Revivify Modulates Healthy Gut Microbiomes and Short Chain Fatty Acids Evaluated by an in vitro model of Gut Microbiome study
Article Information
Ashraf AHMZ1, Ahmed F Pantho2,3, Samee Kamal1, Alyssa Caba4, Syeda H Afroze2,3, Thomas J Kuehl2,3, Liaquat Hossain5, M. Nasir Uddin2,3,5,6*
1The University of Texas at Austin, Austin, 78712, TX, USA
2Orion Institute for Translational Medicine, Temple, 76504, TX, USA
3Emergent Biotechnologies LLC, Temple, 76504, TX, USA
4Texas A&M University-Central Texas, Killeen, 76549, TX, USA
5Advance Pharmaceutical Inc., Holtsville, 11742, NY, USA
6Texas A&M University School of Medicine, Bryan, 77807, TX, USA
*Corresponding Author: M. Nasir Uddin, Orion Institute for Translational Medicine, Temple, 76504, TX, USA
Received: 11 July 2023; Accepted: 19 July 2023; Published: 06 October 2023
Citation: Ashraf AHMZ, Ahmed F Pantho, Samee Kamal, Alyssa Caba, Syeda H Afroze, Thomas J Kuehl, Liaquat Hossain, M. Nasir Uddin. Revivify Modulates Healthy Gut Microbiomes and Short Chain Fatty Acids Evaluated by an in vitro model of Gut Microbiome study. Journal of Food Science and Nutrition Research. 6 (2023): 147-154.
View / Download Pdf Share at FacebookAbstract
Background: The diverse microbial community in our gastrointestinal (GI) helps in the fermentation of metabolites of Short Chain Fatty Acids (SCFAs), mainly acetate, propionate, butyrate, and small number of lactates. Acetate, propionate, and butyrate maintain colonic lining integrity and ideal colon activity. Butyrate is also known to be an energy source and assist in anti-inflammatory response. Maintaining an optimal ratio of these SCFAs reduce the risk of inflammation and promote a healthy colon. We evaluated the effect of Revivify Oral Liquid Gel (composition: US Patent 11 224 636) on gut microbiomes and SCFAs by an in vitro model of gut microbiome study. Gut microbes were cultured in 2 ml 96-well plates and treated with control, SOD, Prefibrotic fiber, Fruit juice, and Revivify gel for 48 hours followed by metaproteomic, chemical analysis (SCFA content), and microbiome profiling.
Results: We found that the amount of SCFAs increased by 2.5 folds, when treated with Revivify gel but the ratio of the SCFAs remain the same across all the treatment groups. Revivify gel promoted the growth of the Firmicutes spp., especially Lactobacillus, a probiotic organism.
Conclusions: Our study demonstrates that Revivify gel promotes a balanced increase of the SCFAs in a consistent manner, supporting a beneficial population of gut microbes for optimal gastrointestinal functionality.
Keywords
Healthy gut microbiomes, Revivify gel, Microbial metabolism
Article Details
Background
The gut microbiota is being increasingly recognized as a key factor in human health and disease [1], and as such, it is an important target for drug therapy [2]. Evidence is mounting that microbial metabolism of drugs can directly affect drug efficacy and toxicity [3,4], and that drugs can, in turn, alter the composition and function of the gut microbiota [5-9], potentially affecting the health of the host. Therefore, the gut microbial ecosystem, specific microbes, and microbial pathways are novel targets in drug discovery. In vitro culture models could be a timely and cost-efficient way to discover microbiome responses to drugs. However, current culture models do not maintain the functional and compositional profiles of the gut microbiome. To simulate an in vivo microbial ecosystem, it is key to conserve both the composition and functional activities of an individual's microbiome. In current in vitro culturing methods, profound shifts in taxa proportions have been frequently described during both batch culturing [10,11] and continuous flow culturing models [12,13] when compared to the inoculum. Importantly, shifts in microbiome composition can alter their functional properties and ecological processes [14], which could affect microbial responses to drug stimuli. To the best of our knowledge, there have been no reported models that maintain microbiome properties like the inoculum. In particular, the preservation of functional activities has not been described elsewhere. Moreover, a high percentage of marketed drugs, and compounds in development, may have off-target effects on the gut microbiome [15,16]. Maier observed that non-antibiotic drugs had extensive antibiotic-like impacts on cultured human gut bacterial strains [17]. However, the response of isolated strains may differ from that of a functionally preserved gut microbiome due to the complex functionality of a microbial community. Therefore, systematic studies of drug effects using functionally maintained individual gut microbiomes, as the in vitro model, is a pressing need. For long-term observations of xenobiotic effects, continuous flow systems [10,12,18] and microfluidic models [19,20] work well. However, these models cannot be readily adapted for high-throughput approaches, partially due to their sizes and the time required for setting up and stabilizing these bioreactors.
Gut microbes can respond to altered environmental factors within a single day [21-23]. To gain a deep insight into a microbiome's responses to drugs, a technique that can precisely quantify microbial functional activities is required. Metaproteomics is a meta-omic tool that directly quantifies the microbial functional responses at the end-product level of expressed proteins [24]. The development and application of mass spectrometry (MS)-based metaproteomic technologies in gut microbiome research has thrived in recent years. The identification coverage and sensitivity of MS-based metaproteomics have increased dramatically, enabling in-depth analysis of microbiome functional activities [25,26]. An advantage of metaproteomics when compared to sequencing-based techniques is that it can combine functional and phylogenic information. Metaproteomics is also more accurate for biomass estimates on a species level [27], and it enables the study of taxon-specific functions through annotation of unique peptides [28]. Comparison of taxon-specific functional profiles of the microbiome is important for determining cultured stability and drug responses. This is because overall functions can remain relatively stable due to the redundancy of functional genes among species in a gut microbiome [29]. As well, genes predicted from metagenomic analyses are not necessarily expressed. Therefore, metaproteomics is a suitable tool for insights into in vitro drug responses of gut microbiome.
In this study, we established a scalable in vitro model for the maintenance of gut microbiome profiles. This model adopted an optimized culture medium and a 96-deep well plate-based format for microbiome culture. The model was first evaluated for its ability to maintain gut microbiome profiles in vitro, followed by testing and evaluation of the model's in vitro correlation with in vivo drug response. This culture model enabled, in combination with metaproteomic analysis, the assessment of drug effects on the microbiome at compositional and functional levels. This model maintained high microbiome compositional stability and greater than 0.83 taxon-function similarity (Pearson's r) over 48 h. In addition, we will demonstrate that our model recapitulated the in vitro effects of prebiotic fiber agents observed on gut microbiomes.
Materials and Methods
Stool Specimen Collection and Processing
Approximately 3 g of fresh stool sample was collected from an individual using a 2.5 ml sterile sampling spoon (Bel-Art, United States). The spoon was dropped into a 50 ml Falcon tube containing 15 ml of sterile PBS pre-reduced with 0.1% (w/v) L-cysteine hydrochloride. The samples were immediately transferred into an anaerobic workstation (5% H2, 5% CO2, and 90% N2 at 37°C). Before homogenization with a vortex mixer, the tube was uncapped for a few seconds to enable gas exchange and in particular oxygen removal. Sample homogenates were filtered using sterile gauze and were immediately inoculated into each medium for static culturing at a final inoculum concentration of 2% (w/v). Culturing of an individual's microbiome was carried out in 1 ml MiPro medium. For the study, 32 replicates were cultured for each medium, allowing for 4 replicates at 8 different time points. Microbiomes were characterized at 0 (immediately after inoculation), 3, 6, 9, 12, 24, 34, and 48 h by measurements of the optical density at 595 nm (OD595), as a proxy of microbial growth and biomass, and by metaproteomic analyses.
Establishing the In Vitro Gut Microbiome Culture Model
Bacterial cells are cultured in 96-deep well and covered with a silicone-gel mat, which was perforated at the top of each well. This cover facilitates gas-exchange with the outer environment in the chamber, to preserve the partial pressure of gases and volatile metabolites in each well, which could subsequently preserve certain levels of dissolved gas molecules in the culture medium.
In Vitro Experiment Using Prebiotic Fiber Agents
The in vitro microbiome drug response was evaluated in the culture model using different prebiotic fiber agents. We hypothesize that the prebiotic fiber agent alters gut microbiota composition and functions [5,6,30]. We, therefore, employed these compounds to validate our culture model by comparing the impact of this agent exposure on gut microbial communities in our in vitro model. Briefly, the culture model was inoculated with the stool microbiome and cultured for 24 h in the presence or absence of the prebiotic fiber agents. The optimal in vitro concentration of the prebiotic fiber agents are as follows: Superoxide dismutase (SOD) (0.1 mg/ml), Prefibrotic fiber (1.3 mg/ml), Fruit Juice (0.46 mg/ml), and Revivify gel (0.56 mg/ml). For the growth study, 1 ml of each of the treatment and control cultures were collected at 0, 3, 6, 9, 12, 24, 34, and 48 hours. Cultured microbiome samples were harvested at 24 hours for metaproteomic analysis by removing 1 ml of the culture in labelled eppendorf tubes.
Maintaining a Taxon-Specific Functional Profile
To evaluate the effectiveness of culture model to simulate the in vivo features of the gut microbiome inoculum, we will use a metaproteomic approach [27,39] to characterize the taxonomic and functional stability of an individuals' gut microbiomes over 9, 24, 34 and 48 h of growth in the media. A minimum of three cultured, technical replicates will be analyzed at each time point by LC-MS/MS. Ninety high-quality MS raw files were obtained with a total of 2,066,069 MS/MS spectra. With an average MS/MS identification rate of 40.8% ± 4.5% (mean ± SD), a total of 58,848 peptides and 16,326 protein groups were identified with a false discovery rate (FDR) threshold of 1%. Bray-Curtis dissimilarity-based approach [46] will be applied for evaluating the variation of species biomass contributions between groups. All cultured microbiomes clustered closely with their corresponding inocula (0 h baseline samples). To assess the stability of functional activities, the identified proteins will be annotated with COG categories and the abundance of each COG category was calculated by summing the LFQ intensities of all the proteins belonging to the same COG category [47]. Principal component analysis (PCA) will be used to assess the relatedness of the samples based on the functional makeup of the metaproteomes. In order to assess the maintenance of taxon-specific functional traits, we carried out a taxon-function-coupled analysis using the iMetaLab platform [48].
Results
Revivify Gel Promotes the Overall Growth of Gut Microbiome
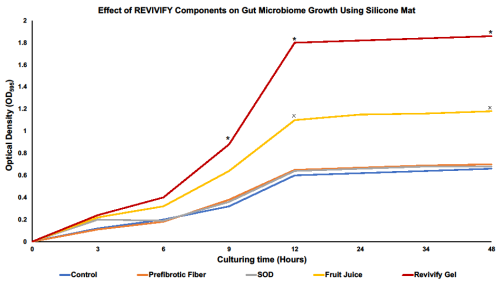
According to our 48-hour growth study, we found that starting after 9 hours of treatment, Revivify gel promotes growth of the overall gut microbiome significantly when compared to other individual prebiotic components of the gel [Figure 1]. Prefibrotic agent and SOD results in approximately an OD595 of 0.6 which is comparable to Control (no treatment) after 48 hours of growth suggesting that these two components do not provide any advantage to gut microbial growth. Cultures treated with Revivify gel has a significantly higher OD595 of 1.9 which is a 2.2-fold-change compared to Control. In addition, it appears that fruit juice, which contain polyphenols, one of the components of Revivify gel, can also significantly promote gut microbial growth after 12 hours of treatment. Therefore, these results suggest that the patented combination of the different components (Prefibrotic fibers, SOD, and Fruit juice) that is used in Revivify gel appears to have a proliferating effect on the gut microbiome.
Figure 1: Revivify gel promotes the growth of gut bacteria. Gut microbiome, when treated with Revivify gel, significantly increases growth 9 hours post treatment *(p < 0.05). Fruit juice, containing polyphenols, a component of Revivify gel also significantly promotes gut microbial growth 12 hours post treatment x(p < 0.05).
Revivify gel promotes the growth of beneficial gut bacteria
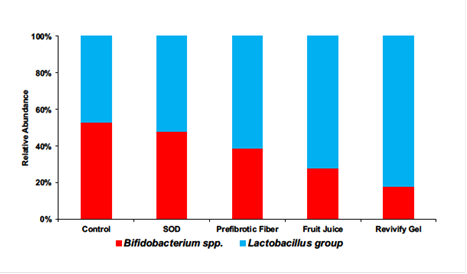
Our study further demonstrated that Prefibrotic fiber, Fruit juice, and Revivify gel promoted the growth of the Firmicutes spp., especially Lactobacillus when compared to the control (no treatment) and SOD [Figure 2]. Lactobacillus is an essential probiotic organism that promotes robust breakdown of food material and aids in the absorption of nutrients for the host. In addition, lactobacillus also ensures that opportunistic pathogenic bacteria in the gut do not outcompete the beneficial flora that may increase the risk of gastrointestinal ailments. Our microbiome profiling result shows the highest increase in lactobacillus group when the gut microbiome is treated with Revivify gel.
Figure 2: Revivify gel promotes the growth of beneficial gut bacteria, lactobacillus group (blue), while suppressing the growth of opportunistic Bifidobacterium spp. (red). Between Bifidobacterium spp. which is harbors opportunistic bacteria, and lactobacillus group, the percentage of lactobacillus is at 45% and then increases to 50%, 60%, 70% and 80% for SOD, Prefibrotic fiber, Fruit juice, and Revivify gel respectively.
Revivify gel increases short-chain fatty acids without changing their relative ratios
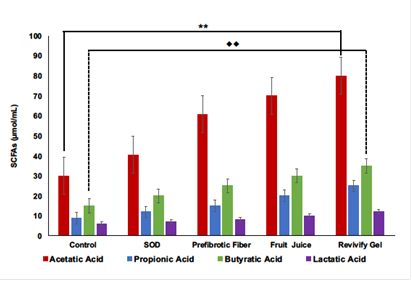
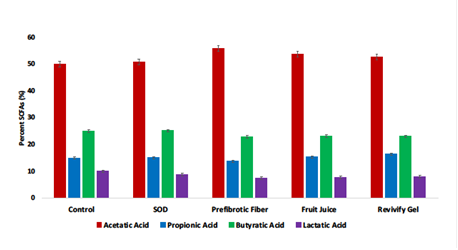
Our study shows the change in gut microbial composition [Figure 2] as well as the SCFAs when they were treated four different compounds: SOD, Prefibrotic fiber, Fruit Juice, and Revivify gel. In response to the treatment with Revivify gel, SCFAs increases significantly [Figure 3] but the ratio of the SCFAs remain the same across all the treatment group including the control [Figure 4]. This demonstrates a beneficial and consistent upregulation in the production of SCFAs when treated with Revivify gel. The concentrations of SCFAs when compared between control and Revivify gel the are as follows: acetate: 30 vs 80 µmol/ml, propionate: 9 vs 25 µmol/ml, butyrate: 15 vs 35 µmole/ml, and lactate: 6 vs 12 µmole/ml. This is a significant increase of approximately more than 2 folds compared to control. Interestingly, the ratio of SCFAs between control and each treatment group relatively remain same and it appears to be as follows: acetate (53%), propionate (15%), butyrate (24%), and lactate (8%). These data suggest that Revivify gel promotes the production of beneficial SCFAs without disrupting their relative amounts.
Figure 3: Revivify gel increases the production of Short Chain Fatty Acids (SCFAs). Revivify gel and its individual components upregulates the production of acetatic acid, propionic acid, butyratic acid, and lactatic acid in the human GI environment. When compared to control, Revivify gel significantly increases the concentrations of acetatic acid **(p < 0.05) and butyratic acid ••(p <0.05).
Figure 4: Revivify gel upregulates the production of Short Chain Fatty Acids (SCFAs) without disrupting their relative ratios. The ratio and of all four SCFAs remains the same even though Revivify gel and its other individual components increase their concentrations upon treatment as shown in figure 3.
Discussion
The gut microbiome constitutes a complex microbial ecosystem in which each microbe may play a different functional role to maintain optimal gut and human health [30]. In addition, these microbes also maintain a good proportion of beneficial species so competition does not result in opportunistic pathogens taking over [30,31]. Changes in diet and susceptibility to diseases, can trigger compositional and functional alterations in the microbiome. For example, Vallianou mentions how patients with non-alcoholic steatohepatitis (NASH) show a higher populace of Proteobacteria, Enterobacteriaceae, and Escherichia spp., while the populations of Faecalibacterium prausnitzii and Akkermansia muciniphila were reduced [32]. Although some studies have shown that the gut bacterial community can be cultured in vitro [10-12,33], these studies have not demonstrated the maintenance of the microbiota's functional activities. While taxonomic profiling is important to characterize an individual's microbiome, biological processes and functions cannot be assessed by compositional analyses only. Elucidating biological processes and functions are essential for gaining deeper understanding of the microbiota's behavior and functional activities and ultimately for designing microbiome-targeted manipulations. The need for functional understanding is even more crucial given that the microbiome's functions can be altered because of compositional changes. For example, sialylated milk oligosaccharides elicit a microbiota-dependent growth in mice, which are used as models of infant undernutrition by inducing the production of microbial metabolites, yet without any significant change in the relative abundance of most microbes [34]. Our study illustrates the overall ratio of the gut bacterial community by taxonomic class is important for the overall health of the organism. Certain imbalances in populations of microbiome populations contribute to elicit different diseases. Hills R mentions how organisms that have a higher ratio of Firmicutes to Bacteroidetes were shown to have higher trimethylamine N-oxide (TMAO) concentrations, indicating a higher risk of cardiovascular disease [35]. Different metabolites tend to shift the microbiota in different ways. To illustrate this further, Liu X [36] showed that glutamic acid appeared to decrease the population of Oxalobacter, leading to potentially causing cardiometabolic and kidney diseases later [36]. Another phylum of bacteria in the gut called Proteobacteria were changed by metabolites like 5-methyltetrahydrofolic acid, alanine, glutamate, and selenium [36]. Jeffery Ian conveys in the results how irritable bowel syndrome (IBS) patients were shown to have an increase of Firmicutes-associated taxa and a decrease in Bacteroidetes-associated taxa which is similar to the findings of Hills R [37]. Jiayu Wu mentions that obesity shows a significant decrease in the populace of Akkermansia, Faecalibacterium, Oscillibacter, and Alistipes [38].
Elucidating the microbiomes’ function requires a comprehensive quantification of the transcripts, proteins and/or metabolites produced by the microbial community. We used a metaproteomic approach [27,39] to characterize the microbiome's population changes in response to treatment with pro-fibrotic agents. To build an in vitro model to assess the microbiome response to pro-fibrotic agents, it is critical to preserve both the compositional and functional profiles of an individual's microbiome. In our in vitro model, the perforated silicone-gel cover allowed the escape of the gas produced by the gut microbiota while minimizing inward gas diffusion from the anaerobic chamber. The 96-deep well format maintained the microbiome composition. One possible explanation could be that the silicone-gel cover may have increased the partial pressure of gasses produced by the gut microbiome, such as H2, CH4, H2S, and NOX, etc. [40] and volatile organic compounds, such as SCFAs to simulate the conditions inside the human gut [41]. These gasses are very beneficial for the metabolic fermentation functions of the gut microbiota [40]. H2 is used by methanogens to produce methane, while acetogens and sulfate reducing organisms produce H2S [40]. This could preserve the levels of dissolved gas in the culture medium. Some dissolved metabolites are important factors for bacteria cross-feeding [41] and the control of pathogens [37], and thus, are presumably essential for the maintenance of an in vitro microbiome. The bacterial cross-feeding is also said to have a major impact on SCFA production and the efficient exploitation of substrates that enter the human gut [41,42]. Other factors may include differences in the surface area to volume ratio and agitation mechanics from cultures in differently shaped vessels. This model will be used for testing the effects of pro-fibrotic agents against different individual's gut microbiomes [43].
Accumulation of bacterial metabolites could be a factor that gradually alters microbiome functionality. For example, adding a bacterial metabolite, namely a secondary bile salt, induced a shift in the microbiota. These bacterial metabolites also have an impact on the pathogenesis of GI diseases like inflammatory bowel disease (IBD) [44]. Petra Louis shows that the Ruminococcus bromii (part of the Firmicutes phylum) population increased in diets rich in resistant starch (dietary starch that is not digested in the small intestine and reaches the colon) and Lachnospiraceae increased in a wheat bran-enriched diet [45]. Therefore, future work will explore whether the formation of secondary and tertiary gut bacterial metabolites during culturing affects the microbiome composition, the functionality of the microbiome, and any additional substances produced by the microbiome. To be specific, it will be important to assess whether the beneficial or harmful bacteria tend to change in amount depending on the effect of the metabolite. Further in-depth investigations on in vitro culturing and drug responses could combine meta proteomics with other omics technologies such as metagenomics and metabolomics to gain deeper insights of the microbiome composition and function. These studies could further help discover the linkages between human health and the microbiome as well as insight on specific diseases correlated with the gut microbiome.
Conclusions
In conclusion, we established the ability of this in vitro model to maintain microbial taxon-function stability and tested the utility of this model for demonstrate the effects of pro-fibrotic agents on the gut microbiome. We will optimize the medium and culture model in terms of time and concentration of pro-fibrotic agent treatment for scalable microbiome culturing in the future. We found that Revivify Oral Liquid Gel supports a beneficial population of gut microbes that can have positive influences in many age-related dysfunctions associated with metabolic, neurological, cardiovascular, and immunity. This study will provide an effective experimental platform for future drug-microbiome interaction studies. Furthermore, this study will impact the understanding of how drugs are absorbed, how the drugs affect the microbiome, and how this interaction overall affects human health.
Declarations
Ethics approval and consent to participate:
All methods were carried out in accordance with relevant guidelines and regulations.
All experimental protocols were approved by the Temple Health and Bioscience District (THBD) Institutional Biosafety Committee (IBC).
Informed consent was obtained from the volunteer participant of this study.
Consent for publication
Not applicable.
Availability of data and materials
Data will be made available upon request.
Competing interests
The authors declare that they have no competing interests.
Funding
Not applicable.
Authors’ contributions
The study proposal was conceptualized by SHA, LH, and MNU. Data curation was done by AZA, AFP, AC, SHA, TJK, and MNU. Formal analysis was performed by AZA, AFP, AC, and MNU. Investigation and methodology were done by AZA and MNU. Project administration was done by MNU. Resources were provided by LH. AZA, SK, MNU contributed to writing the manuscript and SHA, TJK and MNU helped in review and editing. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Patents
Advance Pharmaceutical Inc. has patent for the REVIVIFY gel. (U.S. Patent# 11,224,636)
References
- Kåhrström C, Pariente N, Weiss U. Intestinal microbiota in health and disease. Nature 47 (2016): 535.
- Cani PD, Delzenne NM. The gut microbiome as therapeutic target. Pharmacol Ther 130 (2011): 202-212.
- Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res 179 (2017): 204-222.
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 292 (2001): 1115.
- Wu H. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 23 (2017): 850.
- Zhang X. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep 5 (2015): 14405.
- Maccaferri S. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: an in vitro approach using a continuous culture colonic model system. J. Antimicrob. Chemother 65 (2010): 25-56.
- Xu D. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterology 146 (2014): 484.
- Morgan AP. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. Plos. One 9 (2014): e115225.
- Kim BS, Kim JN, Cerniglia CE. In vitro culture conditions for maintaining a complex population of human gastrointestinal tract microbiota. J. Biomed. Biotechnol 20 (2011): 838040.
- Long W. Differential responses of gut microbiota to the same prebiotic formula in oligotrophic and eutrophic batch fermentation systems. Sci Rep 5 (2015): 13469.
- McDonald JAK. Evaluation of microbial community reproducibility, stability, and composition in a human distal gut chemostat model. J. Microbiol. Methods 95 (2013): 167.
- Auchtung JM, Robinson CD, Britton RA. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome 3 (2015): 42.
- Costello EK, Stagaman K, Dethlefsen L, et al. The application of ecological theory toward an understanding of the human microbiome. Science 336 (2012): 1255.
- Le Bastard Q. Systematic review: human gut dysbiosis induced by nonantibiotic prescription medications. Aliment. Pharmacol. Ther 47 (2018): 332-345.
- Maier L, Typas A. Systematically investigating the impact of medication on the gut microbiome. Curr. Opin. Microbiol 39 (2017): 128-135.
- Maier L. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555 (2018): 623-628.
- Van den Abbeele P. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. Isme. J 7 (2013): 949.
- Shah P. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun 7 (2016): 11535.
- Kim HJ, Li H, Collins JJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. PNAS 113 (2016): E7.
- David LA. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 (2014): 559-563.
- Li L. Evaluating in vitro culture medium of gut microbiome with orthogonal experimental design and a metaproteomics approach. J. Proteome Res 17 (2018): 154-163.
- Faith JJ, McNulty NP, Rey FE, et al. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333 (2011): 101.
- Verberkmoes NC. Shotgun metaproteomics of the human distal gut microbiota. Isme. J 3 (2008): 179.
- Zhang X. Deep metaproteomics approach for the study of human microbiomes. Anal. Chem 89 (2017): 9407-9415.
- Zhang X. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun 9 (2018): 2873.
- Kleiner M. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun 8 (2017): 1558.
- Tanca A. Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome 5 (2017): 79.
- Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol 24 (2016) 402-413.
- Lee H. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes 9 (2017): 155-165.
- Zhao L. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359 (2018): 1151.
- Vallianou N, Christodoulatos GS, Karampela I, et al. Understanding the role of the gut microbiome and microbial metabolites in non-alcoholic fatty liver disease: Current evidence and Perspectives. Biomolecules 12 (2021): 56.
- Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol 35 (1998): 180.
- Charbonneau, Mark R. et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164 (2016): 859-871.
- Hills R, Pontefract B, Mishcon H, et al. Gut microbiome: Profound implications for diet and disease. Nutrients 11 (2019): 1613.
- Liu X, Tong X, Zou Y, et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat. Genet 54 (2022): 52-61.
- Jeffery IB, O' Toole PW, Ohman L, et al. Pyrosequencing reveals irritable bowel syndrome subtype defined by species-specific alterations in the microbial gut environment. Gastroenterology 140 (2012): 235.
- Wu J, Wang K, Wang X, et al. The role of the gut microbiome and its metabolites in metabolic diseases. Protein & Cell 12 (2020): 360-373.
- Cheng K. MetaLab: an automated pipeline for metaproteomic data analysis. Microbiome 5 (2017): 157.
- Pimentel M, Mathur R, Chang C. Gas and the microbiome. Curr. Gastroenterol. Rep 15 (2013): 356.
- Ríos-Covián D. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol 7 (2016): 185.
- Sun Y, O’Riordan MXD. in Advances in Applied Microbiology, 85th (eds. Sariaslani S. & Gadd GM.) (2013): 93-118.
- Li L. RapidAIM: A culture- and metaproteomics-based Rapid Assay of Individual Microbiome responses to drugs (2019).
- Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol 17 (2020): 223-237.
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites, and colorectal cancer. Nat. Rev. Microbiol 12 (2014): 661-672.
- The Human Microbiome Project, C. Structure, function, and diversity of the healthy human microbiome. Nature 486 (2012): 207.
- Zhang X. MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 4 (2016):
- Liao B. iMetaLab 1.0: a web platform for metaproteomics data analysis. Bioinformatics 34 (2018): 3954-3956.