RLSuite: An Integrative R-Loop Bioinformatics Framework
Article Information
Miller HE1,2,3,*, Montemayor D4,5, Levy S1,3,6,7, Sharma K4,5, Frost B1,6,7, Bishop A J R1,2,8,*
1Department of Cell Systems and Anatomy, UT Health San Antonio, San Antonio, TX, USA
2Greehey Children’s Cancer Research Institute, UT Health San Antonio, San Antonio, TX, USA
3Bioinformatics Research Network, Atlanta, GA, USA
4Department of Medicine, UT Health San Antonio, San Antonio, TX, USA
5Center for Precision Medicine, UT Health San Antonio, San Antonio, TX, USA
6Sam & Ann Barshop Institute for Longevity & Aging Studies, UT Health San Antonio, San Antonio, TX, USA
7Glenn Biggs Institute for Alzheimer’s and Neurodegenerative Diseases, UT Health San Antonio, San Antonio, TX, USA
8May’s Cancer Center, UT Health San Antonio, San Antonio, TX, USA
Corresponding Author: Miller H E, Alexander Bishop, Greehey Children’s Cancer Research Institute, Department of Cell Systems and Anatomy, UT Health San Antonio, San Antonio, TX, 78229, USA
Received: 31 August 2023; Accepted: 06 September 2023; Published: 21 December 2023
Citation: Miller HE, Montemayor D, Levy S, Sharma K, Frost B, Bishop AJR. RLSuite: An Integrative R-Loop Bioinformatics Framework. Journal of Bioinformatics and Systems Biology. 6 (2023): 364-378.
View / Download Pdf Share at FacebookAbstract
We recently described the development of a database of 810 R-loop mapping datasets and used this data to conduct a meta-analysis of R-loops. R-loops are three-stranded nucleic acid structures containing RNA:DNA hybrids and we were able to verify that 30% of expressed genes have an associated R-loop in a location conserved manner.. Moreover, intergenic R-loops map to enhancers, super enhancers and with TAD domain boundaries. This work demonstrated that R-loop mapping via highthroughput sequencing can reveal novel insight into R-loop biology, however the analysis and quality control of these data is a non-trivial task for which few bioinformatic tools exist. Herein we describe RLSuite, an integrative R-loop bioinformatics framework for pre-processing, quality control, and downstream analysis of R-loop mapping data. RLSuite enables users to compare their data to hundreds of public datasets and generate a user-friendly analysis report for sharing with non-bioinformatician colleagues. Taken together, RLSuite is a novel analysis framework that should greatly benefit the emerging R-loop bioinformatics community in a rapidly expanding aspect of epigenetic control that is still poorly understood.
Keywords
RLSuite, Bioinformatics, R-loop, RNA:DNA hybrid
RLSuite articles; Bioinformatics articles; R-loop articles; RNA:DNA hybrid articles
RLSuite articles RLSuite Research articles RLSuite review articles RLSuite PubMed articles RLSuite PubMed Central articles RLSuite 2023 articles RLSuite 2024 articles RLSuite Scopus articles RLSuite impact factor journals RLSuite Scopus journals RLSuite PubMed journals RLSuite medical journals RLSuite free journals RLSuite best journals RLSuite top journals RLSuite free medical journals RLSuite famous journals RLSuite Google Scholar indexed journals Bioinformatics articles Bioinformatics Research articles Bioinformatics review articles Bioinformatics PubMed articles Bioinformatics PubMed Central articles Bioinformatics 2023 articles Bioinformatics 2024 articles Bioinformatics Scopus articles Bioinformatics impact factor journals Bioinformatics Scopus journals Bioinformatics PubMed journals Bioinformatics medical journals Bioinformatics free journals Bioinformatics best journals Bioinformatics top journals Bioinformatics free medical journals Bioinformatics famous journals Bioinformatics Google Scholar indexed journals R-loop articles R-loop Research articles R-loop review articles R-loop PubMed articles R-loop PubMed Central articles R-loop 2023 articles R-loop 2024 articles R-loop Scopus articles R-loop impact factor journals R-loop Scopus journals R-loop PubMed journals R-loop medical journals R-loop free journals R-loop best journals R-loop top journals R-loop free medical journals R-loop famous journals R-loop Google Scholar indexed journals RNA:DNA hybrid articles RNA:DNA hybrid Research articles RNA:DNA hybrid review articles RNA:DNA hybrid PubMed articles RNA:DNA hybrid PubMed Central articles RNA:DNA hybrid 2023 articles RNA:DNA hybrid 2024 articles RNA:DNA hybrid Scopus articles RNA:DNA hybrid impact factor journals RNA:DNA hybrid Scopus journals RNA:DNA hybrid PubMed journals RNA:DNA hybrid medical journals RNA:DNA hybrid free journals RNA:DNA hybrid best journals RNA:DNA hybrid top journals RNA:DNA hybrid free medical journals RNA:DNA hybrid famous journals RNA:DNA hybrid Google Scholar indexed journals R-loop articles R-loop Research articles R-loop review articles R-loop PubMed articles R-loop PubMed Central articles R-loop 2023 articles R-loop 2024 articles R-loop Scopus articles R-loop impact factor journals R-loop Scopus journals R-loop PubMed journals R-loop medical journals R-loop free journals R-loop best journals R-loop top journals R-loop free medical journals R-loop famous journals R-loop Google Scholar indexed journals epigenetic articles epigenetic Research articles epigenetic review articles epigenetic PubMed articles epigenetic PubMed Central articles epigenetic 2023 articles epigenetic 2024 articles epigenetic Scopus articles epigenetic impact factor journals epigenetic Scopus journals epigenetic PubMed journals epigenetic medical journals epigenetic free journals epigenetic best journals epigenetic top journals epigenetic free medical journals epigenetic famous journals epigenetic Google Scholar indexed journals RLHub articles RLHub Research articles RLHub review articles RLHub PubMed articles RLHub PubMed Central articles RLHub 2023 articles RLHub 2024 articles RLHub Scopus articles RLHub impact factor journals RLHub Scopus journals RLHub PubMed journals RLHub medical journals RLHub free journals RLHub best journals RLHub top journals RLHub free medical journals RLHub famous journals RLHub Google Scholar indexed journals POS articles POS Research articles POS review articles POS PubMed articles POS PubMed Central articles POS 2023 articles POS 2024 articles POS Scopus articles POS impact factor journals POS Scopus journals POS PubMed journals POS medical journals POS free journals POS best journals POS top journals POS free medical journals POS famous journals POS Google Scholar indexed journals canonical articles canonical Research articles canonical review articles canonical PubMed articles canonical PubMed Central articles canonical 2023 articles canonical 2024 articles canonical Scopus articles canonical impact factor journals canonical Scopus journals canonical PubMed journals canonical medical journals canonical free journals canonical best journals canonical top journals canonical free medical journals canonical famous journals canonical Google Scholar indexed journals
Article Details
1. Introduction
R-loops are three-stranded nucleic acid structures comprised of an RNA:DNA hybrid and displaced single strand DNA. R-loops form during transcription as nascent RNA re-anneals to the template DNA strand, often in regions with high G/C-skew, such as CpG islands [1]. R-loop mapping techniques enable the genome-wide analysis of R-loop locations and dynamics. In 2012, Ginno et al. introduced the first R-loop mapping technique, DNA:RNA immunoprecipitation sequencing (DRIP-Seq) [11]. In the decade since, more than 40 R-loop mapping studies have been described [6]. While these studies have greatly expanded our understanding of R-loop dynamics, they have also revealed pervasive quality control issues that limit the interpretation of their findings [12]. In recent work, we mined 810 public R-loop mapping datasets and developed a robust quality control (QC) approach for R-loop mapping data. Moreover, we built a database of reprocessed and standardized R-loop mapping datasets, and described novel methods for analysing R-loop data [6,13]. With the data and code developed in these studies, we created RLSuite, a collection of software packages for the analysis of R-loop mapping data. There are a growing number of publications that describe bioinformatic approaches for the study of R-loops. However, most have focused on the task of predicting R-loop formation from genomic sequences [14–19]. At present, only two bioinformatic tools for the analysis of R-loop mapping data have been described. The first, footLoop, is designed specifically for the analysis of long-read bisulfite-converted R-loop mapping data (SMRF-Seq) [20], a type of sequencing that is not applicable for most R-loop mapping studies. The second, the DRIP-optimized Peak Annotator (DROPA), facilitates the annotation of DRIP-seq peaks using genomic features and gene expression information [21], but does not provide users with the capability to perform upstream data processing, quality control their data, or analyze and explore their data in the context of public R-loop mapping datasets. To address this critical resource gap, we have developed RLPipes, RLHub, and RLSeq (collectively termed “RLSuite”). RLSuite provides an integrative workflow for R-loop data analysis, including automated pre-processing of R-loop mapping data using a standard pipeline, multiple robust methods for quality control, and a range of tools for the initial exploration of R-loop data. RLSuite also provides the unique capability to compare analysis results with those from the public R-loop datasets that we previously reprocessed [6,13]. Moreover, it provides a user-friendly HTML analysis report which should prove useful for both bioinformaticians and biologists alike.
2. Materials and Methods
Methods are described in the Supplementary Methods which accompany this work.
3. Results
In recent work, we reprocessed 810 R-loop mapping datasets, establishing a robust method of quality control [6]. We then used these data to build RLBase, the largest R-loop mapping database yet described [13]. We also established novel approaches for the analysis of R-loop mapping data, and demonstrated the utility of analysing R-loop mapping data in the context of publicly available R-loop datasets [6]. We then codified and standardized our analysis approaches in the form of three software packages: RLPipes, RLHub, and RLSeq. RLPipes is a Bioconda command line tool for the automated processing of R-loop mapping data from raw files, BAM files, or public data accessions. RLHub is an R/Bioconductor “ExperimentHub” package that facilitates access to the processed datasets stored within RLBase. Finally, RLSeq is an R/Bioconductor package that provides a comprehensive toolkit for quality control, exploration, and visualization of R-loop mapping data. Collectively termed “RLSuite”, these packages provide the first integrative R-loop bioinformatics framework. In the following sections, we describe the workflow for analysis of R-loop mapping data with RLSuite.
3.1 RLPipes: Upstream pre-processing of R-loop mapping data
A typical workflow for RLSuite involves providing raw data files (and relevant metadata) or public data accessions to RLPipes, which then automatically processes the data (Figure S1A). RLPipes is a command-line tool based on Snakemake [22] which implements a uniform workflow for data wrangling, QC, alignment, peak calling, and coverage analysis. This workflow follows standard practices for the analysis of genomic and epigenomic data [23]. Finally, the resulting data is analysed with RLSeq to generate an analysis report (see RLSeq: downstream R-loop data QC, analysis, and visualization). To initiate an RLPipes workflow, a user can invoke the RLPipes build command. After validating the input parameters, RLPipes generates a directory to house the workflow outputs along with a configuration file. Finally, the user invokes the RLPipes run command, which launches all workflow jobs, leading to the generation of BAM files, coverage tracks, peaks, and RLSeq outputs (described in “RLSeq”).
3.2 RLHub: A hub for R-loop data
RLHub is an R/Bioconductor ExperimentHub [24] package that provides a convenient R interface for users to access processed datasets relevant for the analysis of R-loop mapping data (Table S1). These datasets can be loaded via built-in accessor functions. For example, rlbase_samples() loads the manifest of all publicly-available R-loop mapping samples in RLBase. Additionally, rlbps() loads the R-loop binding proteins identified from three recent studies [25–27], with their associated confidence scores. In summary, RLHub provides a convenient R interface for accessing a wealth of processed data relevant for the analysis of R-loops.
3.3 RLSeq: downstream R-loop data QC, analysis, and visualization
RLSeq is an R/Bioconductor package for the downstream analysis of R-loop mapping data. It is run automatically as part of the RLPipes workflow, but it can also be used independently in any standard R session. In a minimal RLSeq analysis workflow, a user will load their data as an RLRanges object with RLRanges(), run the RLSeq() function to perform core analysis steps with default parameters, and then run the report() function to generate a HTML analysis report which can be shared with colleagues (Figure S1B). In the following sections, we will describe each of the steps in this workflow. To illustrate the utility of RLSeq, we analysed DRIP-Sequencing datasets deposited by Jangi et al., 2017 as part of their study of the survival of motor neuron (SMN) gene [28]. We analysed their DRIP-seq data from two SH-SY5Y cell lines, one with short hairpin RNA (shRNA) which is untargeted (shCTR; SRA ID: SRX2187024), and one with shRNA targeting SMN (shSMN; SRA ID: SRX2187025). The following sections will describe the utilities in RLSeq along with the results from running those functions on the SH-SY5Y shCTR dataset and, where indicated, the shSMN dataset as well.
3.4 RLFS analysis
The first step in the RLSeq workflow is to perform R-loop forming sequences (RLFS) analysis using the analyzeRLFS() function. RLFS are computationally-predicted genomic regions that show favourability for R-loop formation [15–18]. In previous work, we demonstrated that high quality R-loop mapping datasets show robust enrichment in these regions, indicating their utility for predicting whether R-loop mapping was successful [6]. In that work, we developed RLFS analysis, a method which employs permutation testing to evaluate the statistical enrichment of R-loop mapping peaks within RLFS [6]. Within RLSeq, this analysis is performed via the analyzeRLFS() function, and the results of the analysis are visualized with the plotRLFSRes() function (Figure 1). In the present study, we ran RLFS analysis on SH-SY5Y shCTR DRIP-seq data and visualized the results (Figure 1). From the resulting plot, we observed that the sample peaks were strongly enriched within RLFS, suggesting the data represents successful R-loop mapping.
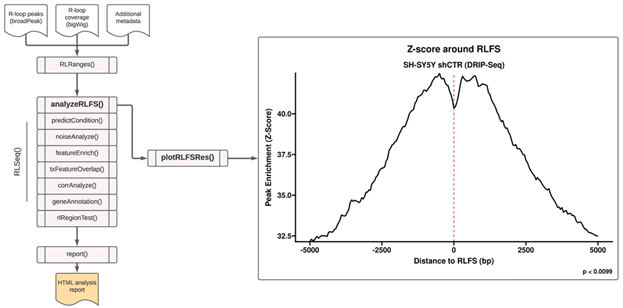
Figure 1: R-loop forming sequences (RLFS) analysis. RLFS analysis was applied to analyze publicly available DRIP-seq data from SH-SY5Y cells with control shRNA (“shCTR”) (SRA ID: SRX2187024). This analysis was invoked with the analyzeRLFS() function and plotted with the plotRLFSRes() function. The analysis involved permutation testing to determine the statistical enrichment of user-supplied R-loop peaks within computationally predicted RLFS. The RLFS analysis plot displays the p value from permutation testing (with the standard 100 permutations, the minimum possible p value is 0.0099). Additionally, the peak enrichment around RLFS is displayed as a Z score.
3.5 Sample quality classification
As demonstrated in our previous work, R-loop mapping data is prone to quality issues which can severely limit interpretability [6]. In that work, we developed a machine learning model which uses the results of RLFS analysis to predict whether a dataset shows evidence of successful R-loop mapping [6]. Samples that are predicted to map R-loops successfully receive a model prediction of “POS” (positive), whereas samples that are predicted to not map R-loops get a prediction of “NEG” (negative). The prediction is performed using the predictCondition() function. For the sample analysed in this example (SH-SY5Y shCTR), the result of this prediction was “POS,” indicating that it robustly maps R-loops as expected.
3.6 Noise analysis
As with all QC methods, the method implemented by predictCondition() has limitations [6] and a comprehensive QC workflow should include additional complementary methods. Towards that end, RLSeq provides multiple additional QC methods. One such approach, termed “noise analysis” (invoked via the noiseAnalyze() function), quantifies the signal-to-noise ratio of R-loop mapping coverage using the method developed by Diaz et al [29]. The results are visualized qualitatively using a “fingerprint plot” (Figure S2A) (named after the deepTools implementation of this same approach [30]. A “hockey-stick” shape (greater inflection) is a sign of good signal to noise ratio because it implies that most signal is found within a subset of the genome (as would be expected for R-loop mapping). From our analysis of the SH-SY5Y shCTR DRIP-seq sample, we observed a strong “hockey-stick” pattern (Figure S2A). This further suggests the high quality of that dataset. RLSeq provides the capability to compare noise analysis results from a user-supplied sample with those from publicly available datasets of the same R-loop mapping modality. This plot, termed a “noise comparison plot” and generated via the noiseComparisonPlot() function, displays the average standardized noise (“Noise index”) for public DRIP-seq samples alongside the user-supplied sample (Figure S2B). Samples are separated based on their “prediction” (from the quality model). This plot enables users to not only quantify the noise within a sample, but also see how it compares to similar samples from previous studies. From the analysis of the SH-SY5Y sample, we observe that it has a relatively low noise index, like other “POS”-predicted DRIP-seq samples (Figure S2B). This further indicates the high quality of this dataset.
3.7 Feature enrichment test
The “feature enrichment test” (invoked via the featureEnrich() function), analyses the enrichment of user-supplied peaks within various genomic features. This test can be used as a QC method because high-quality R-loop mapping peaks are statistically enriched within certain genomic features, including gene body features (e.g., exons), CpG islands, G4 quadruplexes, and regions of high G/C-skew (6). Beyond providing a test of data quality, this analysis also enables the exploration of peak enrichment within a host of genomic features, including repetitive elements, transcription factor binding sites, and cis-regulatory elements. Moreover, by comparing multiple samples analysed with featureEnrich(), users can gain initial insight into the differences between the types of R-loops mapped across experimental conditions. Once analysis is complete, the data is visualized using the plotEnrichment() function (Figure 2A).
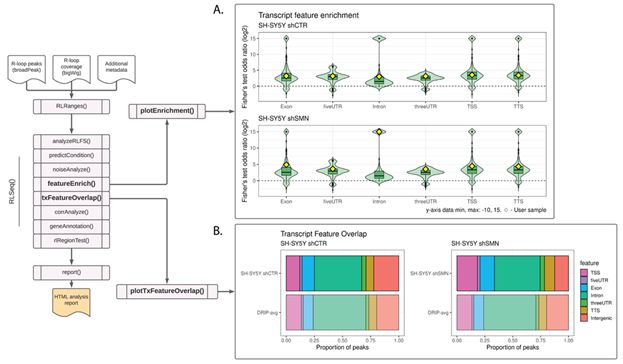
Figure 2: Feature analysis results in SH-SY5Y DRIP-seq data from shCTR (SRX2187024) and shSMN (SRX2187025) cells. (A) A feature plot showing the enrichment of RLBase samples within various transcript features. The Y-axis is the odds ratio (log2 transformed) from Fisher’s exact test. The diamonds represent the odds ratios for the user-supplied sample. The plot was generated using the plotEnrichment() function. (B) A transcript feature overlap plot that shows the proportion of user-supplied peaks overlapping transcript features. The average of all high-quality DRIP-Seq datasets is also shown for comparison. The plot was generated using the plotTxFeatureOverlap() function.
Like previous approaches, this visualization enables users to view their results in the context of similar samples from public datasets and determine whether their results diverge from previous datasets. For the example analysis in this work, we selected two DRIP-seq datasets upon which to perform feature enrichment, SH-SY5Y shCTR and SH-SY5Y shSMN (SMN knock-down) cells. The study from which these data are derived reported that knock-down of SMN (shSMN) caused an increase in intron retention [28]. Moreover, they observed an increase in R-loop formation within gene body features, specifically retained introns and exons. From our analysis of their shCTR and shSMN samples, we likewise observed an overall increase in enrichment within transcriptomic features, particularly within introns (Figure 2A), thus recapitulating the findings of the original study.
3.8 Transcript feature overlap
In addition to the more rigorous statistical analysis described above, it is also useful to broadly analyze the proportion of R-loop mapping peaks which overlap various transcriptomic features, such as exons and the transcription start site (TSS). To facilitate this type of analysis, RLSeq provides the txFeatureOverlap() function. This function finds the overlap of user-supplied peaks with transcript features and summarizes them to determine the proportion of peaks overlapping each type of feature. Finally, the results are then visualized with the plotTxFeatureOverlap() function. This visualization displays the proportion of peaks from the user-supplied sample overlapping various features (Figure 2B). Like the Feature enrichment test analysis above, we applied the Transcript feature overlap analysis to both SH-SY5Y shCTR and shSMN samples. Similar to the prior analysis, we successfully recapitulated the increase in overall gene body R-loops (Figure 2B). While we did not observe a strong increase in introns, this is not unexpected given that this style of analysis enforces a feature priority which disadvantages introns (see Methods). However, we observed an increase in Exon and TSS (Figure 2B), recapitulating the prior study’s findings [28]. In general, RLSeq is not designed for differential analysis of R-loop locations between conditions. However, the findings from the above analyses show that RLSeq is useful for initial exploration of quality and genomic feature enrichment across samples and conditions. They even recapitulated findings from the previous publication regarding the impact of SMN knock-down on R-loop localization [28]. However, deeper insights can be gained from more robust statistical approaches, and, in future work, we plan to introduce tools specifically designed for the differential analysis of R-loop abundance across conditions.
3.9 Correlation analysis
Correlation analysis (invoked via the corrAnalyze() function) is an additional quality control methodology, originally described in the recent work of Chedin et al [12]. The approach involves calculating the pairwise correlation of coverage signal between public R-loop mapping samples and a user-supplied sample [12]. The signal correlation is calculated in the genomic regions surrounding “gold-standard R-loops” (R-loops that were uncovered by single molecule, long-read, bisulfite-based R-loop mapping (SMRF-Seq) [20]. The results are visualized in a correlation heatmap, created with corrHeatmap(). This visualization provides a detailed picture of how well samples agree with one-another and how well a user-supplied sample agrees with similar previous datasets. We applied the correlation analysis method to the SH-SY5Y shCTR DRIP-seq data and plotted the resulting heatmap (Figure S3). We observed that the shCTR SH-SY5Y dataset clustered with similar positive DRIP-seq samples (Figure S3), further supporting that the dataset is of high quality.
3.10 Gene annotation
The gene annotation feature provides a convenient function for identifying the genes with which the user-supplied R-loop mapping peaks overlap. The analysis is performed via the geneAnnotation() function and yields a table of peaks and corresponding gene IDs. We applied this analysis to the SH-SY5Y shCTR data and returned the overlapping genes for each peak (Table S1).
3.11 RL-Regions test
Finally, RLSeq provides the capability to analyze the overlap between user-supplied peaks and R-loop regions (RL-Regions), regions of consensus R-loop formation uncovered in our previous work (6), via the rlRegionTest() function. These regions represent areas of the genome which show conserved R-loop formation across studies and samples. The degree to which the user-supplied peaks overlap these regions is a useful measure of quality. It also indicates the degree to which the user-supplied peaks detect non-canonical R-loops (those not typically detected in mapping experiments). The data are visualized as a Venn diagram using the plotRLRegionOverlap() function (Figure S4). From analysis of the shCTR SH-SY5Y DRIP-seq data, we observe a strong overlap with “canonical” regions of R-loop formation (Figure S4).
3.12 The RLSeq analysis report
Finally, we provide the report() function, which generates a user-friendly HTML analysis report for sharing results with colleagues (Figure 3). The report then shows all the analysis steps and visualizations along with detailed descriptions of the methods used to generate those results. Additionally, interactive data tables are provided to facilitate the exploration of results. Overall, this report provides a valuable interface for non-bioinformaticians to understand the results from RLSeq. To illustrate the utility of the HTML analysis report, we executed the report() function with the SH-SY5Y shCTR dataset (Figure 3). A web URL for this report is provided herein (see Availability).
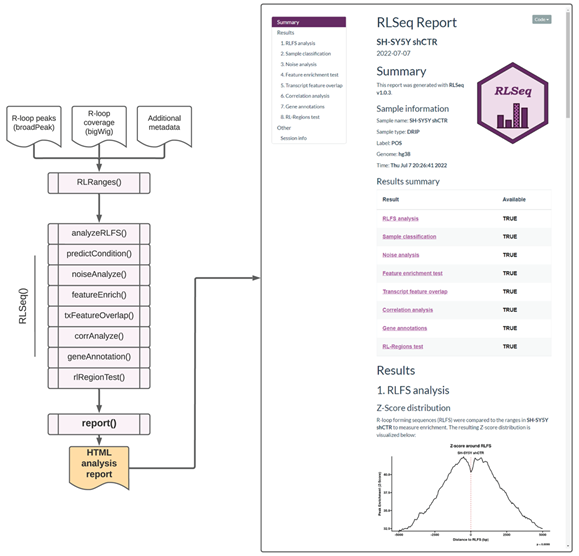
Figure 3: Screenshots of the RLSeq analysis report for SH-SY5Y DRIP-seq data (SRX2187024). The report HTML file was generated by running report() and then opened in a web browser. The figure depicts a screenshot of the overview section of the RLSeq report. This section shows the metadata from the analysis along with the list results available within the report. It also shows the RLFS analysis page, which reveals the results from permutation testing. A link to the full report is provided within this work (see Availability).
4. Discussion
RLSuite is a collection of three software packages (RLPipes, RLHub, and RLSeq) that provides an integrative workflow for R-loop data analysis. RLSuite provides users with the capability to easily process, quality control, and explore their data. Moreover, the workflow produces user-friendly HTML reports which facilitate sharing results with non-bioinformatician colleagues. Finally, by leveraging the wealth of processed data developed previously (accessible via the RLHub package), RLSeq provides users with the capability to both explore their own data and compare their results to those obtained from publicly available samples. It also offers a limited capability to compare results between samples from different biological conditions, as we demonstrated in our analysis of SH-SY5Y DRIP-seq data (Figure 2). However, a full and robust differential R-loop analysis will require additional tools not currently provided by RLSeq, and we intend for those features to be released in future versions of RLSuite. As described above, few studies have addressed the lack of R-loop-specific bioinformatics software packages. Neither DROPA [21] nor footLoop [20] provide quality control tools, the ability to compare a user-supplied sample to public datasets, nor the ability to examine peak enrichment across a wide array of annotations, as RLSeq does. They also do not provide the ability to perform upstream analysis of datasets from common R-loop mapping modalities, as RLPipes does. RLSuite thus represents the first integrative R-loop bioinformatics framework, one which should prove valuable to the field. As new R-loop datasets become available and new features are implemented, we anticipate that the utility of RLSuite will only increase throughout the coming years.
5. Conclusion
Herein we describe RLSuite, an integrative bioinformatics framework for the analysis of R-loop mapping data. RLSuite comprises three primary tools: RLPipes (an automated workflow for upstream processing of R-loop mapping datasets), RLHub (a hub for accessing processed R-loop datasets), and RLSeq (a comprehensive toolkit for the downstream analysis of R-loop mapping data). Taken together, RLSuite (RLPipes, RLSeq, and RLHub) provides a robust workflow for processing, quality control, and downstream analysis of R-loop mapping data. All three packages are accessible from open-source software repositories (Bioconda and Bioconductor) and the source code is available on GitHub (see Availability).
Declarations
Ethical approval and consent to participate
Not applicable
Consent for publication
Not Applicable
Availability of data and materials
- The scripts used to generate the figures in this study are available from GitHub: https://github.com/Bishop-Laboratory/RLSuite-Paper-Miller-2022
- The HTML analysis report for shCTR SH-SY5Y is hosted on AWS at the following URL: https://rlbase-data.s3.amazonaws.com/misc/sh-sy5y__rlseq-report.html
- The datasets generated and/or analysed during the current study are available from the public RLBase AWS repository: s3://rlbase-data/ and https://rlbase-data.s3.amazonaws.com/. The scripts used to generate these data are found in the RLBase-data GitHub repository: https://github.com/Bishop-Laboratory/RLBase-data.
- The data can be accessed via the AWS command line interface, the RLHub Bioconductor package (https://bioconductor.org/packages/release/data/experiment/html/RLHub.html), or the RLBase web database (https://gccri.bishop-lab.uthscsa.edu/rlbase/).
- RLPipes is accessible at https://anaconda.org/bioconda/rlpipes - source code: https://github.com/Bishop-Laboratory/RLPipes. RLHub is accessible at
https://bioconductor.org/packages/release/data/experiment/html/RLHub.html - source code: https://github.com/Bishop-Laboratory/RLHub. RLSeq is accessible at https://bioconductor.org/packages/devel/bioc/html/RLSeq.html - source code: https://github.com/Bishop-Laboratory/RLSeq
Competing interests
None declared.
Funding
NIH/NCI [R01CA152063 and 1R01CA241554], CPRIT [RP150445] and SU2C-CRUK [RT6187] to A.J.R.B and Greehey Graduate Fellowship Award and NIH/NIA [F31AG072902] to H.E.M. DOD [CDMRP PR181598] to K.S.
Authors’ contribution
HEM and AJRB designed the study. HEM was the primary developer on all software and wrote the manuscript. AJRB advised and managed HEM in the development of the software and manuscript. DM and SAL contributed to the software under the supervision of KS and BF. DM, SAL, KS, BF, and AJRB critically reviewed and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We want to thank Lori Kern at Bioconductor for her review of RLSeq and RLHub. We want to also thank Simon Bray at Bioconda for his review of RLPipes. We want to thank the Bioinformatics Research Network for their support in developing RLSuite.
References
- Chédin F. Nascent Connections: R-Loops and Chromatin Patterning. Trends in Genet 32 (2016): 828–838.
- Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Mol Cell 73 (2019): 398–411.
- Gorthi A, Romero JC, Loranc E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nat 555 (2018): 387-391.
- Miller HE, Gorthi A, Bassani N, et al. Reconstruction of Ewing Sarcoma Developmental Context from Mass-Scale Transcriptomics Reveals Characteristics of EWSR1-FLI1 Permissibility. Cancers (Basel) 12 (2020): E948.
- Stork CT, Bocek M, Crossley MP, et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Aguilera A, editor. eLife 5 (2016): e17548.
- Miller HE, Montemayor D, Abdul J, et al. Quality-controlled R-loop meta-analysis reveals the characteristics of R-loop consensus regions. Nucleic Acids Res (2022): 537.
- Pan H, Jin M, Ghadiyaram A, et al. Cohesin SA1 and SA2 are RNA binding proteins that localize to RNA containing regions on DNA. Nucleic Acids Res 48 (2020): 5639-5655.
- Ngo GHP, Grimstead JW, Baird DM. UPF1 promotes the formation of R loops to stimulate DNA double-strand break repair. Nat Commun 12 (2021): 3849.
- Li Y, Song Y, Xu W, et al. R-loops coordinate with SOX2 in regulating reprogramming to pluripotency. Sci Adv 6 (2020): 0777.
- Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol 21 (2020): 167-178.
- Ginno PA, Lott PL, Christensen HC, et al. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45 (2012): 814-825.
- Chédin F, Hartono SR, Sanz LA, et al. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J 40 (2021): 106394.
- Miller H, Montemayor D, Li J, et al. Exploration and analysis of R-loop mapping data with RLBase. Nucleic Acids Research (2022).
- Xu W, Li K, Li S, et al. The R-Loop Atlas of Arabidopsis Development and Responses to Environmental Stimuli. Plant Cell 32 (2020): 888-903.
- Jenjaroenpun P, Wongsurawat T, Yenamandra SP, et al. QmRLFS-finder: a model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res 43 (2015): 27-534.
- Jenjaroenpun P, Wongsurawat T, Sutheeworapong S, et al. R-loopDB: a database for R-loop forming sequences (RLFS) and R-loops. Nucleic Acids Res 45 (2017): 119-127.
- Kuznetsov VA, Bondarenko V, Wongsurawat T, et al. Toward predictive R-loop computational biology: genome-scale prediction of R-loops reveals their association with complex promoter structures, G-quadruplexes and transcriptionally active enhancers. Nucleic Acids Res 46 (2018): 7566-7585.
- Wongsurawat T, Jenjaroenpun P, Kwoh CK, et al. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res 40 (2012): 16.
- Brázda V, Havlík J, Kolomazník J, et al. R-Loop Tracker: Web Access-Based Tool for R-Loop Detection and Analysis in Genomic DNA Sequences. Int J Mol Sci 22 (2021): 12857.
- Malig M, Hartono SR, Giafaglione JM, et al. Ultra-deep Coverage Single-molecule R-loop Footprinting Reveals Principles of R-loop Formation. J Mol Biol 432 (2020): 2271-2288.
- Russo M, De Lucca B, Flati T, et al. DROPA: DRIP-seq optimized peak annotator. BMC Bioinformatics 20 (2019): 414.
- Mölder F, Jablonski KP, Letcher B, et al. Sustainable data analysis with Snakemake. F1000Res 10 (2021): 33.
- Nakato R, Sakata T. Methods for ChIP-seq analysis: A practical workflow and advanced applications. Methods 187 (2021): 44–53.
- Maintainer BP, Morgan M, Carlson M, et al. ExperimentHub: Client to access ExperimentHub resources Bioconductor version: Release 3 (2021).
- Nadel J, Athanasiadou R, Lemetre C, et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 8 (2015): 46.
- Cristini A, Groh M, Kristiansen MS, et al. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep 23 (2018): 1891-1905.
- Wang IX, Grunseich C, Fox J, et al. Human proteins that interact with RNA/DNA hybrids. Genome Res 28 (2018): 1405-1414.
- Jangi M, Fleet C, Cullen P, et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci U S A 114 (2017): 2347-2356.
- Diaz A, Park K, Lim DA, et al. Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol 11 (2012)
- Ramírez F, Ryan DP, Grüning B, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44 (2016): 160-165.
Supplemental Methods:
Preliminary data curation
Prior to data curation, we gathered basic genome information and genomic features. This included: (1) UCSC genome information, (2) R-loop forming sequences (RLFS) predictions, and (3) genomic annotations tailored for R-loop analysis. The following subsections detail the processing steps involved in obtaining these data.
Genome metadata
Prior to running the data generation pipeline, it was necessary to curate a list of all available genomes in the UCSC data repository. For each genome in UCSC, the following information was obtained via the makeAvailableGenomes.R script in the RLBase-data GitHub repository (See Code Availability):
- UCSC organism assembly ID
- Taxonomy ID
- Scientific Name
- Year that the genome assembly was introduced
- Genome length
- Gene annotation availability (TRUE/FALSE)
- Effective genome size at various read lengths (calculated via the Khmer python package).
The resulting data was then packaged for use with RLPipes and RLSeq.
R-loop forming sequences (RLFS)
R-loop forming sequences were generated via methods we described previously [1].
Genomic annotation database
We developed a custom genomic annotation database for use with RLSeq that contained annotations relevant to R-loop biology. This was accomplished via a custom R script, getGenomicFeatures.R, in the RLBase-data GitHub repository (see Code Availability).
RLPipes
RLPipes is a command-line interface (CLI) tool written in python and hosted on Bioconda [2] which is used for upstream processing of R-loop mapping data (see Code availability). It can accept FASTQ and BAM files, and it can also be used with Sequence Read Archive (SRA) or Gene Expression Omnibus (GEO) accessions. The typical workflow for using RLPipes follows the build, check, run pattern.
Build
RLPipes build is a command which processes a “sample sheet” into a configuration file which can be used directly with the underlying workflow. When used with local files (FASTQ or BAM), the build command simply verifies that the files all exist, that all the supplied arguments and options are valid, and it finds the library type (single-end or paired-end) and read length for each sample (using the pysam package [4] for BAM files and the pyfastx package for FASTQ files [4]).
If the samples supplied are SRA or GEO accessions, then the pysradb python package [5] is implemented to query the SRA database and return the following:
- The SRA experiment accession
- The SRA study accession
- The experiment title (author-submitted)
- The organism taxonomy ID
- The SRA run accession(s)
- The library layout (i.e., paired-end or single-end sequencing)
- The total number of bases in the run
- The total number of spots in the run
The organism taxonomy ID is then used to query the available genomes list (see Preliminary data preparation) and return the most recent genome assembly for the organism to be added to the standardized catalogue. Finally, the read length is calculated via floor division:
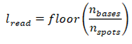
Where is the read length, is the number of bases, and is the number of spots in the sequencing run. Finally, the configuration is saved for use with downstream functions.
Check
The check operation in RLPipes is a useful tool for verifying the configuration with the underlying snakemake workflow manager [6] (equivalent of using the dry-run and dag flags in snakemake). It also provides a visualization depicting all the jobs which will be run as part of the workflow.
Run
All steps in the processing pipeline are run automatically via the Snakemake workflow manager [6] using pre-packaged conda environments. A typical analysis workflow will involve the following procedure: First, raw reads in SRA format are downloaded for each SRA run via the prefetch software from NCBI sra-tools [7]. Then, reads are converted to “FASTQ” format using fastq-dump from sra-tools [7]. Next, technical replicates are merged and interleaved (in the case of paired-end data) using reformat.sh from bbtools [8]. Then, reads are trimmed and filtered with fastp [9], generating a quality report.
For R-loop mapping data, reads are aligned to the appropriate genome using bwa mem [10] or bwa-mem2 [11], a faster implementation that users can enable with the bwamem2 flag. Then, alignments are filtered (minimum quality score of 10 enforced), sorted, and indexed using samtools [12] and duplicates are marked using samblaster (13). Then, peaks are called using macs3 [14] and coverage is calculated using deepTools [15]. Optionally, RLSeq is then called by RLPipes to automatically perform downstream analysis steps (see description below).
RLHub
RLHub is an ExperimentHub [16] R/Bioconductor package which provides convenient access to the processed datasets available in RLBase. It is available in Bioconductor v3.15 and has built-in accessor functions for quickly downloading and caching data. To generate these datasets, the prepRLHub.R script from the RLBase-data GitHub repository was executed (see Code availability). Full documentation of all objects and accessor functions provided through RLHub is provided by the RLHub reference manual (see Code Availability).
RLSeq
RLSeq is an R/Bioconductor package for downstream analysis of R-loop mapping samples. It is available via Bioconductor v3.15 and it depends upon the RLHub package. RLSeq contains three primary functions: (1) RLRanges, (2) RLSeq, (3) report. The following sections detail the functions in the RLSeq package.
RLRanges
The primary class structure used in RLSeq is the RLRanges object, initialized with the RLRanges function. This object is an extension of the GRanges class from the GenomicRanges R package (17) to provide additional validation and storage for the following slots:
- Peaks (the GRanges containing R-loop mapping peaks)
- Coverage (a URL or file path to a bigWig file)
- Mode (the type of R-loop mapping)
- Label (“POS” or “NEG”, the author-assigned data label)
- Genome (the UCSC genome ID)
- Sample name (A sample name used for visualization and reporting)
- RLResults (a list-like class for storing the results of RLSeq analysis)
Analyze R-loop forming sequences (analyzeRLFS)
R-loop forming sequences are regions of the genome with sequences that are favourable for R-loop formation [18]. They are computationally predicted with the QmRLFS-finder.py software program [19] and serve as a test of whether a sample has mapped R-loops [1]. The analyzeRLFS function provides a simple permutation testing method for analysing the enrichment of RLFS within a provided peakset. The full analysis procedure is described in detail in our recent work [1].
Predicting sample condition (predictCondition)
Following R-loop forming sequences (RLFS) analysis, the quality model is implemented for predicting the sample condition (i.e., “POS” if the sample robustly mapped R-loops and “NEG” if the sample resembles a negative control). This is accomplished with the predictCondition function which performs all the steps described in our recent work [1] to render a prediction for each sample. The results of this prediction, along with associated features and metadata, are stored in the RLResults-predictRes slot within the RLRanges object and returned to the user. For more detail, see the RLSeq reference manual (see Code availability).
Analyze noise profile (noiseAnalyze)
The noiseAnalyze function implements a modified version of the noise analysis method described by Diaz et al [20]. Briefly, 1000 random genomic bins of 1Kb in size were generated used the valr::bed_random() function. For a query sample, the coverage is summed within all bins, standardized to the highest bin value. This step was performed for all public datasets in RLBase.
Feature enrichment testing (featEnrich)
A custom list of R-loop relevant genomic annotations was curated for the human (hg38) and mouse (mm10) genomes (see Genomic annotations) and made available via the RLHub R/Bioconductor package (see RLHub annotations). In RLSeq, each annotation type is tested for enrichment within supplied RLRanges, yielding enrichment statistics. The procedure for this testing is described in detail our recent work [1].
Transcript feature overlap analysis (txFeatureOverlap)
The txFeatureOverlap function performs a simplistic overlap analysis via the following procedure: (1) peaks are overlapped with transcriptomic features obtained from RLHub using the bed_intersect function from the valr R package [21]. (2) For each peak which overlapped multiple features, a feature priority was used to assign it uniquely to a feature. The priority order is “TSS”, “TTS”, “fiveUTR”, “threeUTR”, “Exon”, “Intron”, and then “Intergenic”. For example, if a peak overlaps “TSS” and “Exon”, it will be uniquely assigned to “TSS”. (3) Once all peaks were uniquely assigned, they were saved as a table within the RLRanges object.
Correlation analysis (corrAnalyze)
The corrAnalyze function performs a sample-level correlation test that can be used to assess sample-sample similarity by calculating coverage signal (from genomic alignments) around high-confidence R-loop sites [22]. The corrAnalyze function ingests an RLRanges object (with a valid coverage slot) and performs the following procedure: (1) the coverage is quantified within the high-confidence sites and added as a column to the signal matrix (see gs_signal reference in the RLHub documentation to learn more about this matrix), and (2) then the cor function in R is used to calculate the Pearson correlation to yield a correlation matrix. The correlation matrix is saved in the RLResults-correlationMat slot of the RLRanges object and returned to the user.
Gene annotation (geneAnnotation)
The geneAnnotation function provides a simple procedure for annotating RLRanges peaks with gene IDs by overlap. Briefly, gene annotations are automatically downloaded using the AnnotationHub R package [23] and then overlapped with the ranges in the RLRanges object using bed_intersect from valr [21]. The mapping between peak IDs and gene IDs is saved in the RLResults-geneAnnoRes slot of the RLRanges object.
R-loop regions overlap test (rlRegionTest)
R-loop regions (RL regions) are R-loop consensus sites generated during the RLBase-data workflow (see R-loop Regions). The rlRegionTest function uses a simple procedure for finding RL regions which overlap with peaks in the supplied RLRanges object. It also calculates the significance and odds ratio of the overlap using Fisher’s exact test implemented via the bed_fisher function from the valr R package [21]. The results of this test are saved in the RLResults-rlRegionRes slot of the RLRanges object.
RLSeq
The primary workflow in the RLSeq package can be conveniently run in one step with the RLSeq function. This command will run, in order, (1) analyzeRLFS, (2) predictCondition, (3) noiseAnalyze, (4) featureEnrich, (5) txFeatureOverlap, (6) corrAnalyze, (7) geneAnnotation, (8) rlRegionTest. The results are saved in the corresponding RLResults slots of the RLRanges object and returned to the user.
RLSeq plotting functions
RLSeq provides a variety of useful plotting functions which summarize and present analysis results:
corrHeatmap: generates a heatmap of the correlation matrix and annotations produced by corrAnalyze using either the pheatmap [24] or ComplexHeatmap [25] R packages.
plotTxFeatureOverlap: generates a stacked bar plot showing the proportion of user-supplied peaks assigned to various transcriptomic features by the txFeatureOverlap function. In addition, it also shows the average results of running txFeatureOverlap on high-quality, publicly available R-loop mapping samples generated from the same modality as the user-supplied sample.
plotFingerprint: generates a “fingerprint plot” (based on that which was developed by Diaz et al [20] and also implemented in deepTools [26]). The plot displays the results of running the noiseAnalyze function. Specifically, the standardized proportion of maximum bin signal was ranked, and the proportion of bins is plotted against it. Higher signal to noise ratio is indicated by the degree of inflection in the plot (i.e., most signal is contained within relatively fewer bins).
noiseComparisonPlot: generates a jitter+box plot showing the average max-standardized bin signal from noiseAnalyze in the user-supplied sample and the RLBase samples of the same modality. In this visualization, lower average signal across bins indicates better signal-to-noise ratio.
plotEnrichment: generates hybrid violin-box plots showing the Fisher’s exact test odds ratio for each annotation calculated in featureEnrich. Importantly, it displays these results for the user-supplied sample alongside the distribution of results for the public samples within the RLBase database.
plotRLRegionOverlap: generates a Venn diagram showing the overlap between the user-supplied sample peaks and the R-loop regions, along with overlap statistics as calculated by the rlRegionTest function.
plotRLFSRes: plots the Z-score distribution or Fourier transform of the Z-score distribution in a metaplot, based on the analysis results from analyzeRLFS.
Report
The RLSeq report function ingests an RLRanges object which has already been processed by the analysis functions in RLSeq. It then uses RMarkdown templates to automatically build a user-friendly HTML report showcasing all results with summary tables and plots (see the RLSeq reference for an example HTML report).
References:
- Miller HE, Montemayor D, Abdul J, et al. Quality-controlled R-loop meta-analysis reveals the characteristics of R-loop consensus regions. Nucleic Acids Res 27 (2022): gkac537.
- Grüning B, Dale R, Sjödin A, et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods 15 (2018): 475–6.
- Heger A. pysam: pysam (2021). Available from: https://github.com/pysam-developers/pysam
- Du L, Liu Q, Fan Z, et al. Pyfastx: a robust Python package for fast random access to sequences from plain and gzipped FASTA/Q files. Brief Bioinform 22 (2021): bbaa368.
- Choudhary S. pysradb: A Python package to query next-generation sequencing metadata and data from NCBI Sequence Read Archive. F1000Res 8 (2018): 532.
- Mölder F, Jablonski KP, Letcher B, et al. Sustainable data analysis with Snakemake. F1000Res 10 (2021): 33.
- The NCBI SRA (Sequence Read Archive) (2021). Available from: https://github.com/ncbi/sra-tools
- Bushnell B, Rood J, Singer E. BBMerge - Accurate paired shotgun read merging via overlap. PLoS One 12 (2017): e0185056.
- Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (2018): i884–1990.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 (2009): 1754–1760.
- Vasimuddin Md, Misra S, Li H, et al. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In: 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS) (2019): 314–324.
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 (2009): 2078–2079.
- Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30 (2014): 2503–2505.
- Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9 (2008): R137.
- Ramírez F, Dündar F, Diehl S, et al. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42 (2014): 187-191.
- Maintainer BP, Morgan M, Carlson M, et al. ExperimentHub: Client to access ExperimentHub resources (2021). https://bioconductor.org/packages/ExperimentHub/
- Lawrence M, Huber W, Pagès H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9 (2013): e1003118.
- Wongsurawat T, Jenjaroenpun P, Kwoh CK, et al. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res 40 (2012): e16.
- Jenjaroenpun P, Wongsurawat T, Yenamandra SP, et al. QmRLFS-finder: a model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res 43 (2015): 527-534.
- Diaz A, Park K, Lim DA, et al. Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol 11 (2012).
- Riemondy KA, Sheridan RM, Gillen A, et al. Reproducible genome interval analysis in R. F1000Res 6 (2017): 1025.
- Chédin F, Hartono SR, Sanz LA, et al. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J 40 (2021): e106394.
- Maintainer BP, Morgan M, Carlson M, et al. AnnotationHub: Client to access AnnotationHub resources (2021). https://bioconductor.org/packages/AnnotationHub/
- Kolde R. pheatmap: Pretty Heatmaps (2019). https://CRAN.R-project.org/package=pheatmap
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32 (2016): 2847–2849.
- Ramírez F, Ryan DP, Grüning B, et al. DeepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44 (2016): 160-165.
Supplementary Figures:
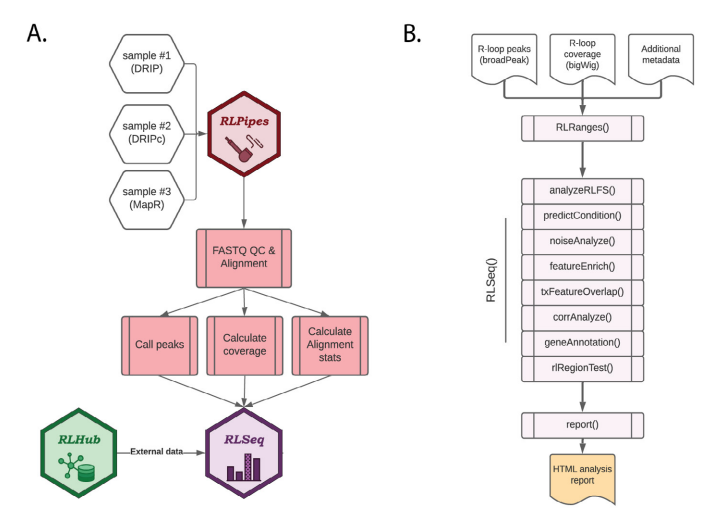
Figure S1: Overview of RLSuite workflow. (A) Flow diagram showing the upstream analysis of R-loop mapping data with RLPipes, terminating in RLSeq analysis (with supporting data from RLHub). Samples of nearly any R-loop mapping modality can be supplied to RLPipes either separately or in combination. The RLPipes workflow performs quality control on the raw read data and aligns it to the genome. Then, alignment statistics, coverage, and peaks are calculated. Finally, the RLSeq analysis workflow is implemented. (B) Flow diagram depicting the RLSeq analysis workflow. Input data are converted to an RLRanges object. In a simplified RLSeq analysis workflow, the RLSeq() function is invoked. This triggers all eight analysis steps, and the results are saved in the RLRanges object. Finally, the report() function is invoked to generate a user-friendly HTML analysis report.
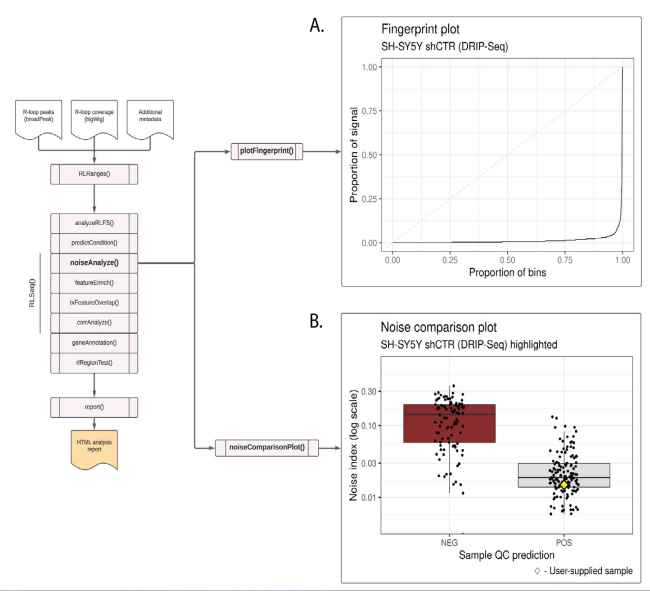
Figure S2: Noise analysis of SH-SY5Y DRIP-seq data (SRX2187024). (A) A fingerprint plot showing the proportion of signal contained in genomic bins. The strong inflection implies that most signal is contained in relatively few bins, indicating a good signal-to-noise ratio. This plot was generated using the plotFingerprint() function. (B) A noise comparison plot showing the noise index for DRIP-Seq samples in the RLBase database. The plot shows that the noise index of the user-supplied sample (SH-S5Y5 shCTR; represented by a yellow diamond) is low, further suggesting good signal-to-noise ratio. This plot was generated via the noiseComparisonPlot() function.
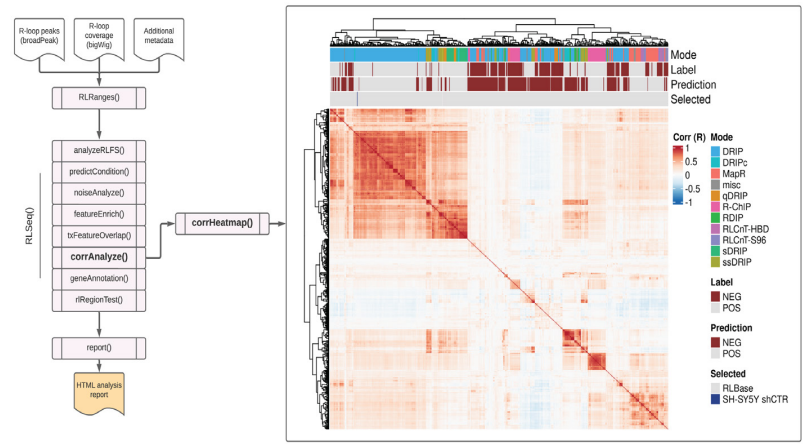
Figure S3: Correlation analysis results from SH-SY5Y DRIP-seq data (SRX2187024). A correlation heatmap showing the pairwise Pearson correlation between the human samples in the RLBase database around “gold-standard R-loops” (see Methods). The location of the user-supplied sample is shown in the “Selected” annotation row on the top of the plot. The plot was generated using the corrHeatmap() function.
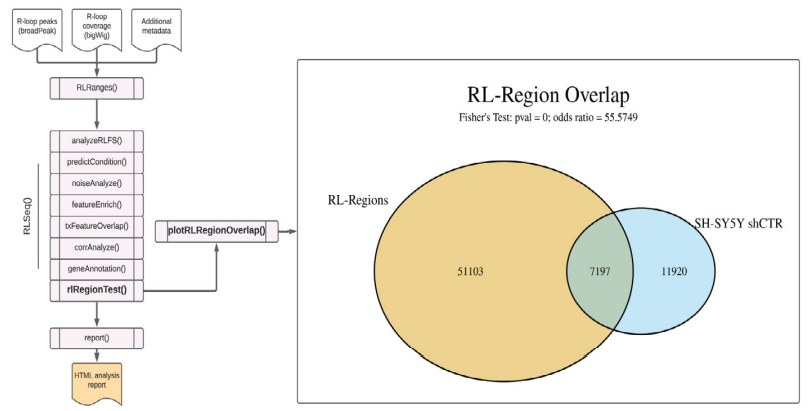
Figure S4: RL Region analysis results from SH-SY5Y DRIP-seq data (SRX2187024). A Venn diagram showing the overlap of user-supplied R-loop peaks with R-loop Regions (RL Regions), regions of conserved R-loop formation (8). This plot was generated via the plotRLRegionOverlap() function.
