SARS-CoV-2 Nosocomial Outbreak in Campinas City, Brazil
Article Information
Daniel A Toledo-Teixeira1†, Luciana S Mofatto1†, Elisa T Mendes2,3†, Mariene R Amorim1, Priscilla P Barbosa1, Welbe O Bragança4, Pierina L Parise1, Karina Bispo-dos-Santos1, Luís G O Cardoso2, Luís F Bachur2, Christian C Hofling2, Gisele A Pedroso5, Kamila C S Krywacz5, José L R Cunha Jr5, Cintia H C Pilenso5, Maria Luiza Moretti6, Mariangela R Resende2, Renata Fagnani2, Eliane Molina2, Amanda T Ferreira2, Tiago Lima2, Rodrigo G Stabeli7, Angelica Z Schreiber5, Magnun N N Santos5, Fernando R Spilki8, Ester C Sabino9,10, Nuno R Faria10,11, William M de Souza12, Rodrigo N Angerami2,13, Fabiana Granja1,14, José Luiz Proenca-Modena1,15,16*
1Laboratory of Emerging Viruses (LEVE), Department of Genetics, Evolution, Microbiology and Immunology, Institute of Biology, University of Campinas (Unicamp), Campinas, Brazil
2Section of Hospital Epidemiology, Hospital of Clinics, University of Campinas (Unicamp), Campinas, Brazil
3Pontifical Catholic University of Campinas (PUC Campinas), Center for Life Sciences, Postgraduate Program in Health Sciences, Campinas, Brazil
4Laboratory of Genomic and Bioenergy (LGE), Department of Genetics, Evolution, Microbiology and Immunology, Institute of Biology, University of Campinas (Unicamp), Campinas, Brazil
5Department of Pathology, Clinical Pathology area, Faculty of Medical Sciences, University of Campinas (Unicamp), Campinas, Brazil
6Division of Infectious Diseases, Faculty of Medical Sciences, University of Campinas (Unicamp), Campinas, Brazil
7Oswaldo Cruz Foundation (Fiocruz), Ribeirão Preto, Brazil
8One Health Laboratory, Feevale University, Novo Hamburgo, Brazil
9Tropical Medicine Institute, Medical School, University of São Paulo (USP), São Paulo, Brazil
10Department of Infectious and Parasitic Disease, Medical School, University of São Paulo (USP), São Paulo, Brazil
11MRC Centre for Global Infectious Disease Analysis, J-IDEA, Imperial College London, London, UK
12World Reference Center for Emerging Viruses and Arboviruses, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, USA
13Campinas Department of Public Health Surveillance, Campinas, Brazil
14Biodiversity Research Center, Federal University of Roraima (UFRR), Boa Vista, Brazil
15Experimental Medicine Research Cluster, University of Campinas (Unicamp), Campinas, Brazil
16Hub of Global Health (HGH), University of Campinas (Unicamp), Campinas, Brazil
†Authors contributed equally to this work
*Corresponding author: José Luiz Proenca-Modena. Laboratory of Emerging Viruses (LEVE), Department of Genetics, Evolution, Microbiology and Immunology, Institute of Biology, University of Campinas (Unicamp), Campinas, Brazil.
Received: 23 June 2023; Accepted: 03 July 2023; Published: 27 July 2023
Citation: Toledo-Teixeira DA, Mofatto LS, Mendes ET, Amorim MR, Barbosa PP, Bragança WO, Parise PL, Bispo-dos-Santos K, Cardoso LGO, Bachur LF, Hofling CC, Pedroso GA, Krywacz KCS, Cunha Jr. JLR, Pilenso CHC, Moretti ML, Resende MR, Fagnani R, Molina E, Ferreira AT, Lima T, Stabeli RG, Schreiber AZ, Santos MNN, Spilki FR, Sabino EC, Faria NR, de Souza WM, Angerami RN, Granja F, Proenca-Modena JL. SARS-CoV-2 nosocomial outbreak in Campinas city, Brazil. Archives of Clinical and Biomedical Research. 7 (2023): 449-458.
View / Download Pdf Share at FacebookAbstract
Coronavirus disease (COVID-19) caused by Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a cause of concern in the hospital environment due to nosocomial transmission among healthcare workers (HCWs) and patients. During the first pandemic wave worldwide, the shortage of personal protective equipment, lack of guidance to manage COVID-19 patients and the rapid spread of the virus were great contributors to nosocomial infections. From May to June 2020 in a tertiary hospital in the city of Campinas, Brazil, we identified an increasing number of HCWs infected by SARS-CoV-2 in wards not intended for the care of COVID-19 patients, which lead us to strengthen nonpharmaceutical interventions to control the virus’s spread. Here we aimed to characterize the occurrence of a nosocomial outbreak in a tertiary reference hospital in Campinas city, Brazil. For this, we combined epidemiological data, universal diagnostic testing and genomic sequencing to define the cluster of infection. We found a proportional rate among total HCWs and infected HCWs, which patient care assistants are the most frequent infected HCWs. We have also shown that epidemiological data, universal diagnostic testing and genomic sequencing as a rapid response managed to characterize a cluster of nosocomial transmission, which allowed us to establish procedures in hospital wards. Our study reinforces the importance of epidemiological and genomic surveillance for the control of SARS-CoV-2’s nosocomial transmission in reference hospitals.
Keywords
Genomic surveillance; Health care worker; Nosocomial infection; Reference hospital
Genomic surveillance articles; Health care worker articles; Nosocomial infection articles; Reference hospital articles
Article Details
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection can cause the Coronavirus disease 2019 (COVID-19), which has been declared a pandemic in March 2020 [1]. Up to March 2022, more than 450 million cases and 6 million deaths occurred worldwide, with 29 million cases and 654 thousand deaths being reported in Brazil [2]. The transmission of SARS-CoV-2 occurs mainly by respiratory droplets, especially by pre-symptomatic carriers [3]. In a pandemic scenario, nonpharmaceutical interventions are important to restrict the virus’s spread being, to some degree, capable of reducing the number of cases and preventing the collapse of public health systems [4-6].
As previously described for other viral respiratory infections, healthcare workers (HCWs) were one of the groups with higher occupational exposure to SARS-CoV-2 [7,8]. Especially in the first wave of COVID-19 worldwide, due to nosocomial transmission, HCWs have been infected by SARS-CoV-2 either functioning as carriers or developing symptoms [9], when vaccines were not yet available. Previous studies have shown that HCWs are at high risk of infection by SARS-CoV-2 compared to the general population in several countries [10,11].
COVID-19 pandemic brought many challenges to health care systems worldwide [12]. In many Brazilian states, the health care system collapsed in 2020, and geographic and temporal fluctuations in COVID-19 in-hospital fatality rates were strongly and primarily associated with geographic inequities and shortages in healthcare capacity [13]. During the first pandemic wave in the world, the novel scenario for practice in patient care, often related to the lack of personal protective equipment (PPE), unavailability of vaccines, and even the absence of guidelines for nonpharmaceutical interventions may have contributed to the initial virus spread [9,14].
The increasing number of HCWs infected by SARS-CoV-2 emphasized the demand for new interventions concerning infection control and prevention in COVID-19 and non-COVID-19 wards [15]. Thus, this study aimed to investigate and track possible distinct and simultaneous hospital outbreaks of SARS-CoV-2 in an important tertiary hospital located in the city of Campinas, Brazil, during the first pandemic wave (from February to October 2020) using epidemiological and genomic data..
2. Material and Methods
2.1 Ethics: This study was approved by the University of Campinas Research Ethical Committee (CAAE 31170720.3.0000.5404). All patient data were anonymized before use, and patients gave informed consent before participation.
2.2 Study’s location: The Clinical Hospital of the University of Campinas (HC-Unicamp) is one of the largest university hospitals in Brazil, located in the city of Campinas. It is a reference for tertiary care and provides medical services for 86 municipalities in the Campinas Metropolitan Region, totalling 6.5 million inhabitants. Data for cases in Campinas city and the Campinas Metropolitan Region were obtained from Brazilian Public Health Care Data Centre (Open Data SUS) (https://opendatasus.saude.gov.br/dataset/srag-2020).
2.3 Sample collection and SARS-CoV-2 detection: Nasopharyngeal samples (combined samples from the nose and throat) were collected using swabs (similar to a long Q-Tip) from all individuals included in this study. All samples were sent to the Laboratory of Clinical Pathology of HC-Unicamp (LPC), where they were submitted to RNA extraction using ZR Viral RNA Kit (Zymo, USA), Extracta RNA Viral Kit MVXA-P096FAST (Loccus, Brazil) or QIAamp Viral RNA Mini Kit (Qiagen, USA), all following manufacturer's instructions. All extracted RNAs were submitted to the SARS-CoV-2 detection assay using the GeneFinder COVID-19 PLUS RealAmp Kit (OSANG Healthcare, South Korea) following the manufacturer’s instructions or RT-qPCR previous described elsewhere using TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems, EUA).
2.4 SARS-CoV-2 genome sequencing: For SARS-CoV-2 genome sequencing, a new RNA extraction was performed for all positive samples by RT-qPCR. The RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Germany), according to the manufacturer’s instructions. Synthesis of cDNA was performed with ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs, USA) with random hexamers (Invitrogen, USA), followed by tiling PCR reaction using V3 primer scheme for SARS-CoV-2 (https://github.com/artic-network/artic-ncov2019/tree/master/primer_schemes/nCoV-2019). Then, Q5 High-Fidelity DNA polymerase (New England Biolabs, USA) was used for amplification, as described before [4,16,17] (https://artic.network/ncov-2019). Clean-up steps were performed using AmpureXP purification beads (Beckman Coulter, USA) and dsDNA was quantified using Qubit 2.0 dsDNA High Sensitivity (Life Technologies, USA). The library was generated using SQK-LSK109 Kit (Oxford Nanopore Technologies, UK), EXP-NBD104 and EXP-NBD114, Native Barcoding Kits (Oxford Nanopore Technologies, UK), and loaded on R9.5.1 flowcells (Oxford Nanopore Technologies, UK). Fast5 files were processed for basecalling, trimming, and demultiplexing using Guppy 2.2.7 software, and Artic guppyplex. The consensus sequences were obtained using Artic Nanopolish pipeline [18].
2.5 Phylogenetic analysis: Representative sequences from Brazil, Italy, Spain, Portugal, the United Kingdom, and the United States of America during the period of this study (from May to July 2020) obtained from GISAID (n = 204, [Supplementary table 1]) and the consensus sequences from the present study with coverage greater than 70% and 20X depth (n = 50) were aligned using MAFFT [19]. UGENE software version 41.0 was used for trimming 5' and 3'-UTR from all sequences. Maximum-likelihood phylogenetic tree was constructed using IQ-TREE web server version 1.6.12 [20]. The best nucleotide model was determined by ModelFinder [21]. Further visualizations of newick files and metadata were performed in the Microreact webserver [22]. All sequences obtained in this study are available at GISAID (Supplementary table 2).
2.6 Data availability: All analyses were conducted using the R project for statistical computing [23]. R packages necessary for analysis and visualization include dplyr, tidyverse, lubridate, rstatix, and ggplot2. No custom code was developed.
3. Results
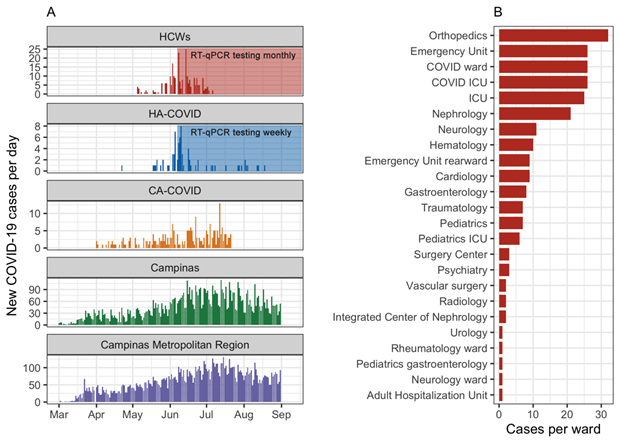
From May to July 2020, 325 people tested positive for SARS-CoV-2 and were enrolled in this study, including 237 HCWs (179 women, mean age of 39.3 years [IQR 31.0 – 45.0], 55 hospitalized patients which tested positive after admission (named as hospital-acquired COVID-19 patients: HA-COVID) (21 women, mean age of 57.3 years [IQR 45.7 – 65.5]) and 33 patients with acute respiratory distress syndrome as the reason for admission (named as community-acquired COVID-19 patients: CA-COVID) (16 women, mean age of 53.9 years [IQR 43.0 – 65.0]) (Figure 1A). On 8th June 2020, the Section of Hospital Epidemiology established universal testing by RT-qPCR in HCWs monthly and weekly for hospitalized patients, which could allow detecting SARS-CoV-2 in both pre-symptomatic and asymptomatic SARS-CoV-2 infected subjects (Figure 1A). During the first couple of weeks of universal testing, the number of HCWs and patients which tested positive had a sharp increase, followed by a slower decline over the next month (Figure 1A).
SARS-CoV-2 rapidly spread across Brazil in early 2020, and it was detected in Campinas city on 13th March 2020 (Open Data SUS), soon to be detected all across the Campinas Metropolitan Region, the second-largest metropolitan region in São Paulo state with 3 million inhabitants [24]. The cases peaked in June and July 2020, characterizing the first wave of circulation of this virus in this region (Figure 1A). During this first wave in Brazil (from February to October 2020), an increasing number of HCWs tested positive for SARS-CoV-2 from May to July 2020 in the HC-Unicamp (Figure 1A), and many of them worked in wards that were not primarily intended for the care and treatment of patients with COVID-19, such as orthopaedics, nephrology, and cardiology wards (Figure 1B).
Figure 1: Epidemiological context during this study and HCWs’ cases distribution in the hospital of study. (A) Epidemiological surveillance of new cases per day during the period of this study, from May to July 2020 in HCWs and in HA-COVID patients, CA-COVID patients, Campinas city and Campinas Metropolitan Region (Campinas city not included) from March to August 2020. The establishment of universal testing by RT-qPCR by the Infection Control Committee is indicated. (B) Distribution of infected HCWs per ward in this study. The establishment of universal testing for HCWs monthly and weekly for HA-COVID patients is indicated. Abbreviations: HCWs, health care workers; HA-COVID, hospital-acquired COVID-19 patients; CA-COVID, community-acquired COVID-19 patients; ICU, intensive care unit.
To characterize this outbreak in the hospital, it was evaluated occupational, clinical, and demographic characteristics of the RT-qPCR SARS-CoV-2 positive individuals, including HCWs, patients admitted to COVID-19 wards (CA-COVID), and patients who were RT-qPCR SARS-CoV-2 positive after being admitted to hospital areas not intended for the treatment of COVID-19 (HA-COVID).
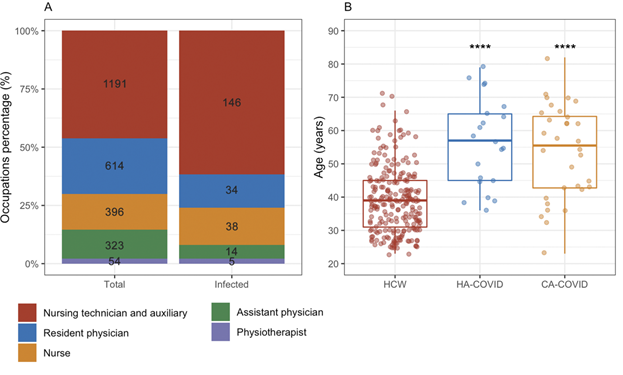
Among all infected HCWs enrolled in this study, the occupation with higher proportion of RT-qPCR positive tests was patient care assistants such as nursing technicians and auxiliaries with 146 individuals, followed by nurses and physicians with 38 and 34 individuals, respectively (Figure 2A). Only 2 HCWs needed hospitalization after the diagnosis of COVID-19 by RT-qPCR (data not shown). In addition, the infected HCWs were younger (mean age of 39.3 years [IQR 31.0 – 45.0]) than HA-COVID (mean age of 57.3 years [IQR 45.7 – 65.5]) and CA-COVID patients (mean age of 53.9 years [IQR 43.0 – 65.0]) (p < 0.001, ANOVA followed by Tukey post-hoc test) (Figure 2B).
Figure 2: HCWs distribution in the hospital and age of study groups. (A) Relative percentage of total and infected HCWs per occupation in the hospital with their absolute numbers in each stack bar. (B) Age distribution of HCWs, HA-COVID patients and CA-COVID patients in this study. Boxplots represent the median, IQR, maximum and minimum whiskers. Statistical significance by ANOVA followed by Tukey post-hoc test comparing to HCW group are represented by asterisks, ****p<0.001. Abbreviations: HA-COVID, hospital-acquired COVID-19 patients; CA-COVID, community-acquired COVID-19 patients.
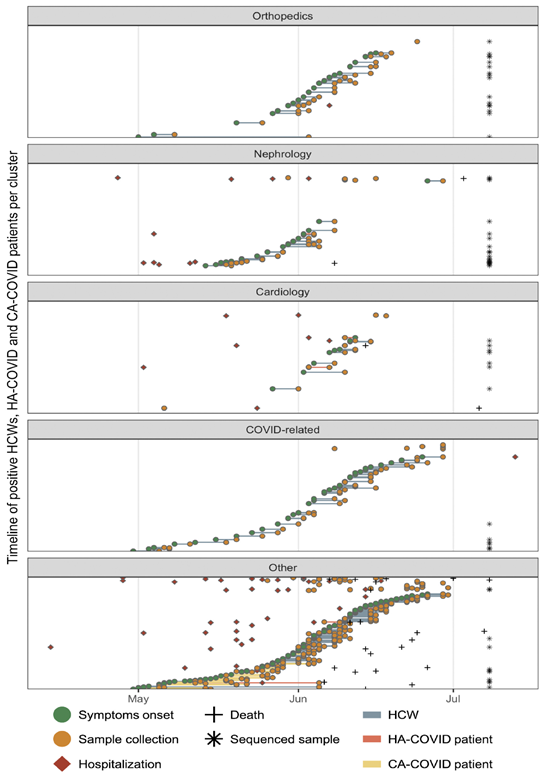
To track the epidemiological link between all infected persons (HCWs, HA-COVID, and CA-COVID patients) we merged several data, such as the onset of symptoms, date of sample collection for molecular diagnosis by RT-qPCR, and date of hospitalization according to the timeline in orthopaedics, nephrology and cardiology wards, and COVID-related wards. We identified a timeframe of events that suggests a potential cluster of SARS-CoV-2 nosocomial transmission between HCWs and areas unrelated to COVID-19 patients, starting in early May 2020 until early July 2020 (Figure 3). Other wards also had diagnosed HCWs during the period of this study (May to July 2020) such as neurology (n = 1 HCW), rheumatology (n = 1 HCW), surgery centre (n = 3 HCWs), paediatrics (n = 7 HCWs), and emergency and intensive care units (ICU) (n = 1 HCW) and others (Figure 1B). However, since the number of RT-qPCR positive HCWs in these facilities was small compared to orthopaedics, nephrology, cardiology and COVID-related wards, we did not perform further investigations in these wards.
Figure 3: Timeline of epidemiological data of HCWs, HA-COVID and CA-COVID patients enrolled in this study grouped by wards. Green circles represent the onset of symptoms, orange circles represent sample collection for molecular diagnosis by RT-qPCR, red diamonds represent hospitalization, crosses represent the day of death, asterisks represent sequenced sample (asterisks positions are not related to the X-axis), line colours connecting onset of symptoms and sample collection represent the study group of HCWs (grey line), HA-COVID (red line) and CA-COVID patients (yellow line). Abbreviations: HCWs, health care workers; HA-COVID, hospital-acquired COVID-19 patients; CA-COVID, community-acquired COVID-19 patients.
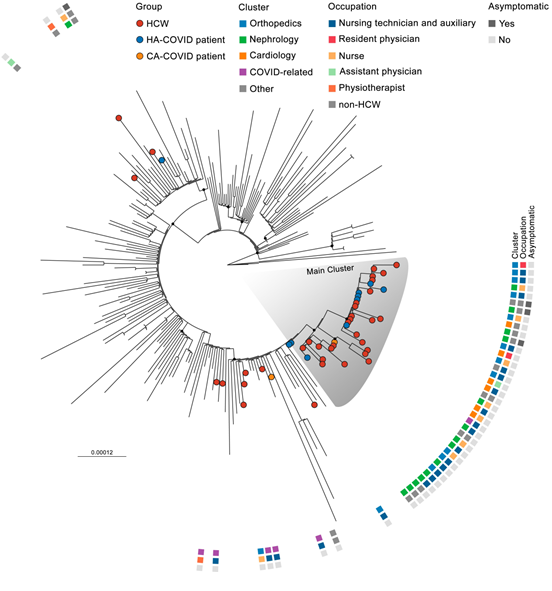
Next, to confirm the nosocomial outbreak of SARS-CoV-2 in HCWs and patients admitted in areas non-COVID-19 wards, we performed SARS-CoV-2 genome sequencing of 36 HCWs samples, 10 HA-COVID patients, and 4 CA-COVID patients. Phylogenetic analysis showed the main cluster of SARS-CoV-2 infections among HCWs and HA-COVID patients (bootstrap value > 70), indicating a possible single origin of nosocomial transmission (Figure 4). The main cluster was composed of 26 HCWs, 8 HA-COVID patients and 1 CA-COVID patient. The 8 HA-COVID patients, who were hospitalized before the molecular diagnosis of COVID-19 by RT-qPCR, ranged from 1 to 59 days before testing and included 3 asymptomatic subjects. Some HCWs’ and patients’ sequences were phylogenetically distant from the main cluster, suggesting a community-acquired infection (Figure 4, Supplementary Tables 2,3).
We sequenced SARS-CoV-2 genome samples obtained from two respiratory specialist physiotherapists, who were potentially highly exposed during the care of patients with various respiratory conditions, including those with severe acute respiratory syndrome, and probably circulating around different hospital wards. Nevertheless, the phylogenetic analysis shows the SARS-CoV-2 sequences from these two physiotherapists were placed into separated clusters, indicating a probable community-acquired infection (Figure 4). In the main cluster, the orthopaedics ward did not host any patients in the main cluster but hosted 11 HCWs including nursing technicians, nurses, and a resident physician. The nephrology ward hosted 8 HCWs, such as nursing technicians, nurses, and physicians, and 5 HA-COVID patients. The cardiology ward hosted 5 HCWs, also nursing technicians, nurses, resident and assistant physicians, and 1 HA-COVID patient in the main cluster (Figure 4). Samples obtained from other wards also had sequences placed into the main cluster, such as the traumatology ward with 1 HCW and 2 HA-COVID patients, and a COVID-related ward with 1 CA-COVID patient. Interestingly, the branch where the CA-COVID patient from the main cluster was placed was also composed of 3 HCWs, each of them from nephrology, cardiology and COVID-related wards. These 3 HCWs showed onset of symptoms ranging from 6 to 10 days after the hospitalization of the CA-COVID patient present in the main cluster (Figure 4).
Figure 4: Genomic characterization of health care workers (HCWs), hospital-acquired (HA-COVID) and community-acquired COVID-19 patients (CA-COVID). Maximum-likelihood tree of sequences of this study (n = 50) and representative GISAID sequences (n=204) from May to July 2020. ML tree was constructed using IQ-TREE version 1.6.12 and alignment of the near-complete genome (without untranslated regions) with the GTR+F+I substitution model and 1000 ultrafast bootstrap replicates. Terminal leaf nodes represent sequenced samples in this study coloured by group. Metadata is indicated by coloured squares beside terminal leaves. Black dots indicate the main nodes with ML bootstrap values > 70% based on 1000 bootstrap replicates. The scale bar indicates the evolutionary distance in numbers of substitutions per nucleotide site.
4. Discussion
During the first wave of the current pandemic of SARS-CoV-2 from March to July 2020, immediate concerns about the availability of PPE for HCWs, the lack of guidelines for infection prevention control, the absence of a vaccine, and the management of COVID-19-positive patients worldwide [9] were raised. The first case of COVID-19 in Latin America was confirmed on 26 February 2020 in the São Paulo metropolis, the most populous city in the Southern hemisphere with around 12 million people [25]. Due to at least 100 introduction events, SARS-CoV-2 spread in São Paulo city and other large cities by airline travellers coming from Italy, USA, France, and other European countries during the beginning of 2020 [4,26]. Many HCWs were infected and became symptomatic, but a proportion has played as asymptomatic carriers and were potential virus disseminators for person-to-person contamination [9,27]. In addition, the transmission between COVID and non-COVID wards has already been described in hospital environments [9,14,28].
The epidemiological monitoring and investigation of the potential link between infected persons combined with viral genomic sequencing can be a critical approach to define real outbreaks, their extent and magnitude, and propose estimable strategies to control viral dissemination [29,30]. In this study, we combined epidemiological and genomic data from HCWs and SARS-CoV-2-infected patients to characterize a cluster of nosocomial transmission in a tertiary hospital in the Campinas Metropolitan Region, Brazil. Our results suggest the occurrence of a nosocomial outbreak, supported by the timeframe of onset of symptoms and RT-qPCR positive tests from HCWs and patients together with genomically-related samples from both HCW and patients among COVID-19 and non-COVID-19 wards. Previous studies described a nosocomial outbreak in a gastroenterology ward, not related to COVID-19 areas [9], and a hospital outbreak caused by symptomatic and asymptomatic HCWs with contact with patients in different wards, highlighting the importance of a cooperative effort for contact tracing, massive testing and the implementation of nonpharmaceutical interventions [14,31,32].
The tracking of COVID-19-positive HCWs and patients in our study was used to guide procedures in the first pandemic wave to prevent further virus dissemination in nosocomial settings, since in that period from May to July 2020, none of the HCWs enrolled had been vaccinated. A retrospective study conducted in a teaching hospital in London showed that 15% of COVID-19 inpatient cases were characterized as definite or probable hospital-acquired infections with possible patient-to-patient contacts [28]. A nosocomial outbreak in Wuhan also demonstrated the high risk of hospital-acquired infection among people with or without symptoms [33]. Here we show that all patients involved in the outbreak were already hospitalized when the infection probably occurred.
In this study, we identified one cluster of subjects with SARS-CoV-2 infection in the hospital, which involved different hospital facilities, such as COVID-19-related wards, orthopaedics, nephrology, cardiology wards, and HCWs from different roles. A study has shown that nursing technicians and nurses are the main frontline HCWs that are more at risk of viral exposure [34]. Our results show a proportional rate between the total number and infected HCWs by occupations, and also suggest that nursing technicians and nurses could be the main carriers and drivers of SARS-CoV-2 transmission during this outbreak, as already observed for HCWs-to-HCWs and HCWs-to-patients transmissions [15,35]. In addition, we identified the sequence from a CA-COVID patient and three HCWs’ sequences related in a single branch, it is plausible to infer that they could be the first introductory nosocomial infections and it suggests they were potential carriers to other wards.During the first wave of COVID-19, the lack of guidance and infrastructure to handle the high number of infected patients was associated with HCWs contamination inside hospitals worldwide and also in Brazil [13,36]. In the cluster observed in this study, we identified genomically-related SARS-CoV-2 samples in different hospital facilities not related to COVID-19, which drew attention to track cases in these wards to rapidly implement changes in routine procedures and human resources contingency. Since the outbreak was identified, response measures such as contact tracing, SARS-CoV-2 testing for asymptomatic HCWs and on admission and weekly for hospitalized patients were readily implemented. The prompt identification of asymptomatic carriers supported the isolation of positive patients, HCW withdrawal from duty, and quarantine for contacts. After identifying a SARS-CoV-2 case a contact tracing was performed and every HCW who had contact with the confirmed case was also tested by RT-qPCR.
Our study reinforces the importance of integrated epidemiological and genomic surveillance in large hospitals to better understand outbreak patterns and distinct transmission chains as tools for timely and effective implementation of infection prevention and control measures and to support the development of guidelines to tackle SARS-CoV-2 and other respiratory viruses. These tools and developments can be used to minimize the transmission of respiratory viruses, contributing to improve safe work conditions for HCWs whilst minimizing the risk of infection for hospitalized patients.
Author contributions
Conceptualization, DATT, LSM, ETM, MRA, RNA and JLPM; Methodology, DATT, LSM, ETM, MRA, PPB, WB, PLP, KBDS and FG; Investigation, all authors; Original Draft, DATT, LSM, ETM, WMS, RNA and JLPM; Review & Editing, all authors. Visualization, DATT, LSM, ETM, MRA, WMS, RNA and JLPM; Funding Acquisition, FRS, ECS, NRF, WMS and JLPM; Resources, FRS, ECS, NRF, WMS and JLPM; Supervision, WMS, RNA, FG and JLPM.
Acknowledgments
We thank the Unicamp-Task-Force against COVID-19 that facilitated this study, Su Yan Ling, Tereza Cristina Faustino, Eliete Boaventura Bargas Zeferino, Michele Neves, Nanci Michele Saita for their support in obtaining clinical and epidemiological information related to the outbreak and for their investigation and operational actions. Lastly, the Brazilian Ministry of Science, Technology, and Innovation (MCTI) and all members of the Corona-ômica network are thanked for their support.
Declaration of conflicting interests
The authors declare no conflict of interest.
Ethical approval
University of Campinas Research Ethical Committee (CAAE 31170720.3.0000.5404)
Funding statement
This study was supported by grants from São Paulo Research Foundation (FAPESP) [grant numbers 2016/00194-8, 2020/04558-0 and 2018/14372-0], Fundo de Apoio ao Ensino, Pesquisa e Extensão from Unicamp (FAEPEX-Unicamp) [grant number 2266/20]. This study was also supported by MCTI through the Rede Corona-ômica Brazil/MCTI funded by the Financier of Studies and Projects (FINEP) [grant number 01.20.0003.00], RedeVírus/MCTI [grant number #01.20.0029.000462/20], and the Brazilian National Council for Scientific and Technological Development (CNPq) [grant number 404096/2020-4). FAPESP also supported KBDS [grant number 2020/02159-0] and PLP [grant number 2017/26908-0]. CNPq also supported JLPM [grant number #305628/2020-8], WMS [grant number 408338/2018-0], DATT [grant number 141844/2019-1], and LSM [grant number 382206/2020-7]. WMS is supported by the Global Virus Network fellowship. The Coordination for the Improvement of Higher Education Personnel (CAPES) supported MRA [grant number 88887.356527/2019-00], and PPB [grant number 88887.661921/2022-00]. NRF was supported by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship [grant number 204311/Z/16/Z]. This project was supported by the Medical Research Council and FAPESP-Brazil-UK Center for (Arbo) virus Discovery, Diagnosis, Genomics, and Epidemiology partnership award [grant number MR/S0195/1] and FAPESP [grant number 2018/14389-0].
References
- World Health Organization Director-General’s opening remarks at the media briefing on COVID-19 (2020).
- World Health Organization Coronavirus (COVID-19) Dashboard.
- Ganyani T. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data. Eurosurveillance 25 (2020): 7201952.
- Candido DS. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 369 (2020): 1255-1260.
- Lai S. Effect of non-pharmaceutical interventions to contain COVID-19 in China. Nature 585 (2020): 410-413.
- De Souza WM. Epidemiological and clinical characteristics of the COVID-19 epidemic in Brazil. Nature Human Behaviour 4 (2020): 856-865.
- Amorim MR. Respiratory Viral Shedding in Healthcare Workers Reinfected with SARS-CoV-2, Brazil, 2020. Emerg Infect Dis 27 (2021): 1737-1740.
- Yang JY. Outcomes of COVID-19 among hospitalized health care workers in North America. JAMA network open 4 (2021): 2035699.
- Pérez-Lago L. Overlapping of Independent SARS-CoV-2 Nosocomial Transmissions in a Complex Outbreak. Msphere 6 (2021): 0038921.
- Huff HV, Singh A. Asymptomatic transmission during the coronavirus disease 2019 pandemic and implications for public health strategies. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 71 (2020): 2752-2756.
- Nguyen LH. Risk of COVID-19 among front-line health-care workers and the general community: a prospective cohort study. The Lancet Public Health 5 (2020): e475-e483.
- World Health Organization COVID-19 continues to disrupt essential health services in 90% of countries (2021).
- Brizzi A. Spatial and temporal fluctuations in COVID-19 fatality rates in Brazilian hospitals. Nature medicine 28 (2022): 1476-1485.
- Lucey M. Whole-genome sequencing to track Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) transmission in nosocomial outbreaks. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 72 (2021): 727-735.
- Paltansing S. Transmission of SARS-CoV-2 among healthcare workers and patients in a teaching hospital in the Netherlands confirmed by whole-genome sequencing. The Journal of hospital infection 110 (2021): 178-183.
- Quick J. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols 12 (2017): 1261-1276.
- Faria NR. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science (New York, NY) 372 (2021): 815-821.
- Loman N. nCoV-2019 novel coronavirus bioinformatics protocol. Nanopore bioinformatics (2020).
- Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research 30 (2002): 3059-3066.
- Nguyen LT. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution 32 (2015): 268-274.
- Kalyaanamoorthy S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature methods 14 (2017): 587-589.
- Argimón S. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microbial Genomics 2 (2016): 000093.
- R Core Team. R: A language and environment for statistical computing 8 (2021).
- Regional Development Secretariat S of DR. Campinas Metropolitan Region (RMC) (2018).
- Resident population estimates with reference date July 1 (2020).
- Candido DDS. Routes for COVID-19 importation in Brazil. Journal of Travel Medicine 27 (2020): 1-3.
- Sikkema RS. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. The Lancet Infectious diseases 20 (2020): 1273-1280.
- Rickman HM. Nosocomial Transmission of Coronavirus Disease 2019: A retrospective study of 66 hospital-acquired cases in a London Teaching Hospital. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 72 (2021): 690-693.
- Meredith LW. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. The Lancet Infectious diseases 20 (2020): 1263-1272.
- Snell LB. Rapid genome sequencing in hospitals to identify potential vaccine-escape SARS-CoV-2 variants. The Lancet Infectious Diseases 21 (2021): 1351-1352.
- De Souza Santos AA. Dataset on SARS-CoV-2 non-pharmaceutical interventions in Brazilian municipalities. Scientific data 8 (2021): 73.
- Li A. SARS-CoV-2 Positivity and mask utilization among health care workers. JAMA Network Open 4 (2021): 2114325.
- Wang D. Clinical Characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323 (2020): 1061-1069.
- White EM. Front-line Nursing home staff experiences during the COVID-19 Pandemic. Journal of the American Medical Directors Association 22 (2021): 199-203.
- Zhao D. Asymptomatic infection by SARS-CoV-2 in healthcare workers: A study in a large teaching hospital in Wuhan, China. International journal of infectious diseases: IJID: official publication of the International society for infectious diseases 99 (2020): 219-225.
- Abbas M. Nosocomial transmission and outbreaks of coronavirus disease 2019: the need to protect both patients and healthcare workers. Antimicrobial Resistance & Infection Control 10 (2021): 7787623.