11 represented in green color stick model to illustrateDNA cleavage point), it loses H-bond and salt bridge network with Arg 128.The carboxyl group of moxifloxacin forms H-bond network with Met127 as it is positioned towards the amino group of Met127. The current pose of quinone moiety of drug facilitates π-π stacking interaction with Tyr93 at 634th frame (63.4ns) as shown in Figure 5.
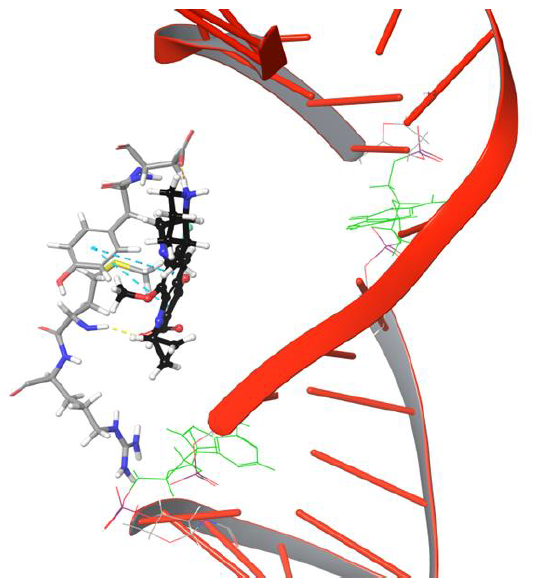
Figure 5: The simulation 634 frame showing Arg128 losing interaction and binding to Met127, Tyr93, Asp94 through various interaction types
Interestingly amino group of Asn121 also forms H-bond with the carboxyl group of moxifloxacin. However, it doesn’t last for long as the drug molecule continues to move towards cleavable region of DNA. Surprisingly, the 634th frame showed drug molecule forming H-bond networks with Asp94 and Met127 for an extended period of time. The amino group of piperidine moiety of the drug showed H-bond and salt bridge interaction with Asp94, whereas quinone moiety formed π-π stacking interaction with aromatic ring of Tyr93 as indicated in Figure 5. The stability of interactions displayed by Tyr93, Asp94, Met127 prevents further movement of drug molecule towards the DNA as they are dynamically stable until 1219th frame. The conformation of the piperidine ring of the drug started flipping towards the DNA cleavable site by losing its binding with Asp94, as shown in Figure 6.
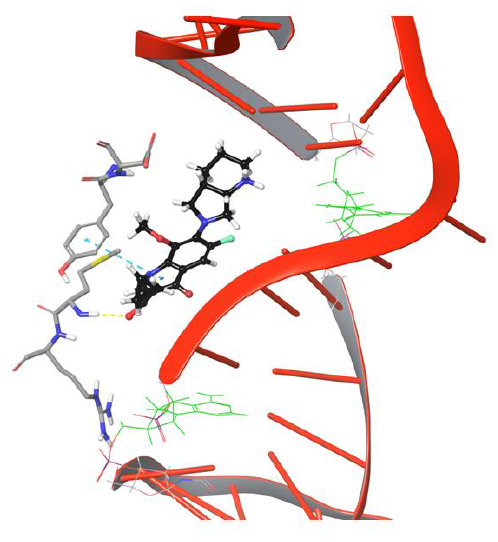
Figure 6: Simulation frame 1531 showing H-bond and salt bridge network broken between drug and Asp94
The π-π stacking interaction between drug and Tyr93 got disrupted further to facilitate passage of drug to bind DNA, as shown in Figure 7.
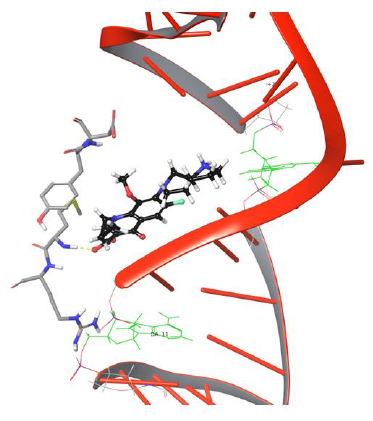
Figure 7: Simulation frame 2204 showing π-π stacking and H-bond, salt bridge network disrupted with Tyr94 and Asp94.
The drug molecule lost old and made new binding interactions consecutively to make its way towards the DNA cleavage site. The bicyclic piperidine ring of drug tends to enter the DNA around the cleavage site but is unable to do so due to the uncleaved state of DNA,leading to amino group of piperidine ring getting pulled by hydroxyl group of Asp94. As the production phase of MD progresses, the drug molecule loses its binding with Tyr93, Asp94 due to the lack of stability in interactions. The overall analysis provided useful insights on Met127 interaction with moxifloxacin as the H-bond evolution started at frame 626 and continued till the last frame 5000 of MD simulations, which indicates that drug molecule will have strong one H-bond network. Our study demonstrated that Met127 plays an important anchor role in entry of drug into cleaved DNA strands as it is the only one stable interaction that is retained under dynamic conditions until the DNA gets cleaved by the enzyme. The critical structural features of drug molecule and catalytic residues Asp94, Met127 facilitate the penetration of the drug into the cleaved DNA strand though it was not selectively developed to target the resealing of the DNA strands. In the present study, drug conformation flipping towards the cleavableDNAstrand is observed very closely in simulation frame 3883, as shown in Figure 8.
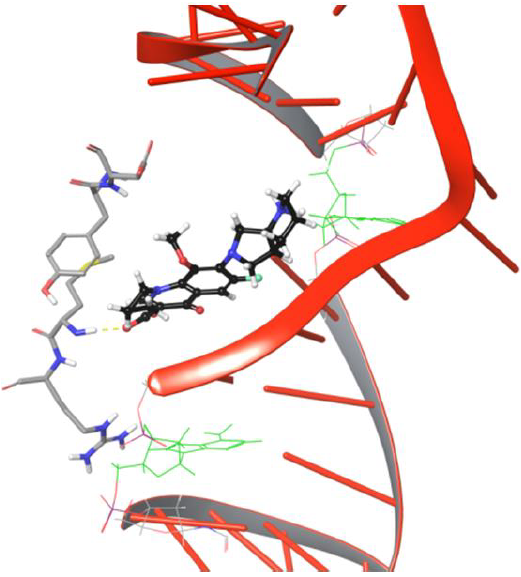
Figure 8: Frame 3883 showing drug conformation towards DNA cleavable site
The entry of drug into cleavage site is restricted by DNA backbone leading to its interaction with Asp94 via H-bond and salt bridge network, as shown in Figure 9. The biochemical study findings from Blower et al. and Mayer et al. demonstrated fluoroquinolone binding at cleaved DNA-gyrase complex and defined it as topoisomerase poison as increases the amount of DNA fragments ‘cleaved complexes’ by blocking DNA resealing. Nagaraja et al., demonstrated from the experimental studies that quinolones will have high drug potency when they selectively bind to gyrase enzyme. However, no research group discovered the chemical path of quinolones from gyrase binding site to the cleaved region of DNA. We conclude that our findings from the DNA-gyrase-drug ternary complex simulation study identified the binding pattern of quinolones before the DNA is cleaved by the enzyme.However, the processneeds to be verified experimentally to getmore meaningful insights thatmay direct the future of quinolone based drug discovery.
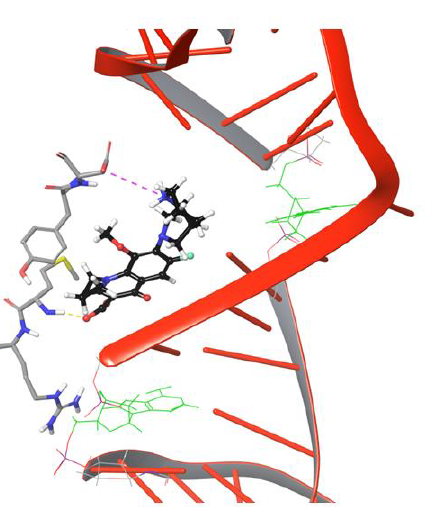
Figure 9: Simulation Frame 4419 showing piperazine moiety of moxifloxacin moving back to catalytic residue Asp94 by forming salt bridge network
3.3 Binding free energy calculations
The MMGBSA technique was employed to calculate the free energy of various simulation frames where significant changes were observed between the ternary moxifloxacin-uncut DNA-gyrase complex using Prime module of Schrödinger. The structures derived from the frame 634, 1531, 2204, 3883, 4119 of second 500ns molecular dynamics simulation studywere subjected to binding free energy calculations to compute binding free energy for moxifloxacin with DNA-enzyme complex.The computed binding free energy for simulation frame 634, 1531, 2204, 3883, 4119 was calculated to be -29.9, -50.5, -38.9, -37.4, -37.9 respectively. The calculations showed significant binding free energy values; however, 634th frame showed less binding free energy as interactions between active site residues and moxifloxacin are not stable but the 1531 frame showed a high free energy value of -50.5kcal/mol as interaction networks are comparatively stable. In the case of 2204, 3883, 4119 frames, binding free energy remains mostly similar as the pose and interactions shown by moxifloxacin to surrounding active site residues varyonly to a small extent.
4. Conclusion
DNA gyrase is a cellular target for fluoroquinolone drug molecules. The continuous and unjust use of fluoroquinolones to treat various infections accelerates the rise of mutations in quinolone binding pocket (QBP) of DNA gyrA complex, leading to a reduction in the fluoroquinolones potency. This facet made us recheck the molecular mechanism of fluoroquinolone binding to gyrase and cleaved part of DNA using molecular dynamics simulation. Our study highlighted that moxifloxacin comfortably bindsto the complex of DNA and gyrase even when the G-segment of DNA is in an uncleaved state. It can be hypothesized that the inhibitor will only enter the cavity by slipping into the space between the two broken ends of the DNA after the enzyme has catalyzed the breaking of DNA.
Credit authorship contribution statement
MES designs the study and GSK performs the study, analysis, manuscript writing, editing, review, methodology with the help of HPS.
Declaration of Competing Interest
The authors declare that they have no potential competing interests
Acknowledgement
The authors thank the National Institute of Pharmaceutical Education and Research (NIPER), Department of Pharmacoinformatics, SAS Nagar, Ministry of Chemicals and Fertilizers, New Delhi, Government of India for providing the facility. And our special thanks to Dr. Prajwal Nandekar, Senior Scientist, Schrödinger, LLC, Bengaluru, India.
References
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Quarterly Reviews of Biophysics 41(2008): 41-101.
- ST Cole et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(1998): 537-544.
- K Mdluli, Z Ma. Mycobacterium tuberculosis DNA gyrase as a target for drug discovery. Infectious Disorders - Drug Targets 7(2007):159-168.
- Gellert M, Mizuuchi K, O'Dea MH, Itoh T, Tomizawa JI. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proceedings of the National Academy of Sciences of the United States of America 74(1977): 4772-4776.
- Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proceedings of the National Academy of Sciences of the United States of America 74(1977): 4767-4771.
- Piton J, Petrella S, Delarue M, Andre-Leroux G, Jarlier V, Aubry A, Mayer C. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PloS One 5(2010): e12245.
- Aldred KJ, Kerns RJ, Osheroff Mechanism of quinolone action and resistance. Biochemistry 53(2014): 1565-1574.
- Anderson VE, Osheroff N. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Current Pharmaceutical Design 7(2001): 337-353.
- Drlica K, Hiasa, H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Current Topics in Medicinal Chemistry 9(2009): 981-998.
- Reece RJ, Maxwell A. DNA gyrase: structure and function. Critical Reviews in Biochemistry and Molecular Biology 26(1991): 335-375.
- Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Applied Microbiology and Biotechnology 92(2011): 479-497.
- Nagaraja V, Godbole AA, Henderson SR, Maxwell A. DNA topoisomerase I and DNA gyrase as targets for TB therapy. Drug Discovery Today 22 (2017): 510-518.
- Petrella S, Capton E, Raynal B, Giffard C, Thureau A, Bonnete F, Alzari PM, Aubry A, Mayer C. Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a CorynebacterialesGyrB-Specific Insert in ATPase Activity. Structure 27(2019): 579-589 e575.
- Tripathi N, Guchhait SK, Bharatam PV. Pharmacoinformatics analysis of merbarone binding site in human topoisomerase IIα. Journal of Molecular Graphics and Modelling 86 (2019): 1-18.
- Schrödinger, Maestro-Desmond Interoperability Tools, Schrödinger Release 2019-1 (2020):Schrödinger, LLC, New York, NY
- Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E watermodels at 298 K. The Journal of Physical Chemistry A 105(2001): 9954-9960.
- Roos K, Wu C, Damm W, Reboul M, Stevenson JM, Lu C, Dahlgren MK, Mondal S, Chen W, Wang L, Abel R, Friesner RA, Harder E. D.OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. Journal of Chemical Theory and Computation 15(2019): 1863-1874.
- Kalibaeva G, Ferrario M, Ciccotti G. Constant pressure-constant temperature molecular dynamics: a correct constrained NPT ensemble using the molecular virial. Molecular Physics 101(2003): 765-778.
- Ryckaert JP, Ciccotti, G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of computational physics 23(1977): 327-341.
- Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. The Journal of chemical physics 98(1993): 10089-10092.
- Chen IJ, Foloppe N. Drug-like bioactive structures and conformational coverage with the LigPrep/ConfGen suite: comparison to programs MOE and catalyst. Journal of Chemical Information and Modeling: JCIM 50(2010): 822-839.
- Schrödinger, LigPrep, Schrödinger Release 2019-1 (2020): Schrödinger, LLC, New York, NY.
- Schrödinger, Protein Preparation Wizard, Epik, Schrödinger Release 2019-1(2016): Schrödinger, LLC, New York, NY.
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry: JCM 47(2004): 1739-1749.
- Halgren TA, Murphy R B, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry: JCM 47(2004): 1750-1759
- Schrödinger, Glide, Schrödinger Release 2019-1(2020): Schrödinger, LLC, New York, NY.
- Hou T, Wang J, Li Y, Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized born surface area methods. II. The accuracy of ranking poses generated from docking. Journal of Computational Chemistry: JCM 32 (2011): 866-877.
- Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery 10 (2015): 449-461.
- Schrödinger, Prime, Schrödinger Release 2019-1(2020): Schrödinger, LLC, New York, N
- Li J, Abel R, Zhu K, Cao Y, Zhao S, Friesner RA. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins 79 (2011): 2794-2812.
- Schrödinger, Desmond Molecular Dynamics System, Schrödinger Release 2019-1 (2020): D. E. Shaw Research, New York, NY.
- Blower TR, Williamson BH, Kerns RJ, Berger JM. Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America 113 (2016): 1706-1713
