Reference Genes for gene expression analysis in Head and Neck Squamous Cell Carcinoma: a Data Science Driven Approach
Article Information
Nehanjali Dwivedi1,4#, Sujan K Dhar2#, Moni A Kuriakose3, Amritha Suresh3, Manjula Das1*
1Molecular Immunology, Mazumdar Shaw Medical Foundation, 8th floor, MSMC, Narayana Health City, Bommasandra, Bangalore 560099, India
2Computational Biology, Mazumdar Shaw Medical Foundation, 8th floor, MSMC, Narayana Health City, Bommasandra, Bangalore 560099, India
3Integrated Head and Neck Oncology, Mazumdar Shaw Medical Foundation, 8th floor, MSMC, Narayana Health City, Bommasandra, Bangalore 560099, India
4Research Scholar, MAHE, Manipal 576104, India
*Corresponding Authors: Manjula Das, Molecular Immunology, Mazumdar Shaw Medical Foundation, 8th floor, MSMC, Narayana Health City, Bommasandra, Bangalore 560099, India; E-mail: manjula.msmf@gmail.com
#Equal contribution by Nehanjali Dwivedi and Sujan K Dhar
Received: 20 May 2022; Accepted 25 May 2022; Published: 17 June 2022
Citation: Nehanjali Dwivedi, Sujan K Dhar, Moni A Kuriakose, Amritha Suresh, Manjula Das. Reference Genes for gene expression analysis in Head and Neck Squamous Cell Carcinoma: a Data Science Driven Approach. Dental Research and Oral Health 5 (2022): 021-037.
View / Download Pdf Share at FacebookAbstract
Objectives
Quantitative real time PCR (qPCR) remains by far the most cost-effective, fast yet sensitive technique to check the gene expression levels in various systems. Traditionally used reference genes over the years were found to be regulated heavily based on sample sources and/or experimental conditions. This paper therefore presents a data science driven -omic approach for selection of reference genes from ~60,000 candidates from The Cancer Genome Atlas (TCGA) and Broad Institute Cancer Cell Line Encyclopaedia (CCLE) for gene expression studies in head and neck squamous cell carcinoma (HNSCC).
Materials and Methods
mRNA-sequencing data of 500 patient samples and 33 cell lines from publicly available databases were analysed to assess stability of genes in terms of multiple statistical measures. A final set of 12 candidate genes were studied in the Indian set of data in Gene Expression Omnibus (GEO) and validated experimentally using qPCR in 35 different types of samples from platforms like drug sensitive and resistant cell lines, normal and tumor samples, fibroblast and epithelial primary culture derived from HNSCC patients from India.
Results
The study lead to the choice of five most stable reference genes –TYW5, RIC8B, PLEKHA3, CEP57L1 and GPR89B across three experimental platforms.
Conclusion
In addition to providing a set of five most stable reference genes for future gene expression analysis experiments across different types of samples in HNSCC, the study also presents an objective framework for assessing reference genes for other disease areas as well.
Keywords
Mouth neoplasms, Data science, Head and Neck neoplasms, Real-Time Polymerase Chain Reaction, Gene expression
Mouth neoplasms articles; Data science articles; Head and Neck neoplasms articles; Real-Time Polymerase Chain Reaction articles; Gene expression articles
Mouth neoplasms articles Mouth neoplasms Research articles Mouth neoplasms review articles Mouth neoplasms PubMed articles Mouth neoplasms PubMed Central articles Mouth neoplasms 2023 articles Mouth neoplasms 2024 articles Mouth neoplasms Scopus articles Mouth neoplasms impact factor journals Mouth neoplasms Scopus journals Mouth neoplasms PubMed journals Mouth neoplasms medical journals Mouth neoplasms free journals Mouth neoplasms best journals Mouth neoplasms top journals Mouth neoplasms free medical journals Mouth neoplasms famous journals Mouth neoplasms Google Scholar indexed journals Data science articles Data science Research articles Data science review articles Data science PubMed articles Data science PubMed Central articles Data science 2023 articles Data science 2024 articles Data science Scopus articles Data science impact factor journals Data science Scopus journals Data science PubMed journals Data science medical journals Data science free journals Data science best journals Data science top journals Data science free medical journals Data science famous journals Data science Google Scholar indexed journals Head and Neck neoplasms articles Head and Neck neoplasms Research articles Head and Neck neoplasms review articles Head and Neck neoplasms PubMed articles Head and Neck neoplasms PubMed Central articles Head and Neck neoplasms 2023 articles Head and Neck neoplasms 2024 articles Head and Neck neoplasms Scopus articles Head and Neck neoplasms impact factor journals Head and Neck neoplasms Scopus journals Head and Neck neoplasms PubMed journals Head and Neck neoplasms medical journals Head and Neck neoplasms free journals Head and Neck neoplasms best journals Head and Neck neoplasms top journals Head and Neck neoplasms free medical journals Head and Neck neoplasms famous journals Head and Neck neoplasms Google Scholar indexed journals Real-Time Polymerase Chain Reaction articles Real-Time Polymerase Chain Reaction Research articles Real-Time Polymerase Chain Reaction review articles Real-Time Polymerase Chain Reaction PubMed articles Real-Time Polymerase Chain Reaction PubMed Central articles Real-Time Polymerase Chain Reaction 2023 articles Real-Time Polymerase Chain Reaction 2024 articles Real-Time Polymerase Chain Reaction Scopus articles Real-Time Polymerase Chain Reaction impact factor journals Real-Time Polymerase Chain Reaction Scopus journals Real-Time Polymerase Chain Reaction PubMed journals Real-Time Polymerase Chain Reaction medical journals Real-Time Polymerase Chain Reaction free journals Real-Time Polymerase Chain Reaction best journals Real-Time Polymerase Chain Reaction top journals Real-Time Polymerase Chain Reaction free medical journals Real-Time Polymerase Chain Reaction famous journals Real-Time Polymerase Chain Reaction Google Scholar indexed journals Gene expression articles Gene expression Research articles Gene expression review articles Gene expression PubMed articles Gene expression PubMed Central articles Gene expression 2023 articles Gene expression 2024 articles Gene expression Scopus articles Gene expression impact factor journals Gene expression Scopus journals Gene expression PubMed journals Gene expression medical journals Gene expression free journals Gene expression best journals Gene expression top journals Gene expression free medical journals Gene expression famous journals Gene expression Google Scholar indexed journals thyroid carcinoma articles thyroid carcinoma Research articles thyroid carcinoma review articles thyroid carcinoma PubMed articles thyroid carcinoma PubMed Central articles thyroid carcinoma 2023 articles thyroid carcinoma 2024 articles thyroid carcinoma Scopus articles thyroid carcinoma impact factor journals thyroid carcinoma Scopus journals thyroid carcinoma PubMed journals thyroid carcinoma medical journals thyroid carcinoma free journals thyroid carcinoma best journals thyroid carcinoma top journals thyroid carcinoma free medical journals thyroid carcinoma famous journals thyroid carcinoma Google Scholar indexed journals Oral Cancer articles Oral Cancer Research articles Oral Cancer review articles Oral Cancer PubMed articles Oral Cancer PubMed Central articles Oral Cancer 2023 articles Oral Cancer 2024 articles Oral Cancer Scopus articles Oral Cancer impact factor journals Oral Cancer Scopus journals Oral Cancer PubMed journals Oral Cancer medical journals Oral Cancer free journals Oral Cancer best journals Oral Cancer top journals Oral Cancer free medical journals Oral Cancer famous journals Oral Cancer Google Scholar indexed journals
Article Details
1. Introduction
Gene expression profiling by qPCR is the most cost-effective and reliable techniques for targeted profiling in in vitro, ex vivo and in vivo systems. However, quantification with reference to normalization controls (a.k.a. reference genes) to negate the inter-experimental variability caused by differences in RNA concentration or variable sample handling processes is paramount [1]. A good reference gene should have constant level of expression in various conditions [2,3]. Previously used qPCR “gold standards” have been shown to change during various cellular processes such as cell cycle, differentiation, cancer progression or by various environmental conditions such as drug exposure, hormonal therapies and chemo or radio therapies [4]. In various disease conditions expression levels of reference genes vary depending on the location [5-9], experimental conditions [10-12], the tissue type under the study [13,14] and the tumor grade [15,16]. Ever since genome-wide expression data was available through high throughput experiments like microarray, there have been many efforts to identify stable genes that could be used for normalization in qPCR experiments [2,3]. Most of the initial work [17] focussed on evaluating a set of known reference gene candidates for stability of expression using several normalization algorithms- geNorm [18], NormFinder [19] and bestKeeper [20]. This has been tried out in several contexts including papillary thyroid carcinoma [21], ovine pulmonary adenocarcinoma [22] and in serum exosome gene expression across multiple cancers pooled from public gene expression datasets [23]. Hoang et.al used geNorm and NormFinder algorithms to identify reference genes for non-melanoma skin cancers from RNA-seq data [24]. Researchers also tried assessing gene stability using bioinformatics approach [25], statistical measures like the Coefficient of Variation (CV) [26] and the difference in DNA entropies in promoters driving the expression of specific genes [27]. Yim et.al attempted to discover reference genes for expression studies in Soybean using two measures – (i) CV and (ii) p-value from a normality test assuming that the true reference genes should follow Normal distribution across samples [28]. An automated workflow called findRG [29] was proposed to find reference genes in different plant species and human cancers using CV as the primary measure. Another study on breast cancer patients [30] shortlisted genes with least variation in TCGA breast cancer dataset and further validated them using qPCR data generated from cell lines. Several studies attempted to find out pan-cancer reference genes through extensive statistical analysis of TCGA datasets through simple heuristics including CV value of the genes and unvarying expression across tumour and normal used more complex scoring systems of correlations of expression with various clinical and pathological characteristics [31-33]. Couple of recent studies (authored by SKD and MD) used a combination of publicly available gene expression data to shortlist reference genes with least variation and further validate them using qPCR data with in-house samples from a plant species [34] and from patients with haematological malignancies [35].
Efforts to identify unregulated set of genes to be used as reference genes has been evident in various cancers like lung [36], rectal [37], ovarian [38], melanoma [39], breast [30,40,41], brain [42,43], heme malignancies [35,44] and liver cancer and other malignancies [40,45-47]. Other attempts have been made to identify a panel of universal reference genes using various established cancerous cell lines [48,49]. Different studies on head and neck carcinoma have validated sets of reference genes on different platforms [50-56]. Though all the efforts focused on identification of genes with least variation across samples, a systematic data science-based approach was not found in HNSCC or oral cancer except the earlier study co-authored by AS and MAK [57] which used in-house microarray data, TCGA RNA-seq data and qPCR data of patient samples to propose a set of reference genes in HNSCC based on simple statistical considerations to shortlist genes with least variations across tumour:normal sets.. The present study focuses on validating reference genes in the cancer of the mouth, using an unbiased -omic approach in all the three major model systems of research (cell lines, patient samples and primary cultures) considering difference in origin and drug resistance. Common list of reference genes across multiple platforms will help researchers to reduce the inter-sample variability and thus arrive at an unambiguous data interpretation.
2. Materials and Methods
2.1 Gene Expression Data Acquisition
As represented in figure 1, statistical analysis for detection of reference gene candidates was carried out based upon data generated by TCGA Research Network [58] and Broad Institute CCLE project [59]. RNA-sequencing fragments per kilobase million (FPKM) values for a set of HNSCC patients (Project ID: TCGA-HNSC) were downloaded from National Cancer Institute (NCI) Genomic Data Commons Portal [60] from which the solid tumor data of 500 patients were selected. RNA-sequencing reads per kilobase million (RPKM) values of various cell lines were downloaded from the CCLE data portal [61] from which the data of 33 cell lines of upper aerodigestive tract origin were selected for analysis. Expression of candidate reference genes were verified in Indian patients from gene expression datasets in GEO. Search on GEO for co-occurrence of search terms “Oral Cancer” or “Head and Neck Cancer” and “India” resulted in nine unique datasets, out of which only two datasets (GSE23558, GSE85195) reported gene expression values from Oral cancer patient samples from India [62,63]. Data in NCBI SOFT format were downloaded from the GEO portal corresponding to the above datasets. Since both datasets used the same microarray platform (Agilent 44K, GPL6480), Log2 expression values from each dataset was merged for analysis. Altogether, both datasets had 61 tumor samples from Oral Cancer and 21 samples corresponding to precancerous lesions or normal samples. Expression data was analysed using R statistical software version 3.5.1.
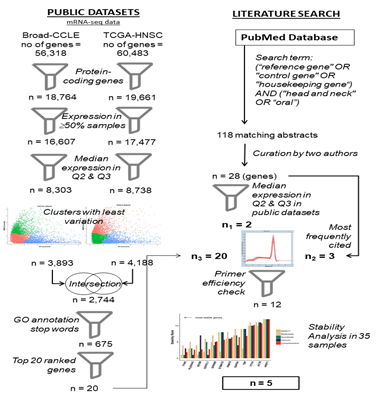
Figure 1: Work flow of the study
2.2 Statistical Analysis of RNA-seq Data
Protein coding genes with non-zero expression values in at least 50% of the samples were exclusively chosen for further analysis. For either cell line or patient dataset, genes were categorized on four standard quartiles based on their median expression value across samples. Genes in the two middle quartiles (Q2 and Q3) were shortlisted avoiding the extreme expression spectrum to enable capturing alteration in gene expression. To assess stability of a gene, two measures were adopted – (i) CV where and are mean and standard deviation of a variable x respectively and (ii) the normality p-value measured by the Shapiro-Wilks Test (p-value < 0.05 indicates the distribution to be away from Normal) [24]. CV, albeit most frequently used, is affected by outliers, and hence fails to be a robust measure. To address this, a third parameter – Median Absolute Deviation (MAD = where is the median of x) [64] was used after normalization with median. MAD is a measure of the spread of the distribution and being based on medians, is less susceptible to deviations by outliers. However, for the patient dataset, the Normality p-value calculated by Shapiro-Wilks Test is <10-4 for most genes, indicating that expression of none of the genes deviate from Normal distribution. Hence only CV and MAD were used as the two parameters for the study. An ideal set of reference genes should have low or similar statistical variation (e.g. CV and MAD) across samples. Therefore, genes were clustered based on their CV and MAD values (normalized to respective z-scores) using the PAM (Partitioning Around Medoids) algorithm originally proposed by Kaufman and Rousseeuw [65]. Optimal number of clusters required is calculated using the Silhouette graphical method of Rosseeuw [66]. For patient and cell line dataset, the cluster having the lowest medoid value for CV and MAD z-scores was selected, and the intersection between the two clusters was identified containing the genes having least CV and MAD values. This list was further pruned by programmatic parsing and eliminating genes based on stop words in their Gene Ontology (GO) annotation such as transcription factors, nuclear receptor or other nuclear localization, DNA binding activity, response to external stimuli, translational and transcriptional activation etc. since genes with such characteristics having dependency on environmental conditions evidently are unsuitable as reference genes candidates. Top 20 genes from the pruned list with least CV and MAD values were selected for and experimental validation.
2.3 Selection of Commonly Used Reference Genes
Most commonly used reference genes were shortlisted by literature based on their frequency of usage in published papers. No unique keywords were used by researchers to report studies on reference genes. Many such articles are not indexed with Medical Sub Heading (MeSH) terms so that the subheadings can be used for disease-based search. Hence a very broad methodology was adopted in which all articles in PubMed were searched for occurrence of any of the terms "reference gene" or "control gene" or "housekeeping gene" along with co-occurrence of the term "head and neck" or “oral” anywhere in the article. Obtained abstracts were manually curated by authors ND and SKD independently to find the relevant articles that described studies on reference genes specifically in the context of oral or head and neck cancer. The shortlisted 28 genes were run on CCLE and TCGA database for expression analysis for their segregation among four standard quartiles.
2.4 Design of primers
Primers were designed (Table 1; supplementary table 1) using Primer Bank Harvard [67] and IDT (Integrated DNA Technologies) by searching NCBI gene symbol for human species. Primers with amplicon size 100-150 base pairs and melting temperature 60-65°C were selected, and synthesized by Juniper Life Sciences, Bangalore, India.
2.5 Cell culture
Eleven different HNSCC cell lines were used in the study. AW13516, SCC047, HSC3, Cal27 and SCC103 were cultured in DMEM medium (Gibco, #11965092) with 10% FBS (Gibco, #10270-106) and 1X PenStrep (Gibco, #15140122). DOK required addition of 500ng/mL of hydrocortisone (Sigma, #H0888) in the basal medium, while SCC029B and SCC040 required addition of non-essential amino acids (Gibco, #111450) along with the basal medium. Cal27 resistant cell lines were cultured with appropriate drugs. Cal 27 Cis R was maintained with Cisplatin (Sigma, #P4394) at a concentration of 3.3µM, Cal 27 Dox R with Docetaxel (Sigma, #01885) at a concentration of 0.2nM and Cal 27 5FU R with 5-flourouracil (Sigma, #F6627) at a concentration of 6 µM in DMEM medium with 10% FBS and 1X PenStrep [68]. The primary cultures MhCT08 and MhCT12 were maintained in RPMI-1640 (Himedia, #AT222A) medium with 20% FBS, 1X GlutaMax (Gibco, #35050061) and 1X PenStrep ([47], manuscript under review).
2.6 Patient samples
All the samples were collected after obtaining prior consent from the patients for the study for primary cultures and RNA isolation. The project was approved by NH Ethical Committee [IRB-12/01/2009; NHH/MEC-CL-2014/216]. Study was done on retrospective samples with inclusion criteria being a matched set of adjacent normal and tumor samples from the same patient.
2.7 RNA extraction and cDNA conversion
Samples were collected in RNA later (Sigma, #R0901) and processed using MN kit (Macherey Nagel, #740955). RNA extraction for primary cultures and cell lines was done using TRIzol reagent (Ambion, #15596018) (70) and quantified using NanoDrop 2000 (Thermo Fisher Scientific) and QUBIT (Thermo, #Q10210). 1µg of total RNA was used for cDNA conversion using AMV Reverse Transcriptase enzyme (NEB, #M0277S) in a 20µl reaction as per manufacturer’s instructions.
2.8 qPCR
qPCR was done on Roche LightCycler 480 II instrument using KAPA SyBr green Universal (Sigma, #KK4600) in triplicates in clear plates with adhesive sealers. 1µl from 1:5 diluted cDNA was used in a total of 5µl reaction volume containing SyBr mix, cDNA template, primers, and water. The reaction conditions followed were – pre-incubation at 95°C for 10 seconds followed by the amplification for 45 cycles (95°C – 1 second; 95°C – 10 seconds; 60°C – 15 seconds and 72°C -15 seconds). For further analysis, primers with single melt curve peak were chosen (Supplementary Figure 1).
PCR efficiency should be taken into consideration as it can lead to bias of stable genes, if ignored [71]. For efficiency check a two-fold five-point dilution of Cal27 Parental cDNA was used as template. Thermo primer efficiency calculator was used to calculate the efficiency of primers using the equation E= 10/-1/slope [72].
2.9 Data analysis
Chosen 12 reference genes were validated across 35 different samples in triplicates. Quantification cycle (Cq) values thus obtained were subtracted by geometric mean of non-template control (NTC) to obtain DCq { Cq(sample) – Geom mean Cq(NTC)} from which the relative expression was calculated as A-DCq for each replicate, where A represents the efficiency of each primer set. Arithmetic mean of expression values of the replicates are plotted for the chosen reference genes across different samples as depicted in results.
3. Results
3.1 Statistical Analysis of RNA-seq data
Among 56,318 genes from cell lines and 60,483 genes from patient data set, 18,764 and 19,661 protein coding genes were selected, respectively. Protein coding genes with non-zero expression values in at least 50% of the samples (in cell lines 16,607 and patient samples 17,477) were exclusively chosen. After assigning the genes into standard quartiles based on median expression value, 8,303 and 8,738 genes were in middle quartiles for cell line and patient datasets, respectively. Clustering results of each dataset based on z-scores of CV and MAD are shown in figure2 (a), (b) for cell lines and patient datasets respectively. Cluster 2 from cell lines and cluster 1 from patient dataset was chosen due to minimum medoid z-score. Totally 3,893 and 4,188 genes were obtained in the selected clusters from cell line and patient dataset respectively, with 2,744 genes common between both clusters. To rank the genes within each cluster, a combined score as average of normalized values of CV and MAD was defined. Comparison of this score for each gene in the cell line and patient dataset shows that they are moderately correlated with r = 0.48 (Supplementary Figure 2). After programmatically pruning the common list of 2,744 genes based on stop-words in their GO annotation to remove DNA binding proteins or transcription factors, a list of 675 candidate reference genes was obtained, from which the top 20 candidates with least value of combined score was selected (Supplementary table 1) for primer design and experimental validation.
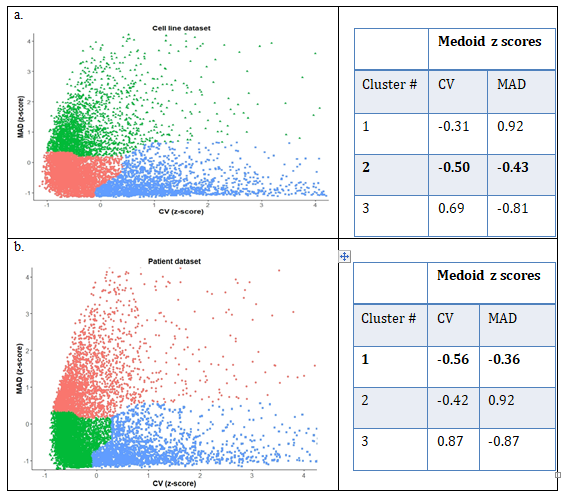
Figure 2: Clustering results for (a) Broad-CCLE cell line dataset, with genes marked in pink with least values of the parameters and (b) TCGA-HNSC patient dataset with corresponding cluster marked in green.
3.2 Selection of Commonly Used Reference Genes
PubMed search yielded a total of 118 unique abstracts which were manually curated by authors ND and SKD independently yielding 28 unique genes from 10 relevant articles. Two genes RNA18SN2 (ribosomal RNA) and MTATP6P1 (mitochondrial RNA) were not captured in TCGA/CCLE mRNA-seq experiments, hence omitted from further analysis. Median expression values of 26 genes when divided into quartiles in patient samples in TCGA (Figure 3(a)) and in cell line data sets (Figure 3(b)) yielded only two reference genes – HMBS and TBP in the middle quartiles. GAPDH, Beta Actin and HPRT were also chosen for further analysis because of their extensive literature based usage not only in head and neck cancer but in other malignancies as well (Supplementary table 2).
3.3 Selection of Primers
From the top 20 selected candidate genes from publicly available data (TCGA and CCLE), melt curve analysis (Supplementary Figure 1) yielded 11 genes with a single amplicon among which 8 genes had primer efficiencies ranging from 90-110% (data not shown). Among the 5 commonly used reference genes 4 had acceptable range of primer efficiency thus making the total number of selected candidate reference genes to 12 (Table 1).
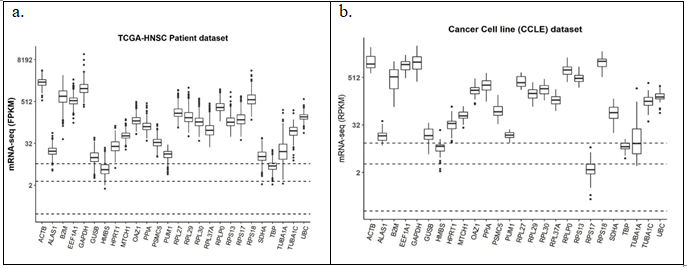
Figure 3: Expression of the commonly used reference genes in literature in (a) TCGA_HNSC patient dataset (n = 500); (b) CCLE cell line dataset (n = 33). Dashed horizontal lines from bottom represent 25%, 50% and 75% quartiles of median expression levels of genes respectively
|
HGNC Symbol |
Forward primer (5’-3’) |
Tm |
Reverse primer (5’-3’) |
Tm |
Amplicon length (bp) |
|
TYW5 |
CAGCATCAAGAGCTGCACAAA |
61.5 |
TGTGTAGGACCATTCGTCGTG |
61.8 |
102 |
|
PLEKHA3 |
ACTGTGACCTCTTAATGCAGC |
60 |
CTCAAGCGTTGTGATGAATGTG |
60.1 |
146 |
|
RIC8B |
ATAGTGTTCAACAGTCAGATGGC |
60.3 |
GCAAGCGCAAGTCAAAGCA |
62.2 |
133 |
|
CEP57L1 |
ATGAACCATCTCAGAATTGCCAT |
60 |
TCTCTCCAGCTCTAAACGATGAA |
60.5 |
137 |
|
GPR89B |
TCCGTGACGTTTGCATTTTCT |
60.8 |
GCAGTAGTCGGATATTGCTCACA |
62 |
184 |
|
STIMATE |
GCTAAGGTGTGATGAGCTAGAA |
62 |
CTCATGCAGGTCTAAGAGGAAG |
62 |
102 |
|
PRMT9 |
GACCTTGCAGACTACTGGATAAA |
62 |
CATTCCAAACCCAAGACACTAATAC |
62 |
107 |
|
GAPDH |
TCGACAGTCAGCCGCATCTTCTTT |
61.2 |
GCCCAATACGACCAAATCCGTTGA |
60.9 |
196 |
|
TBP |
CCACTCACAGACTCTCACAAC |
61.2 |
CTGCGGTACAATCCCAGAACT |
61.2 |
127 |
|
VTI1A |
GAAGAAGCGAAAGAACTGCTTG |
60 |
TAGGCGATCCGTGACCTTTTA |
60.6 |
149 |
|
ACTB |
AGCCATGTACGTTGCTATCCA |
58 |
ACCGGAGTCCATCACGATG |
59 |
120 |
|
HPRT1 |
ACCCTTTCCAAATCCTCAGC |
65 |
GTTATGGCGACCCGCAG |
67 |
125 |
Table 1: Primer sequences, melting temperature and primer efficiency of the genes evaluated in the study. The primers are arranged as per their stability (most to least stable).
3.4 Expression Behaviour of Candidate Genes in Cell Lines
Candidate reference genes when analysed in CCLE dataset (Figure 4 (a)) revealed the expression of GAPDH and Beta Actin (ACTB) to be in the 75% quartiles of median expression level which if used as reference genes will miss out most of the overexpressing genes while over-predicting the down-regulated genes. The spread of both these genes are also larger than the other genes, especially obtained from the unbiased statistical analysis, indicating variations of expression among cell lines. Similar trend was observed in the in-house data (Supplementary Figure 3) though not as pronounced due to small dataset (8 in-house samples against 33 of CCLE samples). As shown in figure 4(b) expression pattern of the candidate genes in drug resistant Cal27 cell lines showed different level of regulation, the least being in RIC8B and maximum in HPRT1. Expression patterns were checked in the established primary cultures [69]. Passage numbers did not have any effect on genes like CEP57L1 and TYW5 whereas some genes like VTI1A showed huge variation (Figure 4(c)). Epithelial and fibroblast cells from the same patient samples expressed CEP57L1 and TYW5 at equal levels whereas VTI1A was regulated (Figure 4 (d)).
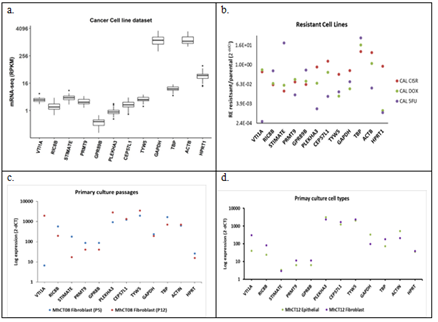
Figure 4: Expression of the candidate reference genes in (a) CCLE cell line dataset (n = 33); (b) Cal27 resistance cell lines; (c) Primary cultures at different passages - P3 vs P10; (d) Epithelial and Fibroblast cell types from the same patient. CAL CIS R - Cal 27 Cisplatin resistant; CAL DOX - Cal 27 Docetaxel resistant; CAL 5FU - Cal 27 5-Flourouracil resistant; P 3/10 - passage numbers
3.5 Behaviour of Candidate Genes in Patient Samples
Analysis of effect of tumor location in 500 TCGA dataset did not show any variation for all the 12 candidate genes (Figure 5(a)). Figure 5(b) with 44 unmatched normal showed similar profile with very high expression of GAPDH and ACTB and moderately high expression for TBP and HPRT1 in TCGA dataset. However, GEO dataset of 61 tumor samples of Indian origin threw a different light pointing out higher variation in some of the stable genes obtained from TCGA (Figure 5(c)). Fold change analysis on a total of 10 matched adjacent normal and tumor samples from the in-house repository showed almost similar variations for all genes (Figure 5(d)). All of these results indicate need of a different reference gene set in the tumor set from Indian population compared to the stable genes found in analysis of Caucasian pool from TCGA.
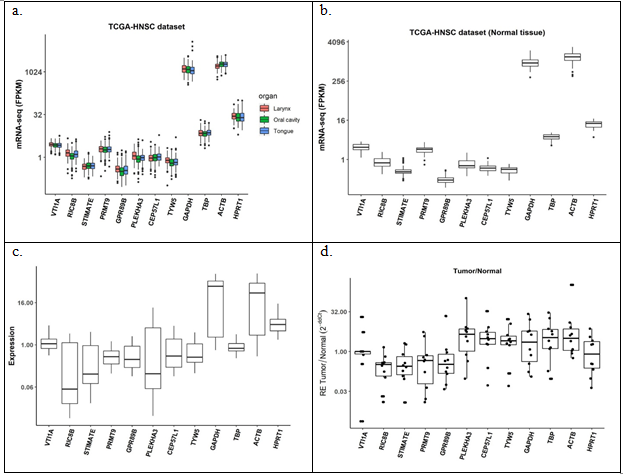
Figure 5: Expression of the candidate reference genes in (a) TCGA_HNSC tumor samples, n = 500; (b) TCGA_HNSC normal samples, n = 44; (c) GEO tumor samples, n = 61; (d) tumor over matched normal in-house samples, n = 10
3.6 Stability Analysis of candidate reference genes
Stability analysis of all 12 candidate reference genes using Cq values from all patient samples (both tumor and normal), cell lines and primary culture was carried out using RefFinder tool [73]. Geometric means of ranks obtained from both algorithms was used to rank the top 5 most stable genes – TYW5 (tRNA-yW synthesizing protein 5), PLEKHA3 (Pleckstrin homology domain containing A3), RIC8B (RIC8 guanine nucleotide exchange factor B), CEP57L1 (Centrosomal protein 57 like 1) and GPR89B (G-protein coupled receptor 89B) (Figure 6b). TYW5 functions in iron binding and the biosynthesis of a hydroxywybutosine (a hyper-modified nucleoside) in tRNA by catalysing hydroxylation [74]. RIC8B guanine nucleotide exchange factor (GEF) can activate some G-alpha proteins by changing bound GDP to free form GTP [75]. PLEKHA3 has several biochemical functions and is involved in golgi apparatus to cell surface trafficking of protein cargo [76]. CEP57L1 has been seen to be required for microtubule attachment to centrosomes [77]. GPR89B lastly is required for proper functioning of Golgi apparatus by maintaining the voltage dependent anion channel [78].
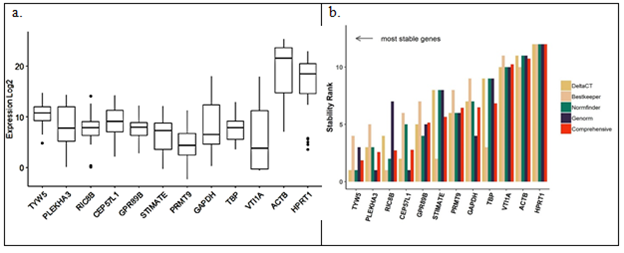
Figure 6: Stability of the candidate reference genes by (a) qPCR analysis in 35 systems (b) as analysed by RefFinder
4. Discussion and Conclusion
The expression levels of genes involved in regular functioning of a cell should remain unaltered. Considering only the functional aspect of the reference genes might lead to erroneous picks. Current study therefore offers an unbiased data science-based approach to shortlist reference genes. Most reliable reference genes should not be regulated across sample types i.e. different cell lines, tumor and normal samples originating from different locations, various primary cultures across different passages or different drug treatment. Extreme expressions of the reference genes can also result in faulty data interpretation. Thus, we have chosen a set of genes with moderate levels of expression across samples from various public databases by rigorous statistical analysis and validated experimentally under different conditions. 12 candidate genes when checked by qPCR in 35 different systems (Figure 6(a)) and subjected to RefFinder, chose 5 genes to be the most stable (Figure 6(b)): TYW5, RIC8B, PLEKHA3, CEP57L1 and GPR89B. This study employs multiple parameters such as CV and MAD to capture variations, and uses clustering approaches in the parameter space to filter out genes with least variations. This is a major improvement over similar approaches found in literature. Some of the improvements include (i) using both patient and cell line datasets to enhance applicability of reference genes in validation experiments, (ii) using a median-based variation parameter (MAD) in addition to the standard deviation based variation to make the analysis less susceptible to outliers often seen with patient samples, and lastly (iii) using PAM clustering approach to identify a set of genes eliciting similar variations. Several studies look specifically at treated cell lines [56] or archival tissues [79,80] or specific population whereas the current study has tried to arrive at a common set of genes by combining multiple platforms from multiple sources like primary culture, cell lines and patient samples of Indian origin and publicly available data of Caucasian origin. However, figure 4 displays different level of regulation in the drug resistant cell lines and/or primary culture and figure 5 points at a different type of HNSCC tumor in Indian population than the Caucasian population represented in TCGA [81]. Thus, the current study despite displaying a robust method is limited by the non-availability of sequence data of various treated cell lines and primary cultures as well as tumor and adjacent normal samples from Indian dataset to find absolute set of ‘invariant’ reference genes, if at all.
Acknowledgements
Funding for this study is provided by MSMF. The authors thank Prof Joy Kuri, Prof Haresh Dagale and Prof Chandramani Singh for critical review of the analysis procedure and Department of Electronic Systems Engineering, IISc, Bangalore for kindly providing computing infrastructure. Cell lines used for the study were procured from various institutes – SCC029B, SCC103 and SCC040 from Dr. Susanne M Gollin, University of Pittsburgh, USA; DOK from Roswell Park Cancer Institute, Buffalo, USA; Cal27 Parental from Dr. Aditi Chatterjee, Institute of Bioinformatics, Bangalore, India; HSC3 from Dr. Shumpei, Tokyo Medical and Dental University, Tokyo, Japan; SCC047 from Dr. Thomas E Carey, University of Michigan, USA and AW13516 from ACTREC, Mumbai, India. We thank all of them for their kind contribution.
Conflicts of interest
None declared
References
- Huggett J, Dheda K, Bustin S et al. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6 (2005): 279-284.
- Kozera B and Rapacz M. Reference genes in real-time PCR. J Appl Genet 54 (2013): 391-406.
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55 (2009): 611-622.
- Thellin O, Zorzi W, Lakaye B, et al.: Housekeeping genes as internal standards: Use and limits. J Biotechnol 75 (1999): 291-295.
- O’Connell GC, Treadway MB, Petrone AB, et al. Leukocyte dynamics influence reference gene stability in whole blood: Data-Driven qRT-PCR normalization is a robust alternative for measurement of transcriptional biomarkers. Lab Med 48 (2017): 346-356.
- Li M, Rao M, Chen K, et al. Selection of reference genes for gene expression studies in heart failure for left and right ventricles. Gene 620 (2017): 30-35.
- Bamias G, Goukos D, Laoudi E, et al. Comparative study of candidate housekeeping genes for quantification of target gene messenger RNA expression by real-time PCR in patients with inflammatory bowel disease. Inflamm Bowel Dis 19 (2013): 2840-2847.
- Arenas-Hernandez M, Vega-Sanchez R. Housekeeping gene expression stability in reproductive tissues after mitogen stimulation. BMC Res Notes 6 (2013): 285.
- Almeida TA, Quispe-Ricalde A, Montes de Oca F, et al. A high-throughput open-array qPCR gene panel to identify housekeeping genes suitable for myometrium and leiomyoma expression analysis. Gynecol Oncol 134 (2014): 138-143.
- Santin AP, Souza AFD, Brum LS et al. Validation of reference genes for normalizing gene expression in real-time quantitative reverse transcription pcr in human thyroid cells in primary culture treated with progesterone and estradiol. Mol Biotechnol 54 (2013): 278-282.
- Kaszubowska L, Wierzbicki PM, Karsznia S, et al. Optimal reference genes for qPCR in resting and activated human NK cells- Flow cytometric data correspond to qPCR gene expression analysis. J Immunol Methods 422 (2015): 125-129.
- Li Y, Xiang GM, Liu LL, et al. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol Med Rep 12 (2015): 4683-4691.
- Caracausi M, Piovesan A, Antonaros F, et al. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol Med Rep 16 (2017): 2397-2410.
- De Campos RP, Schultz IC, De Andrade Mello P, et al. Cervical cancer stem-like cells: systematic review and identification of reference genes for gene expression. Cell Biol Int 42 (2018): 139-152.
- Wierzbicki PM, Klacz J, Rybarczyk A, et al.: Identification of a suitable qPCR reference gene in metastatic clear cell renal cell carcinoma. Tumor Biol 35 (2014): 12473-12487.
- Romani C, Calza S, Todeschini P, et al. Identification of optimal reference genes for gene expression normalization in a wide cohort of endometrioid endometrial carcinoma tissues. PLoS One 9 (2014): e113781.
- Noriega NC, Kohama SG, Urbanski HF. Microarray analysis of relative gene expression stability for selection of internal reference genes in the rhesus macaque brain. BMC Mol Biol 11 (2010): 1-24.
- Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 (2002): 12-15.
- Andersen CL, Jensen JL and Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64 (2004): 5245-5250.
- Pfaffl MW, Tichopad A, Prgomet C et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett 26 (2004): 509-515.
- Javadirad SM, Mokhtari M, Esfandiarpour G et al. The pseudogene problem and RT-qPCR data normalization. SYMPK: a suitable reference gene for papillary thyroid carcinoma. Sci Rep 10 (2020): 1-10.
- Yang H, Zhang L, Liu S. Determination of reference genes for ovine pulmonary adenocarcinoma infected lung tissues using RNA-seq transcriptome profiling. J Virol Methods 284 (2020): 113923.
- Dai H, Yan H, Dong F, et al. Tumor-targeted biomimetic nanoplatform precisely integrates photodynamic therapy and autophagy inhibition for collaborative treatment of oral cancer. Biomater Sci 10 (2022): 1456-1469.
- Hoang VLT, Tom LN, Quek XC, et al. RNA-seq reveals more consistent reference genes for gene expression studies in human non-melanoma skin cancers. PeerJ 12 (2017): e3631.
- Beer L, Mlitz V, Gschwandtner M, et al. Bioinformatics approach for choosing the correct reference genes when studying gene expression in human keratinocytes. Exp Dermatol 24 (2015): 742-747.
- Bahr SM, Borgschulte T, Kayser KJ, et al. Using microarray technology to select housekeeping genes in Chinese hamster ovary cells. Biotechnol Bioeng 104 (2009): 1041-1046.
- Thomas D, Finan C, Newport MJ et al. DNA entropy reveals a significant difference in complexity between housekeeping and tissue specific gene promoters. Comput Biol Chem 58 (2015): 19-24.
- Yim AKY, Wong JWH, Ku YS, et al. Using RNA-seq data to evaluate reference genes suitable for gene expression studies in soybean. PLoS One 10 (2015): “e0236338”.
- Carmona R, Arroyo M, Jiménez-Quesada MJ, et al. Automated identification of reference genes based on RNA-seq data. Biomed Eng Online 16 (2017): 1-23.
- Jain N, Mitre I, Nitisa D, et al. Identification of Novel Endogenous Controls for qPCR Normalization in SK-BR-3 Breast Cancer Cell Line. Genes (Basel) 12 (2021): 1631.
- Rashid M, Shah SG, Natu A, et al. RPS13, a potential universal reference gene for normalisation of gene expression in multiple human normal and cancer tissue samples. Mol Biol Rep 48 (2021): 7967-7974.
- Krasnov GS, Kudryavtseva AV, Snezhkina AV, et al. Pan-Cancer Analysis of TCGA Data Revealed Promising Reference Genes for qPCR Normalization. Front Genet 10 (2019): 97.
- Jo J, Choi S, Oh J, et al. Conventionally used reference genes are not outstanding for normalization of gene expression in human cancer research. BMC Bioinformatics 20 (2019): 13-21.
- Smitha PK, Vishnupriyan K, Kar AS, et al. Genome wide search to identify reference genes candidates for gene expression analysis in Gossypium hirsutum. BMC Plant Biol 19 (2019): 1-11.
- Dwivedi N, Mondal S, Smitha PK, et al. Relative quantification of BCL2 mRNA for diagnostic usage needs stable uncontrolled genes as reference. PLoS One 15 (2020): e0236338.
- Yadav SM, Goyal B, Kumar R, et al. Identification of suitable reference genes in blood samples of carcinoma lung patients using quantitative real-time polymerase chain reaction. J Carcinog 19 (2020): 11.
- Vermani L, Kumar R, Senthil KN. GAPDH and PUM1: Optimal Housekeeping Genes for Quantitative Polymerase Chain Reaction-Based Analysis of Cancer Stem Cells and Epithelial-Mesenchymal Transition Gene Expression in Rectal Tumors. Cureus 12 (2020): 12.
- Yan W, Xie M, Li R, et al. Identification and Validation of Reference Genes Selection in Ovarian Cancer Exposed to Hypoxia. Onco Targets Ther 13 (2020): 7423-7431.
- Razavi SA, Afsharpad M, Modarressi MH, et al. Validation of Reference Genes for Normalization of Relative qRT-PCR Studies in Papillary Thyroid Carcinoma. Sci Rep 9 (2019): 1-11.
- Gorji-Bahri G, Moradtabrizi N, Hashemi A. Uncovering the stability status of the reputed reference genes in breast and hepatic cancer cell lines. PLoS One 16 (2021): e0259669.
- Jain N, Nitisa D, Pirsko V et al. Selecting suitable reference genes for qPCR normalization: a comprehensive analysis in MCF-7 breast cancer cell line. BMC Mol cell Biol 21 (2020): 1-19.
- Dang W, Zhang X, Ma Q, et al. Selection of reference genes suitable for normalization of RT-qPCR data in glioma stem cells. Biotechniques 68 (2020): 130-137.
- Veryaskina YA, Titov SE, Ivanov MK, et al. Selection of reference genes for quantitative analysis of microRNA expression in three different types of cancer. PLoS One 17 (2022): e0254304.
- Lee C, Lee LJ, Chong PP, et.al. Selection of reference genes for quantitative studies in acute myeloid leukaemia. The Malaysian Journal of Pathology 41 (2019): 313-26.
- Gorji-Bahri G, Moradtabrizi N, Vakhshiteh F et al. Validation of common reference genes stability in exosomal mRNA-isolated from liver and breast cancer cell lines. Cell Biol Int 45 (2021): 1098-1110.
- Ahn HR, Baek GO, Yoon MG, et al. HMBS is the most suitable reference gene for RT-qPCR in human HCC tissues and blood samples. Oncol Lett 22 (2021): 1-9.
- Badekila AK, Rai P and Kini S. Identification and evaluation of an appropriate housekeeping gene for real time gene profiling of hepatocellular carcinoma cells cultured in three dimensional scaffold. Mol Biol Rep 49 (2022): 797-804.
- Rácz GA, Nagy N, Tóvári J, et al. Identification of new reference genes with stable expression patterns for gene expression studies using human cancer and normal cell lines. Sci Rep 11 (2021): 1-14.
- Da Conceição Braga L, Gonçalves BÔP, Coelho PL, et al. Identification of best housekeeping genes for the normalization of RT-qPCR in human cell lines. Acta Histochem 124 (2022): 151821.
- Benjamin L, Evrard A, Combescure C, et al. Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Mol Biol 10 (2009) 1-10.
- Yigin AK, Cora T, Acar H, et.al. Selection of reliable reference genes for qRT-PCR analysis on head and neck squamous cell carcinomas. Biomedical Research 0970-938X (2017): 28(5).
- Song W, Li Y, Ren M, et al. Validation of reference genes for the normalization of qRT-PCR expression studies in head and neck squamous cell carcinoma cell lines treated by different chemotherapy drugs (2018).
- Rentoft M, Hultin S, Coates PJ, et al. Tubulin α-6 chain is a stably expressed reference gene in archival samples of normal oral tissue and oral squamous cell carcinoma. Exp Ther Med 1 (2010): 419-423.
- Faibish D, Suzuki M, Bartlett JD: Appropriate real-time PCR reference genes for fluoride treatment studies performed in vitro or in vivo. Arch Oral Biol 62 (2016): 33-42.
- Martin JL: Validation of reference genes for oral cancer detection panels in a prospective blinded cohort. PLoS One 11 (2016): e0158462.
- Song W, Zhang WH, Zhang H, et al. Validation of housekeeping genes for the normalization of RT-qPCR expression studies in oral squamous cell carcinoma cell line treated by 5 kinds of chemotherapy drugs. Cell Mol Biol 62 (2016): 29-34.
- Palve V, Pareek M, Krishnan NM, et al. A minimal set of internal control genes for gene expression studies in head and neck squamous cell carcinoma. PeerJ 6 (2018): e5207.
- The Cancer Genome Atlas Program - National Cancer Institute. Broad Institute Cancer Cell Line Encyclopedia (CCLE). GDC https://portal.gdc.cancer.gov/legacy-archive/search/f
- Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483 (2012): 603-607.
- Ambatipudi S, Gerstung M, Pandey M, et al. Genome-wide expression and copy number analysis identifies driver genes in gingivobuccal cancers. Genes Chromosom Cancer 51 (2012): 161-173.
- Bhosale PG, Cristea S, Ambatipudi S, et al. Chromosomal Alterations and Gene Expression Changes Associated with the Progression of Leukoplakia to Advanced Gingivobuccal Cancer. Transl Oncol 10 (2017): 396-409.
- Pham-Gia T and Hung TL. The mean and median absolute deviations. Math Comput Model 34 (2001): 921-936.
- Kaufman L and Rousseeuw PJ. Finding groups in data: an introduction to cluster analysis. Wiley (2005) https://doi.org/10.1002/9780470316801.
- Rousseeuw PJ, Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20 (1987): 53-65.
- Govindan SV aliyaveeda., Kulsum S, Pandian RS omasundar., et al.: Establishment and characterization of triple drug resistant head and neck squamous cell carcinoma cell lines. Mol Med Rep 12 (2015): 3025-3032.
- Dwivedi N, Charitha G, Pillai V, et al. Establishment and characterization of novel autologous pair primary cultures from two Indian non-habitual tongue carcinoma patients. BioRxiv (2022) https://doi.org/10.1101/2022.01.25.477260.
- Rio DC, Ares M, Hannon GJ et al. Purification of RNA using TRIzol (TRI Reagent). Cold Spring Harb Protoc 5 (2010): pdb-prot5439.
- De Spiegelaere W, Dern-Wieloch J, Weigel R, et al. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS One 10 (2015): e0122515.
- qPCR Efficiency Calculator | Thermo Fisher Scientific – IN https://www.thermofisher.com/in/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/qpcr-efficiency-calculator.html.
- Xie F, Xiao P, Chen D, et al. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80 (2012): 75-84.
- Noma A, Ishitani R, Kato M, et al. Expanding role of the jumonji C domain as an RNA hydroxylase. J Biol Chem 285 (2010): 34503-34507.
- Klattenhoff C, Montecino M, Soto X, et al. Human brain synembryn interacts with Gsα and Gqα and is translocated to the plasma membrane in response to isoproterenol and carbachol. J Cell Physiol 195 (2003): 151-157.
- Godi A, Di Campli A, Konstantakopoulos A, et al. FAPPS control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6 (2004): 393-404.
- Yu H, Tardivo L, Tam S, et al. Next-generation sequencing to generate interactome datasets. Nat Methods 8 (2011): 478-480.
- Maeda Y, Ide T, Koike M, et al. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat Cell Biol 10 (2008): 1135-1145.
- Smith TAD, AbdelKarem OA, Irlam-Jones JJ, et al. Selection of endogenous control genes for normalising gene expression data derived from formalin-fixed paraffin-embedded tumour tissue. Sci Rep 10 (2020): 1-10.
- García-Pérez O, Melgar-Vilaplana L, Córdoba-Lanús E et al. Gene Expression Studies in Formalin-Fixed Paraffin-Embedded Samples of Cutaneous Cancer: The Need for Reference Genes. Curr Issues Mol Biol 43 (2021): 2167–2176.
- Özdemir BC, Dotto GP. Racial Differences in Cancer Susceptibility and Survival: More Than the Color of the Skin? Trends in Cancer 3 (2017): 181-197.
