scRNA-Seq Analysis Immune Changes of Intratumoral Hapten Plus Chemotherapy Drugs for the Treatment of Male Breast Cancer
Article Information
Shuxiao Dong6, Baofa Yu1,2,3,4,5,6*,Feng Gao2, Peng Jing2, Guoqin Zheng2, Peicheng Zhang2, Shengjun Zhou1
1TaiMei Baofa Cancer hospital, Dongping, Shandong Province, China, 271500
2Jinan Baofa Cancer hospital, Jinan, Shandong Province, China, 250000
3Beijing Baofa Cancer Hospital, Beijing, China, 100010
4South China Hospital of Shenzhen University, Shenzhen, China, 518055
5Immune Oncology Systems, Inc, San Diego, CA, USA, 92102
6Shandong Third Hospital, Jinan, China, 271500
*Corresponding Author: Baofa Yu, TaiMei Baofa Cancer hospital, Dongping, Shandong Province, China and Immune Oncology Systems, Inc, San Diego, CA, USA, 92102.
Received: 21 October 2023; Accepted: 01 November 2023; Published: 27 November 2023.
Citation:
Shuxiao Dong, Baofa Yu, Feng Gao, Peng Jing, Guoqin Zheng, Peicheng Zhang, Shengjun Zhou. scRNA-Seq Analysis Immune Changes of Intratumoral Hapten Plus Chemotherapy Drugs for the Treatment of Male Breast Cancer. Journal of Cancer Science and Clinical Therapeutics 7 (2023): 212-223
View / Download Pdf Share at FacebookAbstract
Male breast cancer is a rare studied of disease and the relevant research is also very scarce. Current treatment of male breast cancer is modeled after the treatment of female breast cancer, namely a surgery-based comprehensive treatment, which for men to bear trauma and psychological blow. In this study, we explored the efficacy of hapten enhanced intratumoral chemotherapy for treating male breast cancer. We used scRNA-Seq to profile the changes induced by hapten enhanced intratumoral chemotherapy. We demonstrate that Hapten-enhanced intratumoral chemotherapy (HEIC) for male breast cancer induces an acute immune response, which in turn helps to kill and control cancer cells and may enable immune response as a immunotherapy to advance before any treatment to prevent tumor recurrence and metastasis.
Keywords
<p>Intratumoral injection, Cancer immunotherapy, Drug delivery, Intracellular drug delivery for surgery, Immune surgery</p>
Intratumoral injection articles; Cancer immunotherapy articles; Drug delivery articles; Intracellular drug delivery for surgery articles; Immune surgery articles.
Intratumoral injection articles Intratumoral injection Research articles Intratumoral injection review articles Intratumoral injection PubMed articles Intratumoral injection PubMed Central articles Intratumoral injection 2023 articles Intratumoral injection 2024 articles Intratumoral injection Scopus articles Intratumoral injection impact factor journals Intratumoral injection Scopus journals Intratumoral injection PubMed journals Intratumoral injection medical journals Intratumoral injection free journals Intratumoral injection best journals Intratumoral injection top journals Intratumoral injection free medical journals Intratumoral injection famous journals Intratumoral injection Google Scholar indexed journals Cancer immunotherapy articles Cancer immunotherapy Research articles Cancer immunotherapy review articles Cancer immunotherapy PubMed articles Cancer immunotherapy PubMed Central articles Cancer immunotherapy 2023 articles Cancer immunotherapy 2024 articles Cancer immunotherapy Scopus articles Cancer immunotherapy impact factor journals Cancer immunotherapy Scopus journals Cancer immunotherapy PubMed journals Cancer immunotherapy medical journals Cancer immunotherapy free journals Cancer immunotherapy best journals Cancer immunotherapy top journals Cancer immunotherapy free medical journals Cancer immunotherapy famous journals Cancer immunotherapy Google Scholar indexed journals Drug delivery articles Drug delivery Research articles Drug delivery review articles Drug delivery PubMed articles Drug delivery PubMed Central articles Drug delivery 2023 articles Drug delivery 2024 articles Drug delivery Scopus articles Drug delivery impact factor journals Drug delivery Scopus journals Drug delivery PubMed journals Drug delivery medical journals Drug delivery free journals Drug delivery best journals Drug delivery top journals Drug delivery free medical journals Drug delivery famous journals Drug delivery Google Scholar indexed journals Intracellular drug delivery for surgery articles Intracellular drug delivery for surgery Research articles Intracellular drug delivery for surgery review articles Intracellular drug delivery for surgery PubMed articles Intracellular drug delivery for surgery PubMed Central articles Intracellular drug delivery for surgery 2023 articles Intracellular drug delivery for surgery 2024 articles Intracellular drug delivery for surgery Scopus articles Intracellular drug delivery for surgery impact factor journals Intracellular drug delivery for surgery Scopus journals Intracellular drug delivery for surgery PubMed journals Intracellular drug delivery for surgery medical journals Intracellular drug delivery for surgery free journals Intracellular drug delivery for surgery best journals Intracellular drug delivery for surgery top journals Intracellular drug delivery for surgery free medical journals Intracellular drug delivery for surgery famous journals Intracellular drug delivery for surgery Google Scholar indexed journals Immune surgery articles Immune surgery Research articles Immune surgery review articles Immune surgery PubMed articles Immune surgery PubMed Central articles Immune surgery 2023 articles Immune surgery 2024 articles Immune surgery Scopus articles Immune surgery impact factor journals Immune surgery Scopus journals Immune surgery PubMed journals Immune surgery medical journals Immune surgery free journals Immune surgery best journals Immune surgery top journals Immune surgery free medical journals Immune surgery famous journals Immune surgery Google Scholar indexed journals Male breast cance articles Male breast cance Research articles Male breast cance review articles Male breast cance PubMed articles Male breast cance PubMed Central articles Male breast cance 2023 articles Male breast cance 2024 articles Male breast cance Scopus articles Male breast cance impact factor journals Male breast cance Scopus journals Male breast cance PubMed journals Male breast cance medical journals Male breast cance free journals Male breast cance best journals Male breast cance top journals Male breast cance free medical journals Male breast cance famous journals Male breast cance Google Scholar indexed journals intratumoral chemotherapy articles intratumoral chemotherapy Research articles intratumoral chemotherapy review articles intratumoral chemotherapy PubMed articles intratumoral chemotherapy PubMed Central articles intratumoral chemotherapy 2023 articles intratumoral chemotherapy 2024 articles intratumoral chemotherapy Scopus articles intratumoral chemotherapy impact factor journals intratumoral chemotherapy Scopus journals intratumoral chemotherapy PubMed journals intratumoral chemotherapy medical journals intratumoral chemotherapy free journals intratumoral chemotherapy best journals intratumoral chemotherapy top journals intratumoral chemotherapy free medical journals intratumoral chemotherapy famous journals intratumoral chemotherapy Google Scholar indexed journals Hapten-enhanced articles Hapten-enhanced Research articles Hapten-enhanced review articles Hapten-enhanced PubMed articles Hapten-enhanced PubMed Central articles Hapten-enhanced 2023 articles Hapten-enhanced 2024 articles Hapten-enhanced Scopus articles Hapten-enhanced impact factor journals Hapten-enhanced Scopus journals Hapten-enhanced PubMed journals Hapten-enhanced medical journals Hapten-enhanced free journals Hapten-enhanced best journals Hapten-enhanced top journals Hapten-enhanced free medical journals Hapten-enhanced famous journals Hapten-enhanced Google Scholar indexed journals metastasis articles metastasis Research articles metastasis review articles metastasis PubMed articles metastasis PubMed Central articles metastasis 2023 articles metastasis 2024 articles metastasis Scopus articles metastasis impact factor journals metastasis Scopus journals metastasis PubMed journals metastasis medical journals metastasis free journals metastasis best journals metastasis top journals metastasis free medical journals metastasis famous journals metastasis Google Scholar indexed journals scRNA articles scRNA Research articles scRNA review articles scRNA PubMed articles scRNA PubMed Central articles scRNA 2023 articles scRNA 2024 articles scRNA Scopus articles scRNA impact factor journals scRNA Scopus journals scRNA PubMed journals scRNA medical journals scRNA free journals scRNA best journals scRNA top journals scRNA free medical journals scRNA famous journals scRNA Google Scholar indexed journals
Article Details
1. Introduction
Although male breast cancer (MBC) is a rare studied of disease and accounts for less than 1% of all breast cancer, the incidence has been steadily increasing worldwide. Possible risk factors for MBC include obesity, increased longevity, testicular disease and tumors, and germline mutations in BRCA2 [1]. There is still a significant gap for understanding of the epidemiology of MBC. Several studies have specifically compared the epidemiological risk factors for MBC and female breast cancer (FBC). The proportion of BRCA2 tumors is higher in MBC (occurring in 10%), while the proportion of BRCA1 tumors is lower (only found in 1%). These findings indicate a significant difference in genetic etiology between MBC and FBC [2]. Current treatment of MBC can be inferred from the results of FBC treatment trials.In the absence of evidence from national and international randomised trials, these treatment options appear superficially appropriate [2]. Similar to the treatment of FBC, conservative surgery is effective and safe for MBC. Post-mastectomy chest wall radiation therapy and 5-year tamoxifen therapy also improve survival [3]. Local treatment and new treatment options are the clinical needs to control all types of breast cancer. A new way to target the interaction of FAK and Mdm-2 proteins is crucial for future targeted therapies [4]. Targeted therapies, such as pazopanib, are a promising option and a novel immunotherapy strategy for the treatment of multifocal tumors in breast cancer; Programmed death-1 (PD1) or programmed death ligand-1 (PD-L1) blockers have also had some success and may become effective treatments in the future [5, 6, 7]. Because MBC is a rare disease, there hasn't been a lot of research to explore new treatment options. Ultra-small Incision Individualized Intratuminal Chemotherapy Immunotherapy (UMIPIC) that can be enhanced by hapten is used to treat lung cancer and other solid tumor, now it needed to try for treating male breast cancer [8]. Recent advances in single-cell RNA sequencing (scRNA-seq) have allowed scientists to explore genetic and functional heterogeneity of cellular complexes at the molecular level [9]. Because single-cell transcription profiling offers a suitable alternative, direct comparisons between the same cell types can now be compared before and after treatment. However, current single-cell studies of breast cancer have focused on the tumor microenvironment (TME) [10]. At present, there is no comparative study on the changes of single-cell immune response of MBC before and after hapten enhancement of UMIPIC.
In this study, we used scRNA-seq to study and compare the changes of bone marrow cells, stromal cells, T cells, plasma cells, B cells, platelets, red blood cells, and epithelial cells in tumors before and after the treatment of hemagglutine-enhanced UMIPIC. We found that immune cells are awakened to help with cancer immunotherapy, such as PD1 or PD-l1, to achieve an immune response [11, 12]. Our study provides a detailed overview of how immune cells awaken on a molecular basis against cancer cells. We believe our findings will contribute to the development of a more reliable hapten-enhanced sensitized immunotherapy for the treatment of male breast cancer patients without surgery, which enables immunotherapy to be performed to prevent tumor recurrence and metastasis than earlier of modulation of any treatment.
2. Materials and Methods
2.1 Clinical Specimens
The patient had pathological diagnosis: (Left breast) infiltrating ductal carcinoma, ER: positive (range 90%, intensity: strong), PR: positive (range about 25%, intensity: medium), Ki-67 (positive cell count about 70%), HER2 (2+), FISH test: negative, clinical stage is II stage B T2N1M0. The patient did not have any other therapy before this study. Before receiving treatment at Beijing Baofa Cancer Hospital, the patient’s physical condition was evaluated and determined to meet the indications for UMIPIC. All procedures and protocols in the study has been reviewed and approved by the Ethical Committee of the Beijing Baofa Cancer Hospital (TMBF 0010, 2015). All informed consent forms from patients have been signed prior to the start of the study. The injection combination dosage of Adriamycin (Adr), cytarabine (Ara-C), H2O2 and penicillin as hapten [11, 12].
2.2 Tissue Disassociation and Cells Collection
Small cervical lesions tissues and blood samples collected and immediately stored in the sCelLiVE® Tissue Preservation Solution (Singleron) on ice and marked sample got before treatment as BC_B and sample got after treatme as BC_A. The tissue was prepared into small pieces, transferred to a 15ml centrifuge tube and digested with sCelLiVE® tissue dissociation solution (Singleron) oscillating at 37°C for 15 minutes. The sample was then filtered with a 40µm sterile filter, centrifuged at 1000 rpm at 4 ° C for 5 min, and then added with 2 ml GEXSCOPE® Red cell lysis buffer (RCLB, Singleron) for 10 min. In this study, we used scRNA-seq to study and compare the changes of bone marrow cells, stromal cells, T cells, plasma cells, B cells, platelets, red blood cells, and epithelial cells in tumors before and after the treatment of hemagglutine-enhanced UMIPIC. We found that immune cells are awakened to help with cancer immunotherapy, such as PD1 or PD-l1, to achieve an immune response [11, 12].
2.3 Single-Cell RNA Sequencing
Singleron Matrix® single-cell processing system and onto a microporous chip. Simply put, use the GEXSCOPE® single-cell RNA Library Kit (Singleron) and construct a scRNA-seq library. The library was sequenced on IlluminaHiSeq X platform [13]. Sequencing data processing and quality control were performed as described in previous publications [13, 14, 15].
2.4 Differentially Expressed Genes (DEGs) Analysis
To identify differentially expressed genes, genes were selected that were expressed in more than 10% of the cells and had fold changes of greater than 1. The adjusted P-value is calculated by benjamini-hochberg correction. A P value of 0.05 was used as the criterion to determine the statistical significance of the difference [13].
2.5 Cell Type Annotation
Using the SynEcoSys database, the cell type of each cluster was determined by the expression of typical markers found in deg. The heat map/dot map/violin map for each cell type was generated by the scanpy built-in function and ggplot2 [13].
2.6 Single-Cell Copy Number Variation (CNV) Analysis
Use to detect CNAs in malignant cells. The CNVs of malignant cells were estimated using non-malignant cells (T and NK) as controls. The classification is based on the loci on each chromosome. The relative expression values ranged from the middle to 1, with 1.5 standard deviations of the residual normalized expression values as the upper limit. The sliding window size smooth’s out relative expression on each chromosome to eliminate the influence of gene-specific expression [13].
2.7 Pathway Enrichment Analysis
To investigate the potential function of deg between clusters, Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used [16]. Reference GOES gene sets, including molecular function (MF), Biological process (BP), and Cell component (CC) categories. Channels with p values less than 0.05 after correction are considered to be significantly enriched [13].
2.8 Trajectory Analysis
The Monocle 2 algorithm is adopted, and the dimension reduction method is DDR Tree [13, 17].
2.9 Intratumor heterogeneity (ITH) score was calculated
The ITH score is defined as the average Euclidean distance between a single cell and all other cells, derived from the first 20 principal components based on standardized expression levels of highly variable genes [13].
2.10 Cell-cell Interaction Analysis (CellPhoneDB)
Cell-cell interactions (CCI) between B cells, epithelial cells, fibroblasts, mononuclear phagocytes, mast cells, neutrophils, T cells, and NK cells were predicted using the mobile database v2.1.0 based on known ligand-receptor pairs [18, 19, 20].
3. Results
3.1 Clinical Benefit Characteristics
After treatment UMIPIC to MBC, post treatment pathological diagnosis again: tumor size 1.9 x 1.5 x 1.0 cm, histological type: invasive cancer, non-specific type, and grade: grade II, vascular invasion: (+), lymph node metastasis: left axillary sentinel lymph node A shows macroscopic metastasis, while B and C show no cancer metastasis. No cancer metastasis was found in the axillary lymph nodes (0/15). Follow-up examination every four weeks after the treatment in a six-month period, physical examination and CT of the patient showed no signs of tumor on his breast. Immunohistochemical staining were all set as tissue chip control, the cell display: ER (+80%, strong), PR (+10%, strong), HER (+1), Ki-67 (+40%), AR (+80%, strong), CD8 (+less than 10%), Foxc1 (less than 10%, positive), E-Cad (+), P120 (+) CK5/6, afraid of 6 (-), GATA3 (+), TRPS1 (+), GCDFP15 (-).
3.2 Landscape of single cell transcriptome sequencing before and after lung cancer treatment
There were 10000 cells in the two samples of MBC, including 8 cell types, including endothelial cells, epithelial cells, red blood cells, fibroblasts, mononuclear phagocyte, mast cell, parietal cell and T cells (Figure.1a, c, d).
Compared BC group B to group A, the proportion of endothelial cells, fibroblasts, parietal cell, T cells and mast cell decreased, and the proportion of monocyte phagocyte increased (Figure.1b). The decrease in the proportion of endothelial cells in Group A may lead to a decrease in the nutrient supply of endothelial cells to tumor cells, limiting their growth. The decrease in the proportion of fibroblasts composed of group A may lead to a decrease in the level of fibrosis after treatment.
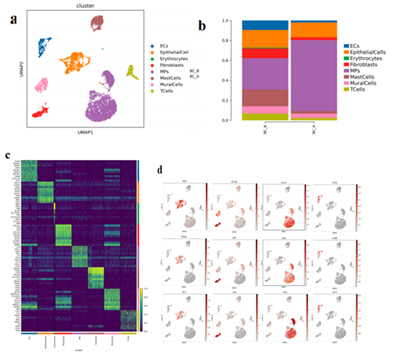
Figure 1:
Cell type reduction map display; b. Histogram of the proportion of cell types from different sample sources; c. Histogram of the proportion of cell types from different sample sources; d.Marker gene expression feature plot (Epithelial cells, fibroblasts, MPs, endothelial cells)
3.3 Changes in tumor cells before and after administration
The inferCNV and CNVscore analysis indicated that all epithelial cells were malignant cells (Figure.2 a, b, c). The total number of EpithelialCells is 1580, and there are 8 different subtypes, including EpithelialCells1-8 (Figure.2 d). Compared with BC_B, the proportion of EpithelialCells1/3/4/8 in BC_A group decreased, and the proportion of EpithelialCells2/5/6/7 increased (Figure.2 e). GO/KEGG enrichment results of differential genes among epithelial cell subtypes suggest that EpithelialCells1 up-regulated gene is mainly enriched in the mRNA catabolism process of nuclear transcription, nonsense mediated decay, SRP-dependent cotranslational protein targeting membrane, cotranslational protein targeting membrane, endoplasmic reticulum targeting protein, and establishment of endoplasmic reticulum localization. This means that these processes may be involved in disease progression (Figure.2 f) [21].
The EpithelialCells2 up-regulated gene was mainly enriched in oxidative phosphorylation, unsaturated fatty acid biosynthesis, fatty acid metabolism, and other pathways (Figure.2 g).
The EpithelialCells5 up-regulated gene was mainly enriched in the MAPK signaling pathway, apoptosis, TNF signaling pathway, and NF-κB signaling pathway (Figure.2 h).
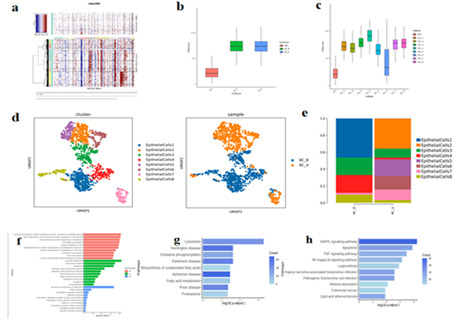
Figure 2:
Chromosome copy number variation (CNV), where genes are arranged from left to right across the entire chromosome. Chromosome amplification is shown as a red block, while chromosome deletion is shown as a blue block. (MP for reference)
Box diagram of b/c.CNV Score. The horizontal coordinate is different samples/modules, and the vertical coordinate is the quantized CNV score value. The larger the CNV score value, the greater the CNV variation of the cell type.
- Epithelial cell subtype vitreogram;
- Histogram of proportion of epithelial cell subtypes;
Bar chart of EpithelialCells1 subpopulation differential gene GO enrichment analysis
g/h. EpithelialCells2/5 subpopulation KEGG enrichment analysis bar chart.
3.4 Changes of Macrophages in immune cell characteristics before and after administration
MPs totaled 5130 cells, and 3 different subtypes were obtained, including Conventional type 2 dendritic cells (cDC2), Macrophages, and Monocytes (Figure.3-1a). Macrophages and Monocytes were subdivided into 5 and 3 subgroups, respectively. Compared with BC_B, the proportion of Macrophages_1 in BC_A group is reduced while Macrophages_2-5 is increased. The proportion of Monocytes_2 decreased, and the proportion of Monocytes_1/3 increased (Figure.3-1b, c). By scoring the characteristic gene set of macrophage subtype, we found that Macrophages_2-5 tends to be macrophages with pro-inflammatory function and lipid metabolism (Figure.3-1 d).
KEGG enrichment results of differential genes among monocyte subtypes suggest that Monocytes_1 up-regulated genes are mainly enriched in cytokines and cytokine receptor interaction, P53 signaling pathway and other pathways (Figure.3-1 e).
In the analysis of transcription factor, Macrophages_1's transcription factor ASCL2 has a function of regulating the tumor immune microenvironment (Figure. 3-1 f). MZF1 activity is high in Monocytes_2 (Figure.3-1 g). MEF2D activity is high in Monocytes_3.
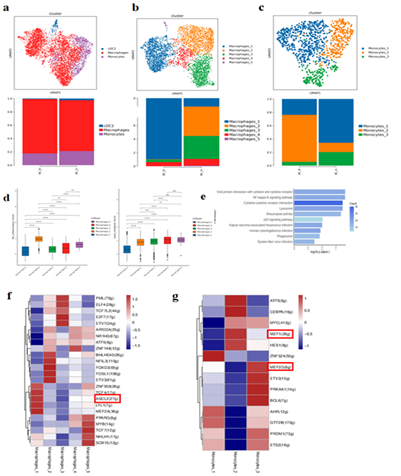
Figure 3-1:
- MPs cell type reduction diagram and MPs cell subtype proportion histogram;
- Dimensionality reduction diagram of macrophage types and histogram of proportion of macrophage subtypes
- Monocyte type reduction chart and monocyte subtype proportion histogram;
- Macrophage subpopulation function assessment gene set score map;
- Bar chart for KEGG enrichment analysis of Monocytes_1 subpopulation;
- Heat map of transcription factor analysis of macrophage subtype Pyscenic, name of behavior regulon, numbers in parentheses represent the number of transcription factor target genes; Listed as cell name;
- Monocyte subtype Pyscenic transcription factor analysis heat map, behavioral regulon name, numbers in parentheses represent the number of transcription factor target genes; Listed as cell name;
3.5 Changes of MasterCells characteristics of immune cells before and after administration
There were 911 MastCells in total, and 7 different subtypes were subdivided, including MastCells1-7 (Figure.3-2 a). Compared with BC_B, the proportion of MastCells1 in BC_A group increased, while the proportion of MastCells2-7 decreased (Figure.3-2 a). The result of differential gene GO enrichment between MastCells subtypes suggested that the up-regulated gene of MastCells1 was mainly enriched in biological processes such as intrinsic apoptosis signaling pathway, regulation of cell adhesion, T cell activation, and leukocyte adhesion (Figure.3-2 b). There were 425 T cells in total, and two different subtypes were subdivided, including CD8+ effector T cells(CD8Teff) and Regulatory T cells(Treg) (Figure.3-2 c). Compared with BC_B, the proportion of CD8Teff in BC_A group increased, and the proportion of Treg decreased (Figure.3-2 c), the enrichment of T cell differential genes in BC_A group was mainly related to reactive oxygen species, regulation of T cell activation, T cell activation, and oxidative stress response. (Figure.3-2 d)
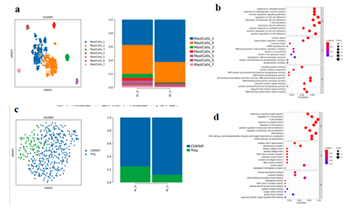
Figure 3-2:
- Mast cell subtype reduction map and mast cell subtype proportion histogram;
- Bubble map of differential gene GO enrichment analysis between MastCells1 subtypes;
- T cell subtype reduction chart and T cell subtype proportion histogram;
- Bubble map of GO enrichment analysis of T cell groups.
3.6 Changes in fibroblast characteristics before and after administration
1Fibroblasts, with a total of 608 cells, showed unsupervised clustering results and were divided into 5 heterogeneous subpopulations, including Fibroblasts1-5(Figure.4 a, c). Compared with BC_B, the proportion of Fibroblasts1/3/5 decreased and the proportion of Fibroblasts2/4 increased in BC_A group (Figure.4 b). KEGG enrichment analysis suggested that the up-regulated genes of Fibroblasts4 in BC_A group were mainly enriched in P53 signaling pathway, endoplasmic reticulum protein processing, IL-17 signaling pathway, TNF signaling pathway, apoptosis, NF-κB signaling pathway, etc (Figure.4 d). The results of gene set scores suggested that Fibroblasts1/3/5 tended to prodce a large amount of collagen in myoFIB cells during tissue repair (Figure.4 f, g).
The results of pseudo-time series analysis suggested that the proportion of fibroblastS1/3/5 at the beginning of differentiation increased after treatment, while the proportion of Fibroblasts1/3 at the end of differentiation decreased. (Figure.4 e, h)
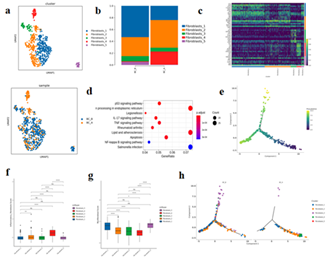
Figure 4:
- Fibroblast subtype vitreogram
- Histogram of fibroblast subtypes
- Heat maps of TOP10 differential genes in each subtype of fibroblasts
- Bubble map of KEGG enrichment analysis of differential genes among fibroblast subtypes;
- Diagram of quasi-sequential sequencing of all cells of fibroblast subtype
- Box map of myofibroblast gene set score;
- Pseudotemporal analysis of fibroblast subtypes -monocle2
- Box map of inflammatory fibroblast gene set scores;
3.7 Analysis of intercellular communication before and after administration
The interaction intensity between malignant epithelial cells and fibroblasts in BC_B group was stronger, while the interaction intensity between malignant epithelial cells and MPs (cDC2, macrophages, and monocytes) in BC_A group was increased after treatment (Figure.5 a, b). CXCL12_CXCR4, TNF_TNFRSF1A /B, and EREG_ERBB4 interact between epithelial cells and MPs cells, and BC_A group has a stronger interaction relationship than BC_B group (Figure.5 c, d).
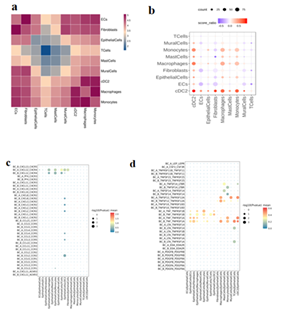
Figure 5:
- Heat map of the number of interactions between cell types in two samples. The darker the color, the more interactions between the two cell types;
- The difference bubble map of the interaction network between cell types before and after treatment to find the cell types with strong interaction with the target cell types. The horizontal and vertical coordinates are the types of interacting cells, the size of bubbles indicates the number of interactions (the number of groups with a large number is displayed), and the color of bubbles indicates the difference in interaction intensity between groups. Red indicates that BC_A group has a high intensity, and blue indicates that BC_B group has a high intensity.
c/d. ligand vs. bubble diagram (chemokine/cytokine), the size of the circle represents the significance of the interaction, the color of the circle represents the intensity of the interaction (gene expression);
3.8 Endothelial cell characteristics before and after administration
A total of 533 ECs cells were subdivided into 4 different subtypes. Arterial endothelial cells (AECs), Capillary endothelial cells(CapECs), Venous endothelial cells(VECs), Endothelial tip cells(TipCells) (Figure.6 a, c). Compared with BC_B, the proportion of AECs and VECs in BC_A group decreased, while the proportion of CapECs increased (Figure.6 b). GSVA enrichment analysis suggested that the differential genes of CapECs subgroup were mainly enriched in vitamin B6 metabolism, lipoic acid metabolism, valine leucine and isoleucine biosynthesis (Figure.6 d).
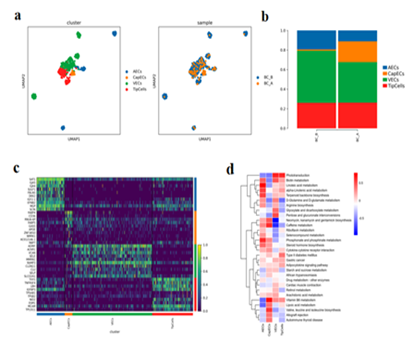
Figure 6:
- Endothelial cell subtype reduction map (by cell type and sample
- Histogram of endothelial cell subtypes
- Heat map of TOP10 differential genes of endothelial cell subtypes;
- Heat map of GSVA enrichment analysis among endothelial cell subtypes;
3.9 Estrogen receptor correlation analysis
Compared with BC_B, the proportion of EpithelialCells1/3/4/8 in BC_A group is reduced, and the proportion of EpithelialCells2/5/6/7 is increased (Figure.7 a, b). ESR1 (estrogen receptor-α gene) encodes estrogen receptor, which is mainly distributed in epithelial cells, and basically does not express ESR1 gene in epithelial cells S3/7. Compared with BC_B, the expression levels of ESR1 and FOXA1 genes in EpithelialCells1/4/8 subtype in BC_A group were decreased (Figure.7 b, c, d).
The gene set of estrogen-related pathway was scored, and the results suggested that the estrogen biosynthesis process and early/late estrogen response pathway had higher scores in EpithelialCells2/5/6 subtypes, and the estrogen inhibition pathway had higher scores in EpithelialCells3/4/7 (Figure.7 e).
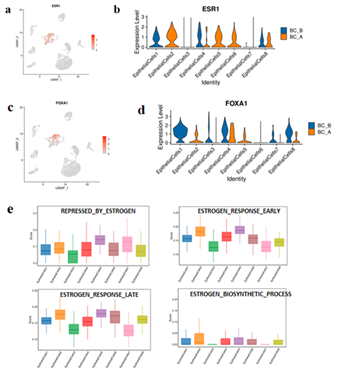
Figure 7:
- feature plot of ESR1 expression in each cell type at the taxonomy level.
- ESR1 expression in each subtype of epithelial cells.
- FOXA1 expression feature plot of each cell type at the taxonomy level.
- Violin diagram of FOXA1 expression in various subtypes of epithelial cells.
- Box map of gene set scores for estrogen-related pathways in epithelial cell subtypes.
3.10 Changes in gene expression associated with breast cancer
Compared with BC_B, BC_A group has a decrease in gene expression of androgen receptors (ARs) (Figure.8-1a), CD8A (Figure. 8-1b), FOXC1 (Figure. 8-1e), GATA3 (Figure. 8-1 d), KI-67 (Figure. 8-1 c), and TRPS1 (Figure. 8-1 f).
Compared with BC_B, BC_A group has decrease expression of PGR (progesterone receptors) (Figure. 8-2 a) while higher the expression of BRCA2 (Figure. 8-2 c). There are no significantly changes for these gene expressions in BC_A:CDH1 (Figure. 8-2 f), CTNND1 (Figure. 8-2 e), ESR1 (Figure. 8-2 b), ERB2 (Figure. 8-2 g), PIP (Figure. 8-2 d).
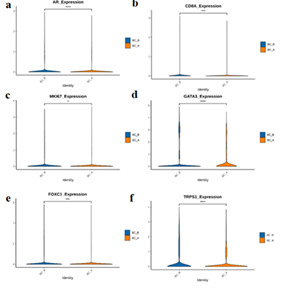
Figure 8-1:
AR: Compared with BC_B, the expression of BC_A group was decreased (Fig.8-1a).
CD8A: Compared with BC_B, the expression of BC_A group was decreased (Fig. 8-1b).
FOXC1: Compared with BC_B, the expression of BC_A was decreased (Fig. 8-1e).
GATA3: Compared with BC_B, the expression of BC_A group was decreased (Fig. 8-1d).
MKI67: Compared with BC_B, the expression of BC_A group was decreased (Fig.8-1c).
TRPS1: Compared with BC_B, the expression of BC_A group was decreased (Fig. 8-1f).

Figure 8-2:
CDH1: Compared with BC_B, the expression of BC_A group was not significantly different (Fig. 8-2f).
CTNND1: Compared with BC_B, the expression of BC_A group was not significantly different (Fig. 8-2e).
ESR1: Compared with BC_B, the expression of BC_A group was not significantly different (Fig. 8-2b).
ERBB2: Compared with BC_B, the expression of BC_A group was not significantly different (Fig. 8-2g).
PIP: Compared with BC_B, there was no significant difference in the expression of BC_A (Fig. 8-2d).
PGR: Compared with BC_B, the expression of BC_A group was decreased (Fig. 8-2a).
BRCA2: Compared with BC_B, the expression of BC_A group was increased (Fig. 8-2c).
4. Discussion
In our study, we mainly provide evidence that hapten-enhanced intratumoral chemotherapy (HEIC) for MBC induces an acute immune response, which in turn helps control the spread of cancer cells. Our study demonstrated that the immune cells can be awakened by HEIC, such the proportion of endothelial cells to tumor cells, Treg cells, fibroblasts, parietal cell is decreased, and the proportion of monocyte phagocyte is increased after treatment. Macrophages_2-5 and the proportion of Monocytes_1/3 are increased after treatment. The proportion of MastCells1 in BC_A group is increased too after treatment. The results of KEGG enrichment of differential genes between monocyte subtypes suggested that the up-regulated genes of monocyte were mainly enriched in the interaction between cytokine and cytokine receptor and P53 signaling pathway. It resulted in pro inflammation of the immune response in tumor microenvironment (TME), also the cytokines are the key proteins of signaling in the TME and have pleiotropic properties. In terms of anti-tumor, tumor necrosis factor (TNF) can specifically identify and kill tumor cells, [22] CXCL1, IL6 and other factors can inhibit cell proliferation and angiogenesis and induce cell apoptosis to delay tumor growth [23; 24]. We also performed estrogen receptor correlation analysis. The result has indicated that the expression levels of ESR1 and FOXA1 genes in EpithelialCells1/4/8 subtype in after treatment were decreased. It has been reported that overexpression of ESR1 and the consequent increase in components of the estrogen pathway contribute to a significant proportion of breast cancer in postmenopausal women [25]. This indicates that the tumor is under control. Estrogen receptor-alpha (ER) is a key feature of most breast cancers, and ER binding to the genome is associated with expression of the forkhead protein FOXA1 [26]. This indicates that the tumor is under control. Estrogen receptor-alpha (ER) is a key feature of most breast cancers, and ER binding to the genome is associated with expression of the fork head protein FOXA1 [27]. FOXC1 plays an important role in breast cancer and can be used as a potential marker for triple-negative breast cancer. Foxc1 is a major regulator of a series of genes involved in cancer cell metastasis, promoting cancer cell growth [28]. GATA3 is a transcription factor for the differentiation of luminal epithelial cells of the breast, the loss of GATA3 function directly leads to basal-like breast cancer, and basal-like breast cancer may also originate from luminal epithelial tumors [29]. The higher the Ki-67 index, the more active the cell division and proliferation, the faster the tumor growth rate, and the anti-proliferative drug chemotherapy effect is also better, so it is very important for the decision of adjuvant treatment in breast cancer surgery [30]. Overexpression of TRPS1 greatly improved the ability of tumor cells to grow and metastasize in xenografted mice [31]. Another result showed decrease expression of PGR (progesterone receptors) while higher the expression of BRCA2 after treatment. There is no significantly a change for these gene expressions of CDH1, CTNND1, ESR1, ERBB2 and PIP. We also noted this novel therapy caused an increase in fibroblasts. The result showed unsupervised clustering results and were divided into 5 heterogeneous subpopulations including Fibroblasts1-5. The increased proportion of Fibroblasts2/4 has the function of promoting epithelial remodeling and restoring tissue integrity [32]. Studies have reported that myofibroblasts play a role in promoting immunosuppressive microenvironment and are associated with poor tumor prognosis [33]. Fibroblasts2/4 is predisposed to overexpress multiple chemokines and interleukins (IL6, IL11, etc.) in inflammatory fibroblasts (iFIB) [34]. It does play an important role in the recruitment and regulation of immune cells. The clinicopathological immunohistochemical results after treatment were found to be consistent with the expression of these genes by the detection of scRNA-seq technology.
Clinical implications
UMIPIC has achieved significant clinical benefits in the treatment of cancer, local treatment can kill local breast cancer by combining hapten and chemotherapy drugs, can also induce an immune response against cancer, and control the expression of several breast cancer-related genes.
Strengths and Limitations for treat MBC with HEIC
Results from previous studies have shown that monotherapy has never been able to induce immune responses such as immunotherapy and control the expression of genes associated with breast cancer. An important limitation of the study is the sample size: only one patient's sample was analyzed, and we will seek additional funding support to expand our study to more patient samples.
Conclusion:
Our clinical study proves that hapten-mediated local chemotherapy is a safe and effective method to induce systemic immunity by initiating the immune response of male breast cancer lesions, and achieve the desired clinical effect.
References
- Stephen Fox, Valerie Speirs, Abeer M Shaaban. Male breast cancer: an update.Virchows Arch 480 (2022): 85-93.
- Ian S Fentiman. Male breast cancer is not congruent with the female disease. Crit Rev Oncol Hematol 101 (2016): 119-124.
- A P Lin, T-W Huang, K-W Tam. Treatment of male breast cancer: meta-analysis of real-world evidence. Br J Surg 108 (2021): 1034-1042.
- Vita M Golubovskaya, Nadia L Palma, Min Zheng. A small-molecule inhibitor, 5'-O-tritylthymidine, targets FAK and Mdm-2 interaction, and blocks breast and colon tumorigenesis in vivo. Anticancer Agents Med Chem 13 (2013): 532-545.
- Laleh Amiri-Kordestani, Antoinette R Tan, Sandra M Swain. Pazopanib for the treatment of breast cancer. Expert Opin Investig Drugs 21 (2012): 217-225.
- Florian Schütz, Stefan Stefanovic, Luisa Mayer. PD-1/PD-L1 Pathway in Breast Cancer. Oncol Res Treat 40 (2017): 294-297.
- Arielle L Heeke, Antoinette R Tan. Checkpoint inhibitor therapy for metastatic triple-negative breast cancer. Cancer Metastasis Rev 40 (2021): 537-547.
- Gao F, Jing P, Liu J, et al. Hapten-enhanced overall survival time in advanced hepatocellular carcinoma by ultro-minimum incision personalized intratumoral chemoimmunotherapy. J Hepatocell Carcinoma 2 (2015): 57-68.
- Ziegenhain C, Vieth B, Parekh S, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 65 (2017): 631-643.
- Si-Qing Liu, Zhi-Jie Gao, Juan Wu, et al. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J Hematol Oncol 15 (2022): 19.
- Baofa Yu, Qiang Fu, Yan, et al. An Acute Inflammation with Special Expression of CD11 & CD4 Produces Abscopal Effect by Intratumoral Injection Chemotherapy Drug with Hapten in Animal Model. J Immunological Sci 6 (2022), 1-9.
- Baofa Yu, Yan Han, Qiang Fu, et al. Awaken Immune Cells by Hapten Enhanced Intratumoral Chemotherapy with Penicillin Prolong Pancreatic Cancer Survival. Journal of Cancer 14 (2023): 1282-1292.
- Baofa Yu, Yan Han, Qiang Fu, et al. Awaken immune cells into Figurehting posture from the primary lung cancer by hapten enhanced chemotherapy: make ready for cancer immunotherapy. J Basic Clin Pharma 14 (2023): 10-16.
- Andrey Kechin, Uljana Boyarskikh, Alexander Kel, et al. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J Comput Biol 24 (2017): 1138-1143.
- Alexander Dobin, Carrie A Davis, Felix Schlesinger, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (2013): 15-21.
- Kechin A, Boyarskikh U, Kel A, et al. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J Comput Biol 24 (2017): 1138-1143.
- Dobin A, Davis CA, Schlesingeret F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (2013): 15-21.
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16 (2012): 284-287.
- Efremova M, Vento-Tormo M, Teichmann SA, et al. CellPhoneDB:inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 15 (2020): 1484-1506.
- Qiu X, Hill A, Packeret J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14 (2017): 309-315.
- Frontiers Editorial Office. Retraction: NEAT1 Overexpression Indicates a Poor Prognosis and Induces Chemotherapy Resistance via the miR-491-5p/ SOX3 Signaling Pathway in Ovarian Cancer. Front Genet 13 (2022): 861109.
- Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 119 (2012): 651-665.
- Miyake M, Furuya H, Onishi S. Monoclonal Antibody against CXCL1 (HL2401) as a Novel Agent in Suppressing IL6 Expression and Tumoral Growth. Theranostics 9 (2019): 853-867.
- Wang H, Lathia JD, Wu Q. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth.Stem Cells 27 (2009): 2393-2404.
- Furth PA, Wang W, Kang K. Overexpression of Estrogen Receptor α in Mammary Glands of Aging Mice Is Associated with a Proliferative Risk Signature and Generation of Estrogen Receptor α-Positive Mammary Adenocarcinomas. Am J Pathol 193 (2023): 103-120.
- Antoni Hurtado A, Holmes KA, Ross-Innes CS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43 (2011): 27-33.
- Witzel I, Loibl S, Wirtz R. Androgen receptor expression and response to chemotherapy in breast cancer patients treated in the neoadjuvant TECHNO and PREPARE trial. Br J Cancer 121 (2019): 1009-1015.
- Huang H, Hu JY, Maryam A. Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat Commun 12 (2021): 2242.
- Bai F, Zheng C, Liu X. Loss of function of GATA3 induces basal-like mammary tumors. Theranostics 12 (2022): 720-733.
- Maranta AF, Brode S, Fritzsche C. Do you know the Ki-67 index of your breast cancer patients? Knowledge of your institution's Ki-67 index distribution and its robustness is essential for decision-making in early breast cancer. Breast 51 (2020): 120-126.
- Yang J, Liu X, Huang Y. TRPS1 drives heterochromatic origin refiring and cancer genome evolution. Cell Rep 34 (2021): 108814.
- Hinz B, Lagares D. Evasion of apoptosis by myofibroblasts: a hallmark of fibrotic diseases. Nat Rev Rheumatol 16 (2020): 11-31.
- Elyada E, Bolisetty M, Laise P. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 9 (2019): 1102-1123.
- Chen Z, Zhou L, Liu L. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun 11 (2020): 5077.
