Role of Inflammasome-Mediated Pyroptosis as a Mechanism of Pathogenicity in Pseudomonas Biofilms
Article Information
Keerthi S1, Vishnu Raj M2, and Maya Nandkumar A1,*
1Div. of Microbial Technology, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Science and Technology, Thiruvananthapuram - 12, Kerala, India
2Div. of Molecular Medicine, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Science and Technology, Thiruvananthapuram - 12, Kerala, India
*Corresponding author: Maya Nandkumar A. Div. of Microbial Technology, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Science and Technology, Thiruvananthapuram - 12, Kerala, India
Received: 15 October 2021; Accepted: 02 November 2021; Published: 14 January 2022
Citation:
Keerthi S, Vishnu Raj M, and Maya Nandkumar A. Role of Inflammasome-Mediated Pyroptosis as a Mechanism of Pathogenicity in Pseudomonas Biofilms. Archives of Microbiology and Immunology 6 (2022): 20-38.
View / Download Pdf Share at FacebookAbstract
Most of the studied mechanisms of pathogenesis hold true for planktonic bacterial infections, while pathogenesis of biofilm is less well understood. Hence, in the current study, we delineate the mechanism of pathogenicity of Pseudomonas biofilms. The model systems used to understand cell-microbial interactions were A549 monolayers representing lung epithelium, THP1 cells representing the immune system, and a combination of both, representing a multicellular system. Our study shows that biofilm of Pseudomonas on endotracheal tube (ETT) reduced reactive oxygen species generation in A549 and THP1. Assessment of morphological changes during infection revealed features of both apoptosis and necrosis in all systems challenged with biofilms of Pseudomonas. Biofilm infection in A549, THP1, and their co-cultures showed increased expression of tumour necrosis factor α (TNFα) but decreased interleukin 8 (IL8) and IL6 expression and induced nitric oxide synthase (iNOS) expression. A reduction in IL8, IL6 and iNOS was observed in all the three model systems challenged with Pseudomonas biofilms on ETT, with a concomitant observation of DNA fragmentation, within an intact nuclear membrane. Taken together, our data suggest that biofilms induce inflammasome activation, as evidenced by the increase in casapase 1 and IL-1β upregulation; downregulation of caspase 3 rules out apoptosis as a mechanism of pathogenicity. This provides an insight into the mechanism of chronic inflammation during biofilm infection.
Keywords
Pseudomonas biofilm, Inflammasome, Pyroptosis, Pathogenicity, cytokine expression, Intubation, Endotracheal tube, COVID-19
Article Details
1. Introduction
Multiple studies have reported the development of VAP among ventilated COVID patients, with Pseudomonas being one of the most commonly isolated organisms [1-3]. Endotracheal tube (ETT) provides respiratory support in mechanically ventilated patients [4]. Despite sterile procedures, the ETT gets colonized by bacteria, leading to biofilm formation and VAP [5]. Pseudomonas is known to form biofilms on ETT, which is difficult to eradicate due to recalcitrance to antibiotics and host immune response [6]. However, the mechanism of biofilm-induced lung injury by Pseudomonas remains elusive.
Host response to Pseudomonas involves a complex interplay between the innate and adaptive immune responses [7]. Both the arms of the immune system are activated in biofilm infections causing increased host tissue damage [8]. Most of the studied mechanisms of pathogenesis hold true for planktonic infections, while pathogenesis of biofilm is less well understood. The interaction of Pseudomonas biofilms with neutrophils and macrophages in cystic fibrosis has been extensively studied [9, 10]. Epithelium is the first line of defense in any organ or organ system. The immune responses elicited in the epithelium dictates the type of immune response. Damaged or disrupted epithelium in intubated patients increased the risk of P. aeruginosa infections [7]. Colonization of ETT by pathogenic organisms such as Pseudomonas and Staphylococcus aureus results in the development of VAP [11].
Inflammasome activation plays a critical role in the clearance of pathogen and infected cells. Inflammasome activation is marked by the cleavage of pro-Caspase 1 to active caspase 1 and increased expression of interleukin 1β (IL1 β) and IL18 [12]. This result in a mechanism of cell death called pyroptosis. Pyroptotic cell death is known to play a key role in chronic inflammatory diseases, such as arthritis [13]. This study provides insights into immune evasion strategies adopted by biofilms and the role of inflammasome-mediated pyroptosis in the pathogenesis of Pseudomonas biofilms.
2. Methodology
2.1 Biofilm Characterization of Pseudomonas Strains
2.1.1 Bacterial Isolates and Culture Conditions
aeruginosa ATCC 27853, was obtained from the American Type Culture Collection. The clinical isolates S373, S402, S125, S253, S135, S268, S45, S277, and S18, sourced from bronchiectasis patients, was a kind gift from Dr. Kavita Raja, Department of Microbiology, Sree Chitra Tirunal Institute for Medical Science and Technology, Thiruvananthapuram, Kerala, India. The isolates were cryopreserved (-80°C) in soyabean casein digest medium (TSB) containing 15% glycerol as cryoprotectant. The cultures were maintained in soyabean casein digest agar (TSA) slants and stored at 4°C. For all experiments overnight cultures of Pseudomonas adjusted to OD 1.0 inTSB was used.
2.1.2 Evaluation of Biofilm Formation
The ability of Pseudomonas isolates to develop biofilms over a period of 24 h, 48 h and 72 h was assessed by Crystal Violet (CV) assay as described by O’Toole [14]. Absorbance was measured at 600nm using Synergy H1 multimode reader (BioTek USA). Uninoculated culture medium was used as negative control.
2.1.3 Evaluation of Biofilms on ETT
The ability of Pseudomonas to colonize the ETT (Rusch®) was assessed. 1 cm pieces of ETT sterilised with ethylene oxide (EtO) at 37°C were used. Overnight culture of Pseudomonas adjusted to OD 1.0 was the inoculum. The biofilms developed on ETT pieces were evaluated by 1% crystal violet and 0.0025% acridine orange staining for 15 min. Biofilms were visualized and imaged using Leica DMR microscope (Leica Microsystems Germany).
2.1.4 Estimation of Pyocyanin
Pyocyanin was estimated from acidified solution of culture supernatant following the method of Essar [15]. After 48 h of incubation at 37 °C , to 1 ml of the culture supernatant, 450 µl of chloroform and 150 µl 0.2 M HCl was added, vortexed and centrifuged at 10,000g. The absorbance of the pink-coloured upper aqueous layer was measured at 520 nm using Synergy H1 multimode reader (BioTek USA).
2.1.5 Estimation of Rhamnolipids
Rhamnolipids were quantified by the method described by Rasamiravaka [16]. The pH of the culture supernatant was adjusted to 2.3 using 1 N HCl. Ramnolipid extraction from 500 µL supernatant was done using 500 µL of ethyl acetate. The extraction was repeated thrice and supernatants pooled, centrifuged at 10,000 rpm for 10 min and air dried to remove ethyl acetate. The air-dried samples were mixed with 1 mL of chloroform containing 100 µL of methylene blue and incubated at 25°C to 28°C for 15 min. 0.2 M HCl was added to the mixture, vortexed, and centrifuged at 10,000 rpm for 10 min. The absorbance of the aqueous phase was measured at 630 nm using Synergy H1 multimode reader (BioTek USA).
2.1.6 Estimation of Proteases
Pseudomonas proteases were estimated in culture supernatants of biofilms by the method described by El-Mowafy [17]. Briefly, 500 µl of cell-free supernatant was added to 1 mL of 1.25% w/v skimmed milk solution and incubated at 37°C for 30 min; the resultant turbidity was measured at 600 nm using Synergy H1 multimode reader (BioTek USA).
2.1.7 Caenorhabditis elegans Killing Assay
Virulence of Pseudomonas isolates was assessed by the C. elegans killing assay as described by Tan, Mahajan-Miklos and Ausubel [18]. Briefly, overnight culture was spread plated on nematode growth medium (NGM) and incubated overnight. Following incubation, 30-40 L4 stage nematodes were added on to each plate and incubated at 25°C. Dead nematodes were counted at 24 h and 48 h post infection. The nematodes that did not respond to touch were considered dead. The nematodes that got stuck at the wall of the plate was excluded from the study. The negative control used in the study was E.coli OP50.
2.2 Understanding cell-microbial interactions
2.2.1 Cell lines and Culture Conditions
Human adenocarcinoma cell lines were purchased from ATCC and were used to represent alveolar epithelial cells. To simulate in-vivo conditions A549 cells were cultured at air-liquid interface using cell culture inserts, using F12K supplemented with 10% FBS (Gibco). THP1 cells (ATCC) were used to represent cells of the immune system. Cells were cultured in RPMI1640 supplemented with 10% FBS at 37°C. To study cell - cell interaction A549:THP1 cells were used in the ratio 105:103.
To understand the effect of planktonic bacterial infection on cells - overnight culture of Pseudomonas was diluted to give a multiplicity of infection (MOI) 100:1and was used to infect A549 cells, THP1 cells and co-cultures. To understand biofilm interaction with A549 cells, THP1 cells and co-cultures Pseudomonas biofilms on ETT tube were used.
2.2.2 Morphological Assessment
Morphological changes in model systems, infected with planktonic and biofilm forms of Pseudomonas, were assessed using 0.5% acridine orange solution in PBS and 3% solution of propidium iodide in PBS after fixing with 4% paraformaldehyde. Changes in nuclear morphology were assessed using Hoechst 33342 stain. Leica DMR fluorescent microscope was used for imaging.
2.2.3 Evaluation of Oxidative Stress
The oxidative stress in Pseudomonas infected cells was measured by 2', 7’-dichlorofluorescin diacetate (DCFDA) method. A549 monolayers were developed in 96-well plates. Upon confluence, the cells were treated with 25 µM DCFDA and incubated at 37°C for 30 min. The cells were then washed with PBS. Overnight culture of Pseudomonas adjusted to an of OD 1.0 was used to infect cells at a multiplicity of infection of 100:1 (planktonic forms). Pseudomonas 48 hour biofilms developed on ETT pieces was placed on A549 monolayers and reactive oxygen species (ROS) generation in response to biofilms was measured at 485 nm/535 nm in Synergy H1 plate reader.
2.2.4 Quantitative real-time PCR
Expression of caspase 1, caspase 3, IL-1 β, IL8, and TNFα in A549 cells exposed to planktonic forms and biofilm forms of Pseudomonas was assessed by real-time PCR. β -actin was the housekeeping gene. RNA was extracted from the infected cells using Trizol®. Relative gene expression was analysed by ΔΔCt method.
2.2.5 Quantification of cytokines produced by enzyme-linked immunosorbent assay (ELISA)
The cytokine protein expression was quantified using ELISA. Prior to infection, the cell culture medium was replaced with serum-free medium without antibiotics. The medium was removed 2 h and 6 h post-infection and centrifuged at 1000 ×g for 20 min. The supernatant was used for protein estimation by Bradford method. Equal amount of protein was used for ELISA. The ELISA Kits for TNFα, IL8 and IL1β was purchased from Origin Diagnostics. ELISA was performed as per the kit protocol.
2.2.6 Statistical Analysis
The results were analysed by Student’s T test and 2-way analysis of variance (ANOVA) using Graph pad Prism 6. For all analyses p<0.05 was considered statistically significant.
3. Results
3.1 Biofilm Characterization of Pseudomonas Isolates
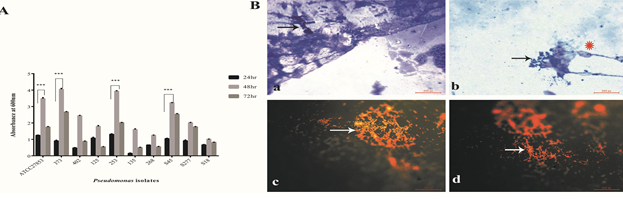
Initially, potential biofilm producers that were identified by crystal violet assay were used as candidates for developing biofilms on ETT. Both ATCC 27853 and clinical isolates of Pseudomonas showed increased biofilm formation at 48 h compared with 24 h. Biofilm formation reduced at 72 h (Fig.1A). The isolates differed in their ability to develop biofilms. A significant increase in biofilm formation was observed in ATCC 27853, S373, S253, and S45 isolates. S268, S125, and S18 showed reduced biofilm formation at 48 h.
Figure 1: Biofilm characterisation of Pseudomonas isolates (A) Quantification of biofilm formation by Pseudomonas isolates by crystal violet assay. (B) Morphological analysis of biofilms developed on ETT. (Ba and Bb) Crystal violet stained images of Pseudomonas biofilms (48hr) on ETT (Magnification 1000x), Bacterial aggregates (White arrow heads) within exopolysaccharide fibers (indicated by white Δ) was observed. Bc and Bd - acridine orange (AO) stained biofilms of Pseudomonas on ETT (Magnification 1000x). AO stained bacteria within the biofilms orange. Bacterial aggregates were observed (White hollow arrows). Statistical analysis was carried out by students T test, p<0.01 was considered significant.
Pseudomonas biofilms developed on ETT at 48 h were evaluated using crystal violet staining. Fig 1Ba and Bb show small aggregates of Pseudomonas within the exo-polysaccharide matrix fibers (Fig Ba and Bb). Thin fibers of exopolysaccharide matrix were observed on ETT biofilms (Fig 1B). Crystal violet stained both Pseudomonas and the exopolysaccharide matrix. Acridine orange staining revealed large bacterial aggregates (Fig 1Bc and Bd; see white arrows). The bacteria appeared as yellowish orange fluorescent rod-shaped bodies. The exopolysaccharide matrix fibers were not visualized by acridine orange staining.
3.2 S373, S253, and S45 Showed Increased Expression of Virulence Factors
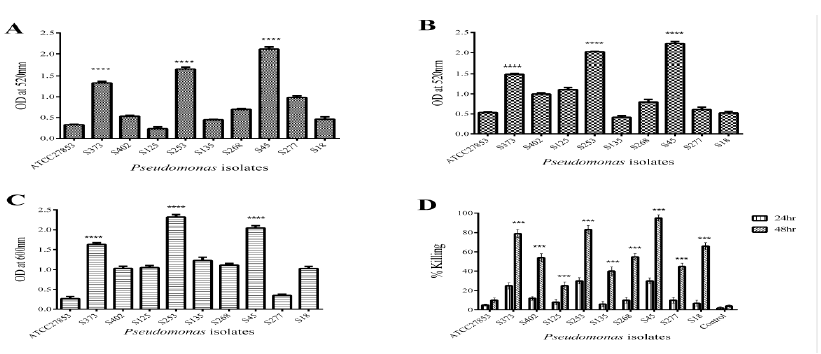
Pyocyanin is one of the major virulence factors produced by Pseudomonas with multiple roles in pathogenicity. Cytotoxic effect of pyocyanin is through the disruption of mitochondrial membrane potential and inhibition of protein synthesis. It is also known to regulate biofilm formation in Pseudomonas. Pyocyanin production in biofilms of ATCC 27853 and clinical isolates of Pseudomonas at 48 h was assessed by acidified chloroform method. It was observed that the ability to produce pyocyanin was isolate specific (Fig 2A). Biofilms of clinical isolates S373, S253, and S45 showed increased expression of pyocyanin in comparison to ATCC 27853 that showed reduced pyocyanin production (Fig 2A).
Rhamnolipids are virulence factors secreted by Pseudomonas and play an important role in early invasion of the airway epithelium. Rhamnolipids help to maintain the biofilm architecture. It is also involved in bacterial adhesion to surfaces and microcolony formation. Figure 2B indicates that rhamnolipid production was also strain specific. Except S135, rest of the clinical isolates - S373, S253, and S45 showed increased rhamnolipid production compared with ATCC 27853 isolate. The clinical isolate S135 produced the least amount of rhamnolipids among all tested isolates (Fig 2B).
Proteases play a key role in initiating and controlling tissue invasion by Pseudomonas. Proteases also have an important role in biofilm dispersion and exopolysaccharide production. Protease activity showed a clear isolate-specific variation in protease expression (Fig 2C). S373, S253, and S45 showed increased expression of proteases compared with the ATCC 27853 strain and the other isolates. Nematode killing was assessed at 24 h and 48 h post infection. Fig. 2D, depicts the percentage killing of C elegans by ATCC 27853 and clinical isolates of Pseudomonas. It was evident from the graph that clinical isolates exhibited increased killing of C. elegans in comparison to ATCC 27853 strains. The isolates differed in their ability to kill C.elegans. S373, S253, and S45 showed increased killing of C.elegans at 80%, 85%, and 90%, respectively. S402, S268, S277, and S135 showed 60%, 63%, 51%, and 45% killing, respectively. ATCC 27853 induced 12% death (Fig.2D).
Figure 2: Understanding virulence of Pseudomonas isolates. Secreted virulence factors, like Pyocyanin (A), Rhamnolipids (B) and Proteases (C) was estimated in 48hr biofilm supernatants of Pseudomonas isolates. C. elegans killing assay was used to assess pathogenicity (D). Compared to the ATCC27853 isolate S373, S253 and S45 showed an increase in virulence factors production. S373, S253 and S45 also showed an increase in C. elegans killing at 48hr. Statistical analysis was carried out by student T test, p<0.01 was considered significant.
3.3 Biofilms of Pseudomonas Reduced ROS Generation in A549 and THP1 Cells
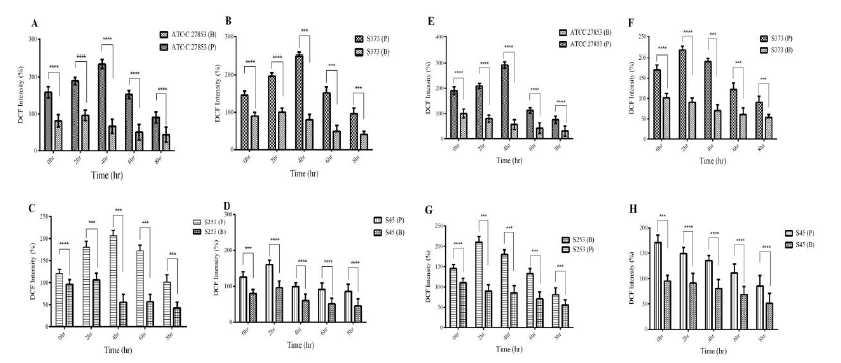
Elevated levels of intracellular ROS often result in oxidative stress in cells. Both planktonic and biofilm forms of Pseudomonas induced ROS response in A549 cells (Fig 3). A time-dependent increase in ROS generation was observed until 4 h post infection in A549 cells exposed to planktonic (P) Pseudomonas. A reduction in ROS response was observed at 6 h and 8 h post infection (Fig.3A-D). Biofilms (B) significantly reduced ROS generation in A549 cells. Biofilms of both ATCC 27853 and clinical isolates showed a 2 to 2.5-fold decrease in ROS generation in A549 monolayers.
THP1 cells exposed to planktonic (P) cells (both ATCC 27853 and clinical isolates) showed increased generation of ROS. A time-dependent increase was observed till 4 h post infection, following which a reduction was observed at 6 h and 8 h post infection, probably due to the reduction in cell viability (Fig. 3E-H). Among the clinical isolates, planktonic forms of S45 isolate induced maximum during the initial phases of infection, followed by a reduction. Biofilms (B) of both ATCC 27853 and clinical isolates reduced ROS generation in THP1 cells. A 2.5 to 3-fold decrease was observed in ROS generation.
Figure 3: Analysis of oxidative stress I cells. The oxidative stress induced by Pseudomonas planktonic (P) and Biofilm (B) forms was analysed by DCFDA method. The data has been represented as percentage intensity. ROS generation in A549 cells exposed to (A) ATCC 27853, (B) S373, (C) S253 and (D) S45. ROS also was quantified in THP1 cells exposed to (E) ATCC 27853, (F) S373, (G) S253 and (H) S45. Biofilms significantly reduced ROS generation in cells.
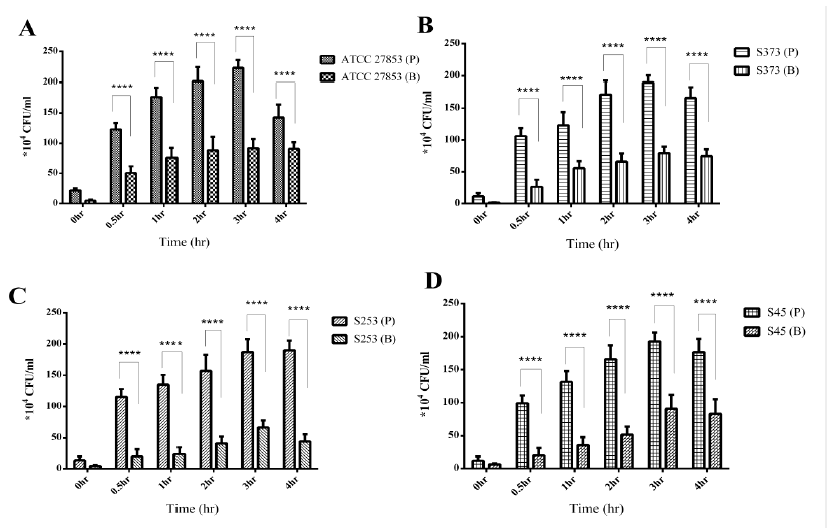
3.4 Biofilm Pseudomonas was less Internalized than Planktonic Pseudomonas
Gentamicin protection assay (GPA) is commonly used to understand the internalization of bacteria by eukaryotic cells. The GPA also reveals the ability of bacteria to survive within the host cells. The ability of planktonic and biofilm Pseudomonas to invade and survive within THP1 cells was assessed by GPA. It was observed that planktonic (P) Pseudomonas was more internalized than Pseudomonas biofilms (Fig4 A-D). A time-dependent increase was observed in the internalization of planktonic Pseudomonas up to 3 h post infection, followed by a decrease at 4 h. Biofilm (B) bacteria was also internalized by THP1 cells; however, no significant increase in internalized bacteria was observed over time, infact 2 to 2.5-fold decrease in internalized bacteria was observed.
Figure 4: Gentamicin Protection Assay: Internalization of planktonic (P) or biofilm (B) forms of Pseudomonas isolates. A) ATCC 27853 B) S373 C) S253 and D) S45 by THP1 cells. Biofilm bacteria were less internalized than the planktonic bacteria. No significant increase in biofilm bacteria internalization over a period of time was observed.
3.5 Biofilm-Infected A549 and THP1 cells Showed both Apoptotic and Necrotic Changes
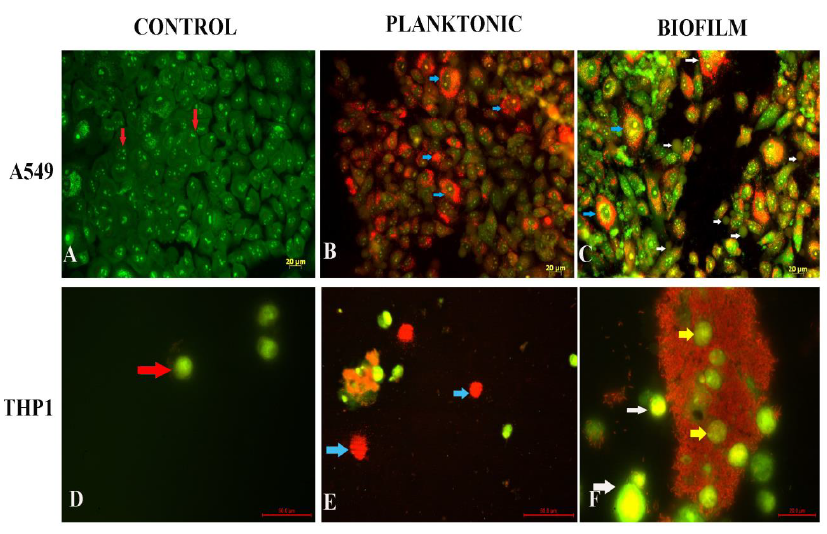
Fig. 5 (A-C) depicts the acridine orange/propidium iodide stained images of A549 cells infected with Pseudomonas planktonic forms and biofilm forms. Control cells appear green in colour (Fig 5A). The cells showed an intact epithelial morphology. Planktonic Pseudomonas-infected cells showed changes in cell morphology at 8 h post infection. The cells appeared spherical in shape. A decrease in cell membrane integrity, indicated by accumulation of propidium iodide in the cytoplasm, was noted in A549 cells 4 h post infection (Fig 5B). Such cells appeared yellow in color (blue arrow). A few cells also appeared swollen pointing towards necrosis as a mechanism of cell death. A549 cells challenged with biofilms of Pseudomonas showed cellular blebbing (white arrows) at 8 h post infection, as seen in early stages of apoptosis (Fig 5C). Dead cells appear red in colour. Accumulation of propidium iodide was found in the cytoplasm of the cells, indicating loss of membrane integrity (Blue arrow). Taken together, both apoptotic and necrotic cell deaths were observed in cells infected with biofilms of Pseudomonas.
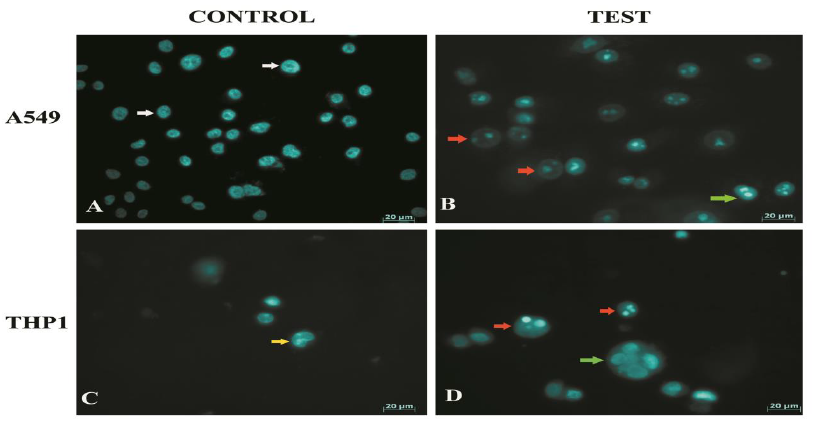
Morphological changes in THP1 monocytes were also assessed by acridine orange staining (Fig 5 D-F). Monocytes exposed to planktonic Pseudomonas showed necrotic cell death, 8 h post infection (Fig 5E). On the other hand, monocytes exposed to biofilms (Fig 5F) showed cellular blebbing (white arrow). The cells lost their spherical morphology and showed altered morphologies. Monocytes were found entrapped in biofilms (yellow arrows). Nuclear changes in A549 and THP1 cells exposed to biofilms of Pseudomonas were assessed by staining with Hoechst 33342. Control cells showed an intact nuclear morphology (Fig 6A and C). Biofilm-infected cells showed nuclear fragmentation with an intact nuclear membrane (red arrow). Multiple fragments were observed in majority of cells. A few cells also appeared to have condensed nuclei (green arrow), indicative of apoptosis. Nuclear fragmentation is one of the key features of apoptotic cells. In apoptotic cells, nuclear fragmentation is accompanied by the dissolution of nuclear membrane. Nuclear membrane remained intact in the cells infected by Pseudomonas biofilms (Fig 6B).
Figure 5: Assessment of mechanism of cells death induced by planktonic and biofilm forms of Pseudomonas on A549 cells and THP1 cells by Acridine orange / Propidium iodide staining. A. uninfected A549 control appeared green and showed intact epithelial morphology. (B). A549 cells infected with planktonic forms of Pseudomonas. Excessive accumulation of propidium iodide was observed (Blue arrow). (C) A549 cells infected with biofilm showed features of both apoptosis and necrosis. Cellular blebbing (white arrows) and accumulation of propidium iodide was observed (blue arrow). (D)Control THP1 cells showed intact morphology. (E) Cells infected with planktonic Pseudomonas appeared due to excessive accumulation of propidium iodide. (F) Monocytes were found entrapped within biofilms (yellow arrow). Cells also appeared to have blebs (white arrows)
Figure 6: Assessment of nuclear change in A549 cells and THP1 infected with biofilms of Pseudomonas (A - D). (A) Nucleus of control A549 cells appeared intact. (B) Biofilm infected cells showed DNA fragmentation within intact nuclear membrane (Red arrow). (C) Nucleus of THP1 control cells appeared intact. Kidney shaped nuclei was visualised. (D) Nucleus of biofilm infected THP1 cells appeared to have fragmented nuclei within intact nuclear membrane (atypical features of apoptosis)
3.6 Cytokine expression in model systems exposed to planktonic and biofilms of Pseudomonas
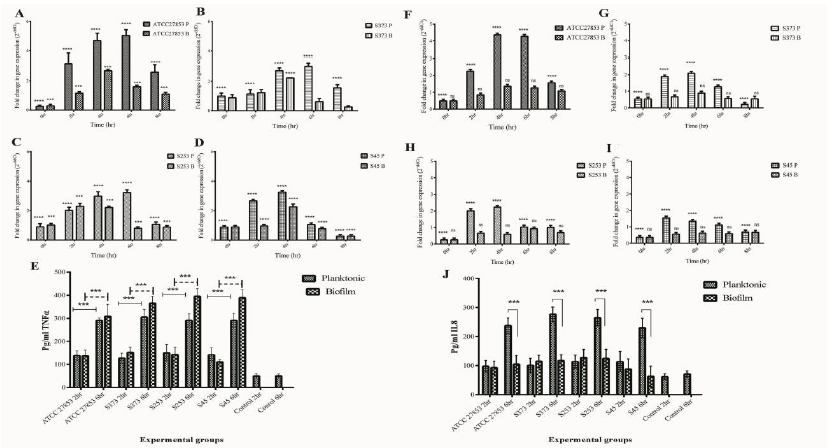
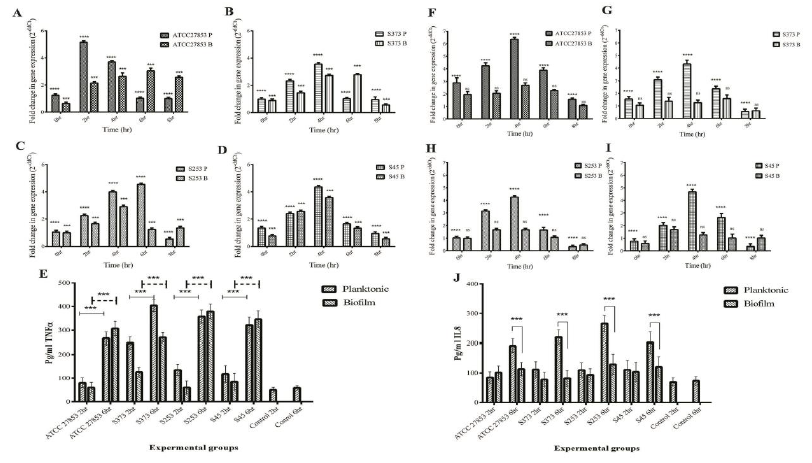
The pro-inflammatory cytokines analyzed were TNF-α and IL-8. TNFα produced by epithelial cells, monocytes, or macrophages is an acute phase response to a microbial challenge. They have an important role in immune response to bacterial pathogens. IL8, a pro-inflammatory cytokine is also known as neutrophil chemoattractant protein. The major function of IL8 is in the recruitment of neutrophils to the site of infection and their activation. IL8 therefore plays an important role in pathogen clearance. It is evident from figure 7 that, both planktonic (P) and biofilm (B) forms of Pseudomonas up regulated TNFα in A549 cells (Fig 7 A-D). A time dependent upregulation was observed till 6 h post infection. It can also be observed that planktonic forms of clinical isolates showed reduced TNFα expression when compared to the ATCC 27853 isolate. Alternatively, it was also observed that biofilms were capable of reducing TNFα expression when compared to their planktonic counterparts. TNFα protein expression also showed similar trend (Fig.7E).
On the other hand, IL8 expression was significantly up regulated only in A549 cells infected with planktonic forms of Pseudomonas (Fig 7 F-I). it can also be observed that clinical isolates reduced IL8 expression when compared to ATCC 27853 strain. Biofilms significantly reduced IL8 expression by A549 cells. A time dependent increase was observed in IL8 protein expression in A549 cells at 6 h post infection (Fig 7 J). Similar observations were made in monocyte cultures exposed to planktonic and biofilm exposed Pseudomonas. Both planktonic and biofilm forms of Pseudomonas induced TNFα expression by THP1 cells (Fig 8 A-E). Biofilms failed to induce an upregulation in IL8 expression in infected THP1 cells (Fig F-J). A549:THP1 cocultures exposed to planktonic forms of Pseudomonas significantly upregulated TNFα expression (Fig 9A-E), but failed to induce IL8 expression (Fig 9F-J).
Figure 7: Pro-inflammatory cytokine gene expressions in A549 cells exposed to planktonic (P) and biofilm (B) forms of Pseudomonas. TNFα gene expression in A549 cells exposed to (A) ATCC 27853, (B) S373, (C) S253 and (D) S45. (E) Quantification of TNFα protein in cells exposed to Pseudomonas. IL8 gene expression in cells exposed to (F) ATCC 27853, (G) S373, (H) S253 and (I) S45. (J) Quantification of IL8 protein expression in A549 cells.
Figure 8: Pro-inflammatory cytokine gene expression in THP1 cells exposed to planktonic (P) and biofilm (B) forms of Pseudomonas. TNFα gene expression in A549 cells exposed to (A) ATCC 27853, (B) S373, (C) S253 and (D) S45. (E) Quantification of TNFα protein in cells exposed to Pseudomonas. IL8 gene expression in cells exposed to (F) ATCC 27853, (G) S373, (H) S253 and (I) S45. (J) Quantification of IL8 protein expression in THP1 cells.
Figure 9: Pro-inflammatory cytokine gene expression in A549:THP1 co-cultures exposed to planktonic (P) and biofilm (B) forms of Pseudomonas. TNFα gene expression in A549 cells exposed to (A) ATCC 27853, (B) S373, (C) S253 and (D) S45. (E) Quantification of TNFα protein in cells exposed to Pseudomonas. IL8 gene expression in cells exposed to (F) ATCC 27853, (G) S373, (H) S253 and (I) S45. (J) Quantification of IL8 protein expression in A549:THP1.
3.7 Biofilms of Pseudomonas Upregulate Inflammasome Markers
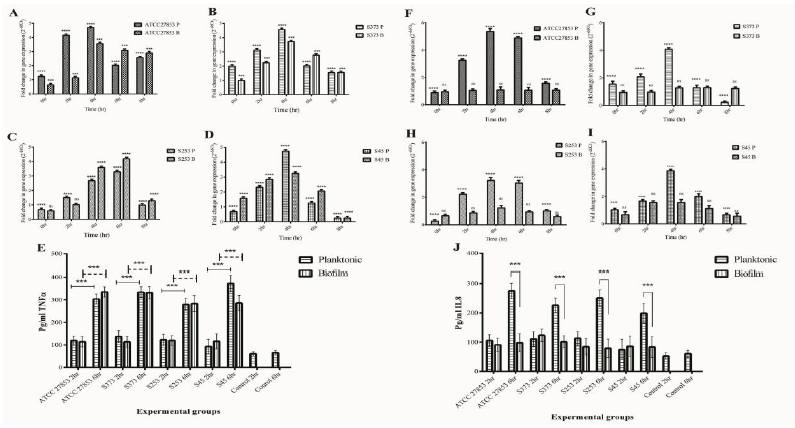
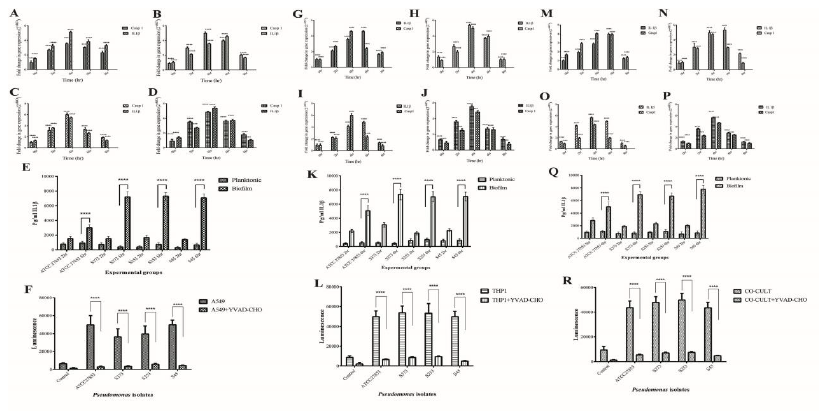
Inflammasomes are cytosolic multiprotein oligomers of the innate immune system responsible for the activation of inflammatory responses. Activation and assembly of the inflammasome promotes proteolytic cleavage, maturation, and secretion of pro-inflammatory cytokines IL-1β and induce a pro-inflammatory form of programmed cell death distinct from apoptosis, referred to as pyroptosis. The inflammasome gene expression in A549 cells exposed to biofilms of Pseudomonas was evaluated. Caspase 1 and IL-1β are important modulators of inflammasome response. A time-dependent increase in Caspase 1 and IL-1β gene expression was observed in A549 cells exposed to biofilms of both ATCC 27853 (Fig 10A) and clinical isolates (Fig 10B, C, and D). Caspase 3, which is the executioner caspase in the apoptotic pathway, was found to be expressed only at basal levels (data not shown).
IL-1β protein expression also showed significant increase at 6 h post infection in A549 cells (Fig 10E) exposed to biofilms of Pseudomonas. The protein expression correlated with the gene expression data. Planktonic infection did not significantly increase IL-1β. Caspase 1 expression showed significant upregulation in A549 cells exposed to biofilms of both ATCC 27853 and clinical isolates of Pseudomonas (Fig 10F). The influence of biofilms on inflammasome response in THP1 cells was also evaluated. Biofilms significantly increased IL-1β and caspase 1 expression. A 5 to 6-fold upregulation of IL-1β and Caspase 1 was observed (Fig 10 G-J). Biofilms of both ATCC 27853 and clinical isolates showed similar responses. A significant increase in IL-1β protein expression was observed at 6 h post infection in biofilm-infected THP1 cells (Fig 10K). Caspase 3 expression was found only at basal levels in the cells exposed to biofilms of Pseudomonas. No significant increase in caspase 3 was observed in THP1 cells exposed to planktonic forms clinical isolates. IL-1β expression showed an increase at 6 h post infection in THP1 cells exposed to biofilms, whereas planktonic forms of Pseudomonas failed to induce IL-1β production in THP1 cells (Fig 10 G-K). An upregulation was observed in caspase 1 activity in THP1 cells exposed to biofilms of Pseudomonas (Fig 10L)
A549:THP1 co-cultures showed an increase in caspase 1 and IL-1β expression. ATCC 27853 biofilms induced a 3 to 5-fold increase in IL1β expression and 4-fold increase in caspase 1 expression (Fig 10 M). On the other hand, clinical isolates induced a 5 to 6-fold increase in caspase 1 and IL-1β expression (Fig 10N-R). IL1β protein expression was in concordance with the gene expression data (Fig 10 Q). Biofilms induced significant upregulation in caspase 1 activity in co-cultures (Fig 10R). No significant changes were observed in caspase 3 expression in A549:THP1 co-cultures infected with Pseudomonas biofilms up to 8 h post infection. A significant decrease was observed in caspase 1 and IL-1β expression at 8 h post infection. This could be due to a cell death due to infection.
Figure 10: Inflammasome markers in model systems exposed to planktonic (P) and Biofilm (B) forms of Pseudomonas. Caspase1 and IL 1β gene expression in A549 cells (A.ATCC 27853, B.S373, C.S253 and D.S45), (E) IL1β protein expression in A549 cells exposed to Pseudomonas planktonic and biofilm forms. (F) Caspase 1 activity in biofilm infected cell was quantified. Addition of YVAD-CHO (inhibitor of caspase1) reduced the expression. Caspase1 and IL 1β gene expression in THP1 cells (G.ATCC 27853, H.S373, I.S253 and J.S45) and A549:THP1 co-cultures (M. ATCC 27853, N. S373 O.S253 and P.S45) was assessed by RT-PCR. K, Q Quantification of IL1β expression by THP1 and A549:THP1 co-cultures exposed to Pseudomonas. L, R Quantification of caspase1 activity by THP1 and A549:THP1 co-cultures exposed to Pseudomonas
4. Discussion
Pseudomonas is one of the most commonly isolated organism from VAP [19]. The mechanism underlying Pseudomonas associated VAP induced lung injury remains elusive. Most of the elucidated immune evasion mechanisms hold good for acute infection caused by planktonic bacteria [19]. This paper discusses the mechanisms of pseudomonas biofilm pathogenesis. The expression of Pseudomonas virulence factors important in biofilm pathogenesis like Pyocyanin, rhamnolipids, and proteases were higher in clinical isolates compared to the expression by ATCC 27853 strain (Fig 2). The different clinical isolates used in the study differed in their ability to produce these virulence factors. Among the clinical isolates, S373, S253, and S45 produced comparatively higher amounts of virulence factors.
elegans is a widely accepted model to study host pathogen interaction [20]. Showed (Fig 4) S373, S253, and S45 to be more virulent and they also had higher expression of virulence factors pyocyanin, rhamnolipids, and proteases. These strains were used to study the interaction of Pseudomonas biofilms with the alveolar epithelium and its effects on monocytes and co-cultures. Innate immune responses to infection are elicited by the epithelial layer initially followed by the activation of polymorphonuclear leucocytes (PMNs). In-vitro system representing both the epithelial layer as A549 monolayers and PMNs - THP1 were used in isolation and in combination to understand the complicated cellular immune machinery. Cellular responses to Pseudomonas biofilms was analysed by morphological, biochemical, and molecular methods. ROS generation in A549 and THP1 cells was reduced in biofilm-infected cells compared with their planktonic counterparts. The reduction in ROS could be due to alginate [21] and super oxide dismutase expressed by biofilms of Pseudomonas as they have scavenging effect. The reduction observed in bacterial internalization of biofilm of Pseudomonas (both ATCC 27853 and clinical isolates) by THP1 cells compared with their planktonic counterparts was comparable to the observations of Mc. Calsin et al who reported that the alginate component in the biofilm of bacteria protects them from internalization and phagocytosis by neutrophils and macrophages [22].
Pseudomonas employs multiple mechanisms to induce cell death in invading host cell. This is accomplished by the use of virulence factors [23]. Apoptosis and necrosis are the most extensively studied mechanism of cell death induced by Pseudomonas. We observed that exposure to Pseudomonas biofilms induced both apoptotic and necrotic changes in A549 and THP1 cells (Fig 5). This can be attributed to the production of pyocyanin. Pyocyanin, produced by Pseudomonas was found to induce both apoptotic and necrotic cell death [24]. Some atypical features of apoptosis were also observed. Nuclear changes revealed the presence of fragmented DNA within an intact nuclear membrane (Fig 6).
We further analysed the cytokine expression in A549, THP1 and their co-cultures exposed to planktonic and biofilm forms of Pseudomonas. It was observed that planktonic forms of Pseudomonas induced an upregulation of pro-inflammatory cytokines IL8 and TNFα in A549, THP1 and their co-cultures. Epelman and his colleagues observed that Exoenzyme S (ExoS) induced the upregulation of pro-inflammatory cytokines and chemokines [25]. On the contrary, we observed that biofilms of Pseudomonas induced and up-regulated only TNFα and not IL8. IL8 was expressed only at basal levels. IL8 plays a key role in recruitment of innate immune cells such as neutrophils, monocytes, and other immune cells to the site of infection [26]. In the present study, biofilms significantly reduced IL8 expression, indicating that infiltration of neutrophils and other immune cells is reduced during biofilm infection.
TNF alpha is known to play vital roles in the progression of Pseudomonas infection. It is also recognised as a key molecule that is involved in apoptosis. Tsay and his colleagues observed that, blocking of TNFα in a burn mice infection model increased the mortality rates due to Pseudomonas infection [27]. Biofilm forming Pseudomonas specifically upregulate the expression of TNFα and IL6 in monocytes [28]. We observed an upregulation in TNFα but not IL6.
Evaluation of inflammasome markers revealed upregulation of caspase 1 and IL-1β. Executioner caspase 3 was found to be expressed only at basal levels, indicating that biofilms did not induce cell death via apoptosis (fig 10). [29]. observed that extracellular vesicles of S. aureus induce upregulation of caspase 1 and IL-1β in monocytes, resulting in inflammasome-mediated pyroptotic cell death [30]. Probably a similar mechanism is in operation in Pseudomonas biofilm pathogenicity. Fragmentation of DNA, nuclear fragmentation is indicative of apoptotic cell death. DNA fragmentation in pyroptosis is marked by the presence of an intact nuclear membrane and figure 6 clearly denotes such observations in A549 and THP1 cells exposed to biofilm of Pseudomonas. These data strongly suggest the involvement of pyroptotic mechanism of cell death in biofilm-infected cells. Pyroptotic cell death often results in localised inflammatory response, tissue disruption and dissemination of the pathogen [13]. We believe that Pseudomonas biofilms use the pyroptotic machinery to cause chronic and persistent infection in the host.
5. Conclusion
Our data suggest that pyroptotic cell death is the mechanism of pathogenesis of Pseudomonas biofilms and not apoptosis. A reduction in IL8 expression also suggested a reduction in the infiltration of immune cells to the site of infection. Taken together, this leads to chronic inflammation and persistence of infection within the host. This understanding is critical in the management of the Current Covid pandemic where secondary infections were caused by biofilm forming variants of Pseudomonas [2]. Intubation of critical Covid patients also increased the risk of Pseudomonas biofilm infection. Activation of pyroptosis under such conditions might worsen the patients’ prognosis. Alternatively, pyroptotic cell death also helps enhancing the adaptive immune responses. Therefore, understanding the complex interplay of pyroptosis mediated adaptive responses is critical in ensuring positive therapeutic outcome.
Conflicts of interest
All the authors declare that they have no conflicts of interest and no affiliation with or involvement in any organization or entity with any financial interest related to this study.
Acknowledgements
The Director SCTIMST, Head BMT Wing for the facilities provided. Dr. Anoopkumar Thekkuveetil, for providing facilities for C.elegans killing assay. DST INSPIRE for fellowship and funding and DBT for funding of part of the project.
References
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X & Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet 395 (2020): 507-513.
- Fu Y, Yang Q, Xu M, Kong H, Chen H, Fu Y, Yao Y, Zhou H & Zhou J. Secondary Bacterial Infections in Critical Ill Patients With Coronavirus Disease 2019. Open Forum Infectious Diseases 7 (2020): ofaa220.
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S & Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. The Lancet Respiratory Medicine 8 (2020): 475-481.
- Jordan P, Ham WT & Fataar D. Endotracheal tube verification in adult mechanically ventilated patients. Southern African Journal of Critical Care 31 (2015): 20-23.
- Danin PE, Girou E, Legrand P, Louis B, Fodil R, Christov C, Devaquet J, Isabey D & Brochard L. Description and Microbiology of Endotracheal Tube Biofilm in Mechanically Ventilated Subjects. Respiratory Care 60 (2015): 21-29.
- Moser C, Jensen PØ, Thomsen K, Kolpen M, Rybtke M, Lauland AS, Trøstrup H & Tolker-Nielsen T. Immune Responses to Pseudomonas aeruginosa Biofilm Infections. Frontiers in Immunology 12 (2021): 625597.
- Maurice NM, Bedi B & Sadikot RT. Pseudomonas aeruginosa Biofilms: Host Response and Clinical Implications in Lung Infections. American Journal of Respiratory Cell and Molecular Biology 58 (2018): 428-439.
- Jensen PØ, Givskov M, Bjarnsholt T & Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunology and Medical Microbiology 59 (2010): 292-305.
- Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M & Høiby N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatric Pulmonology, 44 (2009): 547-558.
- Kolpen M, Hansen CR, Bjarnsholt T, Moser C, Christensen LD, van Gennip M, Ciofu O, Mandsberg L, Kharazmi A, Döring G, Givskov M, Høiby N & Jensen PØ. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65 (2010): 57-62.
- Hotterbeekx A, Xavier BB, Bielen K, Lammens C, Moons P, Schepens T, Ieven M, Jorens PG, Goossens H, Kumar-Singh S & Malhotra-Kumar S. The endotracheal tube microbiome associated with Pseudomonas aeruginosa or Staphylococcus epidermidis. Scientific Reports 6 (2016).
- Malik A & Kanneganti TD. Inflammasome activation and assembly at a glance. Journal of Cell Science, 130 (2017): 3955-3963.
- Bergsbaken T, Fink SL & Cookson BT. Pyroptosis: Host cell death and inflammation. Nature Reviews. Microbiology 7 (2009): 99-109.
- O’Toole GA. Microtiter dish biofilm formation assay. Journal of Visualized Experiments: JoVE, 47 (2011).
- Essar DW, Eberly L, Hadero A & Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. Journal of Bacteriology 172 (1990): 884-900.
- Rasamiravaka T, Vandeputte OM & Jaziri ME. Procedure for Rhamnolipids Quantification Using Methylene-blue. Bio-Protocol 6 (2016): e1783-e1783.
- El-Mowafy S a, Shaaban M i & Abd El Galil Kh. Sodium ascorbate as a quorum sensing inhibitor of Pseudomonas aeruginosa. Journal of Applied Microbiology 117 (2014): 1388-1399.
- Tan MW, Mahajan-Miklos S & Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America 96 (1999): 715-720.
- Ramírez-Estrada S, Borgatta B & Rello J. Pseudomonas aeruginosa ventilator-associated pneumonia management. Infection and Drug Resistance 9 (2016): 7.
- Marsh EK & May RC. Caenorhabditis elegans, a Model Organism for Investigating Immunity. Applied and Environmental Microbiology 78 (2012): 2075-2081.
- Learn DB, Brestel EP & Seetharama S. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infection and Immunity 55 (1987): 1813-1818.
- McCaslin CA, Petrusca DN, Poirier C, Serban KA, Anderson GG & Petrache I. Impact of alginate-producing Pseudomonas aeruginosa on alveolar macrophage apoptotic cell clearance. Journal of Cystic Fibrosis?: Official Journal of the European Cystic Fibrosis Society 14 (2015): 70-77.
- Grassmé H, Jendrossek V & Gulbins E. Molecular mechanisms of bacteria induced apoptosis. Apoptosis: An International Journal on Programmed Cell Death 6 (2001): 441-445.
- Hassett DJ, Schweizer HP & Ohman DE. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. Journal of Bacteriology 177 (1995): 6330-6337.
- Epelman S, Bruno TF, Neely GG, Woods DE & Mody CH. Pseudomonas aeruginosa exoenzyme S induces transcriptional expression of proinflammatory cytokines and chemokines. Infection and Immunity 68 (2000): 4811-4814.
- Sokol CL & Luster AD. The Chemokine System in Innate Immunity. Cold Spring Harbor Perspectives in Biology 7 (2015).
- Tsay TB, Yang MC, Chen PH, Lai KH, Huang HT, Hsu CM & Chen LW. Blocking TNF-α enhances Pseudomonas aeruginosa-induced mortality in burn mice through induction of IL-1β. Cytokine 63 (2013): 58-66.
- Ciornei CD, Novikov A, Beloin C, Fitting C, Caroff M, Ghigo JM, Cavaillon JM & Adib-Conquy M. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immunity 16 (2010): 288-301.
- Wang X, Eagen WJ & Lee JC. Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proceedings of the National Academy of Sciences 117 (2020): 3174-3184.
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X & Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet 395 (2020): 507-513.
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S & Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. The Lancet Respiratory Medicine 8 (2020): 475-481.