Recent Developments in High Spatial-Temporal Image-Based Tracking of Proteins in Subcellular Spatial Proteomics Applications
Article Information
Wenfa Ng
Department of Biomedical Engineering, National University of Singapore, Singapore
*Corresponding author: Wenfa Ng, Department of Biomedical Engineering, National University of Singapore, Singapore
Received: 09 August 2023; Accepted: 16 August 2023; Published: 22 August 2023
Citation: Wenfa Ng. Recent Developments in High Spatial-Temporal Image-Based Tracking of Proteins in Subcellular Spatial Proteomics Applications. Journal of Biotechnology and Biomedicine. 6 (2023): 294-312.
View / Download Pdf Share at FacebookAbstract
Dawn of the omics revolution in biological sciences meant that we seek to understand more and in greater detail the molecular constituents of cells and biological systems. While we have gained significant insights from conventional omics tools, we now seek to understand the spatial dimensions of the data where subcellular localisation may impact on cellular physiology and phenotype. This review paper seeks to address current questions in the new field of image-based spatial proteomics as well as outline future challenges of the field. At first glance, spatial proteomics offers enormous potential to expand our understanding of different cell types in different disease and cell states. But limitations in types of fluorophores and issues with spectral overlap significantly hampers the practical implementation of the technique. On the other hand, while we have super-resolution microscopy techniques such as STED, PALM and STORM able to achieve 10 to 20 nm spatial resolution in single molecule localisation, problems with slow image acquisition limits high temporal resolution tracking of multiple protein targets in live cell imaging. Hence, the field of spatial proteomics is a mix of promises and challenges where we could image, in multi-colour, upwards of 10 well-chosen proteins that could inform on the molecular mechanisms of selected biological processes, but, at present, the method could not tackle larger scale questions. In essence, current implementation of image-based spatial proteomics is useful, but it is unable to fulfil the mission of large-scale projects such as the Human Protein Atlas or Human Cell Atlas. Future challenges in the field includes the development of more fluorophores (especially photo switchable and photoactivable ones) for single molecule localisation microscopy, as well as seeking to improve temporal resolutions to the sub-millisecond range.
Keywords
<p>spatial proteomics, spatial resolution, temporal resolution</p>
spatial proteomics articles spatial proteomics Research articles spatial proteomics review articles spatial proteomics PubMed articles spatial proteomics PubMed Central articles spatial proteomics 2023 articles spatial proteomics 2024 articles spatial proteomics Scopus articles spatial proteomics impact factor journals spatial proteomics Scopus journals spatial proteomics PubMed journals spatial proteomics medical journals spatial proteomics free journals spatial proteomics best journals spatial proteomics top journals spatial proteomics free medical journals spatial proteomics famous journals spatial proteomics Google Scholar indexed journals spatial resolution articles spatial resolution Research articles spatial resolution review articles spatial resolution PubMed articles spatial resolution PubMed Central articles spatial resolution 2023 articles spatial resolution 2024 articles spatial resolution Scopus articles spatial resolution impact factor journals spatial resolution Scopus journals spatial resolution PubMed journals spatial resolution medical journals spatial resolution free journals spatial resolution best journals spatial resolution top journals spatial resolution free medical journals spatial resolution famous journals spatial resolution Google Scholar indexed journals temporal resolution articles temporal resolution Research articles temporal resolution review articles temporal resolution PubMed articles temporal resolution PubMed Central articles temporal resolution 2023 articles temporal resolution 2024 articles temporal resolution Scopus articles temporal resolution impact factor journals temporal resolution Scopus journals temporal resolution PubMed journals temporal resolution medical journals temporal resolution free journals temporal resolution best journals temporal resolution top journals temporal resolution free medical journals temporal resolution famous journals temporal resolution Google Scholar indexed journals analytical chemistry articles analytical chemistry Research articles analytical chemistry review articles analytical chemistry PubMed articles analytical chemistry PubMed Central articles analytical chemistry 2023 articles analytical chemistry 2024 articles analytical chemistry Scopus articles analytical chemistry impact factor journals analytical chemistry Scopus journals analytical chemistry PubMed journals analytical chemistry medical journals analytical chemistry free journals analytical chemistry best journals analytical chemistry top journals analytical chemistry free medical journals analytical chemistry famous journals analytical chemistry Google Scholar indexed journals biomedical engineering articles biomedical engineering Research articles biomedical engineering review articles biomedical engineering PubMed articles biomedical engineering PubMed Central articles biomedical engineering 2023 articles biomedical engineering 2024 articles biomedical engineering Scopus articles biomedical engineering impact factor journals biomedical engineering Scopus journals biomedical engineering PubMed journals biomedical engineering medical journals biomedical engineering free journals biomedical engineering best journals biomedical engineering top journals biomedical engineering free medical journals biomedical engineering famous journals biomedical engineering Google Scholar indexed journals biotechnology articles biotechnology Research articles biotechnology review articles biotechnology PubMed articles biotechnology PubMed Central articles biotechnology 2023 articles biotechnology 2024 articles biotechnology Scopus articles biotechnology impact factor journals biotechnology Scopus journals biotechnology PubMed journals biotechnology medical journals biotechnology free journals biotechnology best journals biotechnology top journals biotechnology free medical journals biotechnology famous journals biotechnology Google Scholar indexed journals cell biology articles cell biology Research articles cell biology review articles cell biology PubMed articles cell biology PubMed Central articles cell biology 2023 articles cell biology 2024 articles cell biology Scopus articles cell biology impact factor journals cell biology Scopus journals cell biology PubMed journals cell biology medical journals cell biology free journals cell biology best journals cell biology top journals cell biology free medical journals cell biology famous journals cell biology Google Scholar indexed journals biochemistry articles biochemistry Research articles biochemistry review articles biochemistry PubMed articles biochemistry PubMed Central articles biochemistry 2023 articles biochemistry 2024 articles biochemistry Scopus articles biochemistry impact factor journals biochemistry Scopus journals biochemistry PubMed journals biochemistry medical journals biochemistry free journals biochemistry best journals biochemistry top journals biochemistry free medical journals biochemistry famous journals biochemistry Google Scholar indexed journals microscopy articles microscopy Research articles microscopy review articles microscopy PubMed articles microscopy PubMed Central articles microscopy 2023 articles microscopy 2024 articles microscopy Scopus articles microscopy impact factor journals microscopy Scopus journals microscopy PubMed journals microscopy medical journals microscopy free journals microscopy best journals microscopy top journals microscopy free medical journals microscopy famous journals microscopy Google Scholar indexed journals molecule articles molecule Research articles molecule review articles molecule PubMed articles molecule PubMed Central articles molecule 2023 articles molecule 2024 articles molecule Scopus articles molecule impact factor journals molecule Scopus journals molecule PubMed journals molecule medical journals molecule free journals molecule best journals molecule top journals molecule free medical journals molecule famous journals molecule Google Scholar indexed journals
Article Details
Subject areas:
biochemistry, cell biology, biotechnology, analytical chemistry, biomedical engineering,
Glossary
- Diffraction limit - Theoretical and practical limit for the spatial resolution of light microscopy without using super-resolution imaging methods.
- Fluorophores - Biomolecules that exhibit fluorescence behaviour after excitation by a laser light source of defined wavelength.
- FRET - Shortform for fluorescence resonance energy transfer, which is a fluorescence-based method for measuring proximity of two labelled biomolecules or protein domains.
- Genomics - The study of the ensemble of genes in an organism using the tools of genome sequencing and bioinformatics.
- Photoactivable fluorophores - Fluorescent molecules or proteins able to exhibit fluorescence characteristics upon light irradiation through a range of bond cleavage or formation.
- Photoactivable localisation microscopy (PALM) - A super-resolution microscopy that uses repeated selective activation of selected fluorophores and imaging to stitch together a sub-diffraction limit image of the field of view of the specimen.
- Proteomics - The study of the ensemble of proteins in an organism using the tools of biochemical fractionation, mass spectrometry and other emerging techniques with data analysis support from genomics and bioinformatics.
- Point spread function - A theoretical formalism to understand the experimental observation of distribution of light intensity for light rays coming from a point source that passed through a particular objective lens.
- Super-resolution microscopy - A set of microscopy techniques that could surpass the theoretical resolution limit of traditional light microscopy.
- Spatial resolution - The limit at which features could be resolved (with clarity) in the x, y, z plane.
- Structured illumination microscopy (SIM) - A type of super-resolution microscopy where a phase mask with fine-grained gratings controls the spot size of laser excitation, and which in turns help improve the spatial resolution achievable.
- Stimulated emission depletion (STED) microscopy - A type of super-resolution microscopy that uses point spread function engineering to improve spatial resolution.
- Stochastic optical reconstruction microscopy (STORM) - A type of super-resolution microscopy with single molecule localisation capability that relies on photo switchable fluorophores for imaging at the nanoscale.
- Temporal resolution - The limit at which features could be resolved (with clarity) in the time domain.
Introduction
Biologists have long sought to understand, in aggregate, the molecular constituents of the cellular milieu, and how it impacts on phenotype and cellular behaviour that could be observed under the microscope. This christened the idea of developing methodologies and techniques to profile the aggregate ensemble of nucleic acids, proteins, metabolites, and signalling molecules within the cellular confines. In modern parlance, this is what we now know as omics where genomics profile nucleic acids, proteomics profile proteins, metabolomics profile metabolites, and interactomics profile signalling molecules.
Research in the above fields have advanced our understanding of how molecular events impacts on observable phenotypes and cellular behaviour that holds implications for how we understand cells and treat diseases. But it must be mentioned forthright that omics techniques are data and technology intensive. Take, for example, genomics. While we may think that genomics profiling and sequencing is getting cheaper, it took massive investment of manpower, time, effort, and money to get to the contemporary era where a bacterial genome is not of exorbitant cost and is accessible. Typically, modern genomics experiments generate massive amount of data that places inordinate demands on data storage and processing, the latter often requiring the support of high-performance computing. However, the possibilities that such collections of genomics information bring is bountiful. For example, not only could genomics experiment inform on the mutant variants that correlate with higher disease burden, but it could also offer insights into population patterns in evolution of resistance to diseases such as malaria and dengue fever.
This review paper focuses on proteomics, in particular, the sub-field of spatial proteomics otherwise also known as subcellular proteomics. Specifically, it sought to understand the protein localisation pattern, and protein-protein interactions pattern of proteins within the cells [1, 2]. Such profiling exercise could offer hitherto unknown insights into (i) how individual types of proteins migrate within the cells on dynamic timescale, [3 4] (ii) on the possibility of observing the migration of proteins in the protein secretion pathway, [3 4] (iii) compartmentalisation and localisation of different proteins and how it impacts on their functionalities, [5] and (iv) how different proteins interact with each other that manifests as liquid-liquid phase separation or biomolecular condensates.
While it is more common to understand proteomics in the aggregate level such as the profiling of the population-based ensemble of proteins from a given cell line, technological advances have helped realise sub-cellular proteomics. Two technologies, in particular, helped provide high throughput detection of proteins at the sub-cellular level. One of which is mass spectrometry imaging (MSI) where proteins at a location in the cells are gently ionized into a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF MS) for detection [6-8] Using raster scanning, a mass spectrometry map of the cellular proteomics at each location could be produced for unbiased and label-free detection of proteins [9 10]. This technique will not be covered in this review as it could not be used in live-cell imaging. On the other hand, fluorescence microscopy based large field of view imaging of subcellular proteins in a field known as imaging based spatial proteomics is another approach that can be used to answer the questions posed in the previous paragraph. Specifically, advent of super-resolution microscopy is a key development that enabled fluorescence microscopy based spatial proteomics that helped image clusters of proteins in protein-protein interaction networks. In addition, availability of high fluorescence, low blink rate, and lower phototoxicity fluorophores have also brought forth live cell imaging spatial proteomics that provided critical insights in protein localisation pattern and their dynamics in live cells.
Super-resolution microscopy may be the key enabling tool for contemporary spatial proteomics research, but what is the key challenge hampering real-time tracking of individual or cluster of proteins in live cells is high temporal resolution fluorescence imaging. Specifically, while major advances in low blink rate, high temporal resolution fluorescence imaging has been achieved in recent years, there remain critical technological gaps in both inexpensive and facile image acquisition and data processing. For example, high definition and high-speed fluorescence imaging necessitates the use of expensive dedicated research grade epifluorescence microscopes equipped with special filter cubes and objective lens. On the other hand, the computational task at deconvoluting the trajectories of hundreds to thousands of proteins moving in space and time within a live cell in the imaging field of view is immense and would require new advances in image processing algorithms and hardware parallel computing acceleration. Latter sections of this review will explore the above issues, questions, and future research directions in greater detail.
This review will survey recent developments that have enabled high spatiotemporal resolution image-based tracking of proteins in spatial proteomics of fixed and live cells. Topics that would be covered include: (i) technology developments that enabled high spatial temporal resolution tracking of proteins in cells, (ii) current state of the art in subcellular spatial proteomics, (iii) pushing the envelope to single molecule protein imaging with super-resolution microscopy, (iv) super-resolution microscopy methods are inherently information intensive and are slower techniques; hence, calling the need for high temporal resolution methods, (v) technology developments that can enable high temporal resolution imaging of proteins with less phototoxicity to cells in live cell imaging and are there caveats?, (vi) review on the high spatial resolution imaging of microtubule, cytoskeletal network, endoplasmic reticulum, Golgi apparatus to pinpoint the subcellular milieu, and (vii) future directions.
2. Technology developments that enabled high spatial temporal resolution tracking of proteins in cells
2.1 Evaluating the theoretical feasibility of image based spatial proteomics and what are the likely challenges and limitations in data interpretation
The nature of modern proteomics research typically involves probing the relative abundance of about 25 to 35 proteins across a population of cells. While single-cell proteomics is a recent research topic, most proteomics data are still derived from population level assays comprising at least hundreds or thousands of cells. This is important as current proteomics experiments still lack sensitivity at the level of protein abundance equivalent to tens of cells. Hence, most proteomics experiment still relies on the biochemistry route where the standard workflow involves the isolation of the cells, obtaining whole cell lysate and downstream chromatographic or electrophoretic separation, and final mass spectrometric or colorimetric and fluorometric detection of target proteins.
Image-based proteomics, on the other hand, has emerged as a viable alternative to conventional biochemical proteomics experiment due to the advent of a palette of fluorophores that afford the facile detection of different types of proteins at numbers resembling a low coverage proteomics experiment. Importantly, potential spectral overlap between fluorophores has been partially solved with narrow spectrum cut-off filters as well as fluorophores with narrow excitation and emission spectrum properties. Despite this, it is still impossible to interrogate 1000s of proteins in a cell simultaneously using the fluorometric approach. This ranks as one of the significant handicaps of image-based fluorescence tracking of proteins in cells compared to the label-free mass spectrometric imaging approach or mass cytometry approach where different proteins are labelled with a metal, and their individual masses are profiled by a mass spectrometer.
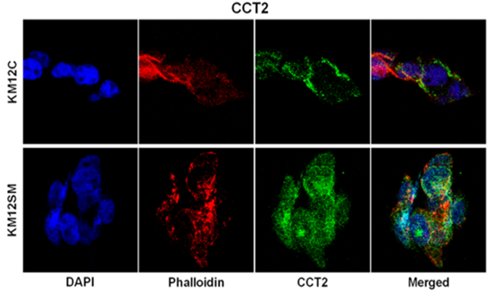
Figure 1: Confocal microscopy imaging of different colour fluorophore and merging of images from different channels. Image taken from: Marta Mendes et al., (2017), Mapping the Spatial Proteome of Metastatic Cells in Colorectal Cancer.
Early spatial proteomics efforts could only profile a couple of target proteins through either fluorescent protein fusion or antibody-fluorophores (Figure 1) with confocal laser scanning microscopy as imaging tool. As can be seen in Figure 1, there is indeed possibility of imaging different fluorophores and merging them together to obtain what is misconstrue as co-localisation maps of different proteins. Noting that conventional confocal microscopy is a diffraction limited imaging technique with spatial resolution of 200 to 250 nm; thus, each speck of fluorescence on the final image may be clusters of tens to low hundreds of labelled protein molecules. Hence, conventional confocal fluorescence microscopy could not image single molecule co-localisation pattern, and the merged images must be interpreted with caution. Specifically, in areas where there is mixing of colours of two fluorophores, the image meant that there is homogenous mixing of the two labelled proteins. Different hues would indicate different relative abundance of the two labelled proteins.
Another useful feature of image-based proteomics is the possibility of using fluorescence resonance energy transfer (FRET) to measure different conformation of proteins during association and dissociation of different proteins from each other, as well as measuring the proximity between two partner proteins in a labelled pair during protein localisation experiments. Overall, while FRET is a sensitive technique able to yield exquisite information and specificity on the protein-protein binding partner and their proximity, it nevertheless remains a low-throughput method requiring heterologous expression of the FRET biosensor via recombinant DNA technology, and judicious choice and engineering of different aspects and partner of the FRET binding pair. But, before one move into the field of sub-cellular or spatial proteomics, one must understand the theoretical possibilities of the technique. In essence, the overarching goal of spatial proteomics experiment is to use the fluorescence imaging approach to first locate as many types of proteins in the cell as possible without losing contextual information on their subcellular compartmentalisation. Naturally, such experiments require different fluorophores for each type of protein. One main approach for conjugating fluorophore to individual proteins is through fluorophore conjugated antibody that recognise the protein through partner binding. However, it must be realised that in a typical small research group, it is impossible to access the microscopy instrumentation for interrogating a large palette of available fluorophores. In addition, except for the well-known protein targets, it is currently difficult to find commercially available antibodies for each protein in the human proteome. This then set one technical limitation of the spatial proteomics approach.
Another challenge revolves around our ability to discriminate closely spaced proteins in the cellular milieu. Current limit in spatial resolution for super-resolution microscopy techniques such as stimulated emission depletion microscopy (STED) is about 50 to 60 nm. Such spatial resolution limit thus prevents the direct interrogation of protein-protein interactions except for large aggregates of different proteins. It also means that current instrumentation and optical techniques may not access the single molecule/protein limit except in the case of single molecule localisation microscopy. In this case, the spatial resolution limit would be 10 to 20 nm, which may allow the visualisation of 1:1 protein-protein interaction at a very high imaging and computational cost, which will be elaborated later in the review. In terms of temporal resolution, high speed imaging requires sufficient amount of light to be collected, which indirectly limits the temporal resolution of the experiment. Except in cases where a highly fluorescent fluorophore is used, it is hard to decouple the trade-off between high temporal resolution and high spatial resolution, where improvement in one means poorer resolution for the other. Current scientific CMOS detectors could reach 2000 x 2000 pixels imaging with 100 frames per second, which means a 10-millisecond temporal resolution. What does this mean in terms of protein dynamics and conformational flexibility?
Electron paramagnetic resonance is one important contemporary technique for probing protein dynamics at the domain level. Using Cu (II) labelling approach, it has been determined that protein conformational dynamics occur at the pico-second (bond rotation) to nano-second (domain movement) level [11]. In another approach, two-dimensional fluorescence lifetime correlation spectroscopy was used to assess the conformation dynamics of cytochrome C between its three reported conformational states, with results indicating micro-second conformation dynamics for the protein [12]. Overall, it is difficult for current commercially available image detector to probe the conformational dynamics of proteins at the micro-second level. More importantly, inability to image the fluorescence from single protein meant that the fluorescence image must be interpreted as population-based averages.
2.2 What is the super-resolution microscopy method that enabled spatial proteomics?
The optical diffraction limit has hindered the exploration of molecular processes in living cells due to the inability to discriminate molecules or cellular features beyond the diffraction limit of 200 to 250 nm. However, extensive research over the past 3 decades in various areas of optics engineering and chemical biological assembly of fluorescence molecules have progressively unveiled the mysteries of the sub-diffraction limit to the modern light microscope equipped with super-resolution optics and specially designed fluorophores. Known as super-resolution microscopy, the field of sub-diffraction limit light and fluorescence microscopy has blossomed in the past 15 years with the effervescent of increasingly refined and sophisticated techniques able to probe the sub-cellular space with tens of nanometres (20 to 60 nm) spatial resolution [13]. Approaches that could yield useful sub-diffraction limit molecular information include stimulated emission depletion (STED) microscopy, photoactivated localisation microscopy (PALM), and stochastic optical reconstruction microscopy (STORM).
In STED microscopy, careful use of two laser excitation light beams help realise point spread function (PSF) engineering that enabled reduce PSF of the central excitation spot, which is where visualisation of the molecular details is implemented. Examples of STED use in spatial proteomics include the visualisation of protein trafficking in plant cell vesicles,4 localisation of cross-linked functionalized proteins in mitochondria [14] visualization of transmembrane proteins at the neuronal synapse [13] and understanding the nanoscale localisation of proteins of the silt diaphragm of the kidney glomerular unit [15]. Overall STED microscopy has afforded biologists an unprecedented view of the subcellular localisation of different labelled proteins, which opened our understanding of how differences in protein localisation pattern in different subcellular microcompartments could impact on pathobiology and disease. However, STED is an expensive technique, and is not readily accessible to most bioimaging researchers. On the other hand, there are two related single molecule localisation microscopy techniques: PALM (photoactivated localisation microscopy) and STORM (stochastic optical reconstruction microscopy) that have revolutionized the field of single molecule localisation microscopy (SMLM). Photoactivable fluorophores and new approaches to reconstructing time-resolved fluorescence images are critical enabling tools for SMLM. In general, SMLM uses computational approaches to deduce the localisation of single molecule in the field of view. By repeated imaging the same area with short temporal resolution, it is possible to use computational reconstruction to obtain an overall image of the sample.
Hence, what is the achievable resolution of the PALM and STORM? Typically, the spatial resolution would be around 20 to 60 nm depending on the size of the fluorophore used, its brightness, and the microscope optics. In a typical spatial proteomics experiment, it would be necessary to interrogate the relative abundance and localisation of about 6 to 10 proteins. Given that it is necessary to label each protein with a fluorophore of different colour, a successful spatial proteomics experiment using SMLM technique depends on the availability of appropriate antibodies conjugated to the correct fluorophore. More importantly, imagine multi-colour imaging of protein-protein interactions in close proximity in a subcellular compartment, it would be necessary to choose two fluorophores with small point spread function so that their Airy disk do not overlap too much during imaging in order to computationally reconstruct the localisation of the two fluorophores (and proteins) with reasonable level of accuracy. What is even more critical to realise is that SMLM is a slow technique as it takes many frames of the same field of view to reconstruct the overlapped image of the different fluorophores dotting the specimen. Imaging time to obtain a sampling level appropriate to the Nyquist Shannon sampling theorem would depend on the relative abundance of the protein, size of specimen, and magnification level. Firstly, relative abundance of proteins affects the ease of detecting both proteins, particularly those that co-localise to the same locale in the cell. Specifically, high abundance protein would deliver a stronger signal to the detector compared to a lower abundance protein with fluorophore of similar fluorescence emission intensity. Hence, it may be more difficult to detect the lower abundance protein, and with stronger fluorescence and higher number of photons reducing the size of the point spread function in SMLM, spatial resolution that could be achieved is dependent on the amount of fluorescence signal and, by extension, abundance of protein. Naturally, lower fluorescence signal would require a longer imaging time with more images (frames) to be taken.
Specimen size is an important factor particularly for microscopy systems that rely on raster scanning such as laser confocal scanning microscopy. In SMLM, activation of fluorophores is random, which meant that a significant number of frames is needed to obtain fluorescence signal from fluorophore that line, for example, a cell membrane. Hence, the number of frames (and time needed) scales with the size of the specimen. Whether the scaling relationship is linear or non-linear would depend on the distribution of the fluorophore per unit area. If the scaling is linear such as the case for fluorophore that illuminate receptors on the cell membrane, the dependence of imaging time would be directly proportional to the specimen size. On the other hand, if the fluorophore is detecting a lysosomal protein or enzyme, then the scaling would be non-linear and, by inference, the dependence of imaging time for the fluorophore on specimen size would be non-linear. Finally, magnification level affects the level of details available in the final image, as well as the amount of light and fluorescence collected. In general, the higher the magnification level, the greater the number of details that needs to be captured to render visible the target object such as an organelle, which means that more image pixels need to be captured in the final reconstructed image. Using this logic, the imaging time would also increase.
2.3 Advent of high refresh rate, high speed camera system for tracking fast protein dynamics
One of the important questions that biologists wish to answer concerns the migration and trafficking of proteins in the cellular milieu. Whether as part of the pathway for modifying synthesized proteins to yield the final version with desired post-translational modifications or for understanding the movement of proteins in and out of subcellular compartment as part of cellular response or defence, tools are needed for tracking protein movements. Such tools came in the form of different types of dye-based fluorophore, some of which are photoactivable. In the case of photoactivable fluorophores, biologists now have the tools to reversibly activate and switch off fluorophores, which provide better control over the imaging process and allow more high-quality data to be obtained over the useful lifetime of the fluorophores, some of which could be very expensive.
But fluorophores could not implement tracking of protein dynamics without the advent of modern high refresh rate and high-speed camera and detector systems. One such high speed imaging detector for microscopy is scientific CMOS (sCMOS), where, in each pixel element, there is a dedicated signal amplifier and analog-to-digital converter. This differs significantly from previous generation electron multiplying charged coupled device (EMCCD), which is slower in readout because there is only one amplifier and analog-to-digital converter for each segment of pixels. One caveat of high speed or high temporal resolution imaging is that the time available for exposure is shorter, which would limit the spatial resolution and penetration depth achievable [16]. In the case of spatial resolution, higher imaging speed reduces the signal obtained, which increases the size of the point spread function (PSF) as the ratio between signal and noise degrades. On the other hand, high speed imaging that comes with high-speed camera also reduces penetration depth as the amount of time available for photons in the depths of the camera to travel to the detector is shorter.
2.4 Development and discovery of fluorophores with low blink rate and less vulnerability to photobleaching and more photostable
Spatial proteomics experiments aim to obtain wide-field high content images of the distribution, relative abundance and localisation of different proteins at the subcellular and compartmental level. To do this, fluorophores must have low phototoxicity, more photostability, and less vulnerability to photobleaching. Utility of each of the above requirements will be explained in detail in the following paragraphs. Low phototoxicity is a key requirement for live cell imaging as some dye-based fluorophores exert a toxicological effect on live cells. Once the phototoxicity effect crosses a threshold, cellular metabolism, cellular processes, and physiology are affected, which will result in imaging artefacts and erroneous interpretations. Does low phototoxicity equals low quantum yield or reduced fluorescence emission? The answer is no. This comes about because there are a variety of chemistries available to engineer a high quantum yield dye-based fluorophore. What is important is to do careful selection of functional groups on the fluorophore dye to reduce its cytotoxicity effect on cells. Such selection is increasingly possible given our expanding knowledge base of the roles of different functional groups in fluorophores that exert toxicity effects on cells. Another requirement is the need for higher photostability and less vulnerability to photobleaching in fluorophores for long-duration or time-lapse imaging. In general, higher photostability allows a consistent level of fluorescence emission, which lends the data obtained suitable for quantitative spatial proteomics where the relative abundance of different proteins in the subcellular space could be estimated. It also allows stable tracking of individual proteins across the subcellular space to gain a deeper understanding of the migratory dynamics and behaviour of different proteins. Finally, higher photostability means a longer imaging duration which affords longer term study of cellular adjustments or regulatory control after a perturbation.
2.5 Development of fast, efficient fluorophore conjugation methods that does not affect protein function and molecular diffusion significantly.
In a typical biological experiment, the researcher aims to obtain a readout of the cellular processes and phenotype, whether at the macroscopic or microscopic level, without disturbing cellular processes at the genetic or protein level. This requirement may not be fully fulfilled in most biological experiments in the contemporary era. But we are getting better with reducing the amount of disturbance that we cause to the cells when we manipulate them to obtain signal readout using either microscopy methods or more modern instrumentation. To date, microscopy methods remain the least intrusive method for signal readout at the cellular level. Expression of fluorescence protein fusion is not discussed here as it requires genetic engineering, and is quite intrusive, and may result in phenotypic artefacts that can be misconstrued as genuine biological phenomena.
To put the above discussion in context for the field of spatial proteomics would mean that we need efficient method to label (or conjugate) the fluorophores to the protein. Conjugation should be facile, fast and does not need significant chemical biological tools or expose the cells to toxic chemicals. One popular approach is the use of click chemistry, which is biorthogonal and relatively easy to implement given that commercial kits are available. However, click chemistry reactions require the use of Cu(I) as catalyst, and this may introduce some toxicity to cells. By far, the most commonly used labelling technique for fluorophores in cells is the antibody-based technique, which is also known as immunofluorescence. While this approach is well-documented and research, there is significant difficulty and cost involved in developing an antibody tag specific to the protein of interest. Thus, researchers in the field would need to rely on protein targets that are well researched in order to procure antibodies that could target these proteins of interest. However, there are also other more direct methods for labelling protein of interest with fluorophore. For example, one more conventional method is direct reaction with side chains of native amino acids. While useful, this approach often yields heterologous products, making quantification of the fluorescence signal in the cellular context difficult [17]. An evolution of this approach is the more recent single site and dual site, site-specific protein labelling approach that allows more reproducible labelling, which translates into more predictable quantitative readout of fluorescence signal [18]. One example is the use of sortase-mediated ligation where engineered improved variants of sortase enzymes could enhance the kinetics of attaching fluorophores to the N or C-terminus of heavy or light chains of antibodies [19].
3. Current state of the art in subcellular spatial proteomics
3.1 Current paradigms in the imaging approach to spatial proteomics
As mentioned in the Introduction, there are two main approaches for pursuing spatial proteomics experiments: mass spectrometry-based, and fluorescence imaging-based. In general, the mass spectrometry-based approach to spatial proteomics inquiry is more mature and yields larger dataset, where there are increasing number of artificial intelligence and machine learning tools for data analysis and visualisation. On the other hand, fluorescence imaging-based approach is less well developed due in large part to its need for labelling as well as our current inability to introduce too many fluorophores for target proteins without significant spectral overlap. Current state-of-the-art in fluorescence imaging based spatial proteomics could detect up to approximately 100 different proteins [20].
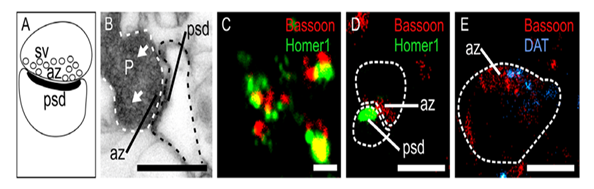
Figure 2: Comparison in localizing proteins in different microscopy modes ranging from a) theoretical, b) electron microscopy, c) confocal microscopy, d) dSTORM, e) localization of proteins as small specks of light in single molecule localization microscopy. Image taken from: Andras Gabor Miklosi et al., (2018), Resolution Matters: Correlating Quantitative Proteomics and Nanoscale-Precision Microscopy for Reconstructing Synapse Identity.
Before delving into the imaging details of spatial proteomics, it is important to appreciate the conceptual underpinnings of the imaging process. In Figure 2, a series of theoretical and real images from different microscopy techniques serve to showcase our progress towards single molecule localisation microscopy needed in spatial proteomics applications. For example, in Figure 2a, the theoretical problem of imaging the localisations and potential protein-protein interactions of two types of proteins was showcased. Figure 2b reveals an electron microscopy perspective of the localisations of the two types of protein. Blurred images from conventional confocal microscopy depicted in Figure 2c reveals the difficulty of achieving single molecule resolution in conventional point-scanning fluorescence microscopy. Improved images were obtained with dSTORM as shown in Figure 2d. Finally, Figure 2e showed achievement of single molecule localisation that revealed the interactions and distribution of the two types of proteins.
With the large amount of protein localisation and spatially resolved relative abundance data in spatial proteomics experiments, there is a call for more cell biologists to use spatial proteomics approaches in their enquiries [20]. In particular, subcellular single cell proteomics offer a glimpse into protein localisation and dynamics [21]. However, a key bottleneck in fluorescence imaging spatial proteomics is the inability to simultaneously probe more than 10 different proteins in a single imaging experiment without incurring exorbitant cost and running into serious technical challenges requiring the support of senior subject matter experts. Imaging based subcellular spatial proteomics could provide cellular level compartmentalisation readout of protein abundance and localisation, which provides new insights on non-genetic based cellular heterogeneity [22]. One alternative method for performing fluorescence imaging based spatial proteomics is through detecting mRNA transcripts which, under certain circumstances, could serve as proxies for the relative abundance of particular proteins. In this approach, DNA probes with fluorophores are used to profile a mixture of mRNA transcripts to detect specific mRNA transcripts of target proteins. Upon hybridisation of the DNA probes with target mRNA transcripts, there would be a detectable fluorescence signal. One example is a study on how DNA-barcoded fluorescence microscopy could be used for spatial proteomics via fluorescence microscopy imaging [23].
The large and multivariate datasets that emanates from either mass spectrometry or fluorescence imaging based spatial proteomics experiments call for sophisticated data analysis tools to extract the maximum amount of information and knowledge available in the multidimensional datasets. One popular contemporary approach is the use of machine learning and artificial intelligence methods. Of more significance is that machine learning has become essential to analyse the large datasets from either mass spectrometry or imaging-based subcellular spatial proteomics studies [24]. It is anticipated that there will be future innovations in the application of machine learning and artificial intelligence tools in fluorescence based spatial proteomics. Finally, it must be remembered that current spatial proteomics approaches are not a cure-all for detecting important pathological states of diseases. Although it is commonly presumed that pathological states in the cells could be due to mislocalisation of proteins or proportioning of wrong types of proteins to wrong subcellular compartments, this paradigm may shift with our expanding knowledge of how protein misfolding or mutant protein variants partake in the disease process. Such processes may not be adequately surveyed by current mass spectrometry or imaging-based spatial proteomics approaches. Hence, while imaging and mass spectrometry-based techniques for surveying the subcellular proteomes exists, there remains important limitations in current technologies for understanding the full dimension and repercussions of human disease pathologies [25].
3.2 Development of molecular probes and tools
A couple of techniques, both biochemical and cell biology (imaging)-based exists to characterize the spatial proteomes of cells and tissues [26]. A review article in 2003 describes foundational fluorescence imaging technologies that underpins current capabilities in using fluorescence microscopy for multiplex analysis of protein-protein interactions [27]. While significant achievements have been made, in situ subcellular spatial proteomics for single cells has not been achieved [28]. One challenge is the design of probes for specific target proteins. A new study outlines a computational algorithm and software that allow facile design of DNA sequence probes for targeted proteins [28]. Another approach, albeit indirect method, is to use fluorescence in situ hybridisation (FISH) approach to probe the type and location of mRNA that encodes proteins during translation. For example, fluorescence in situ hybridisation based techniques have been used to characterize the tumour microenvironment [29]. But, caution must be exercised as existence of mRNA need not mean that the protein would ultimately be produced in similar proportion as the relative abundance of the detected mRNA transcripts. The above is a brief summary of the technological trends in spatial proteomics approach which there is a detectable shift from detecting protein to detecting the mRNA that encode the protein. Such a shift potentiates the development of mRNA probes as described above. Consensus has not been reached in the spatial proteomics field on whether the shift towards detecting mRNA will take hold. On the other hand, a lot of the cutting-edge knowledge on development of protein conjugation probes is in the biotechnology companies and remain out-of-reach of the publicly accessible scientific literature.
3.3 Technology limit on number of proteins probed.
Perhaps, one of the first demonstrations of a subcellular proteomics approach was the study seeking to use green fluorescent protein fusion to study the localisation pattern of enzymes of different lipid biosynthetic pathways in Saccharomyces cerevisiae [30]. In more recent times, with a modest budget, it is now possible to probe a panel of 24 biomarkers in lung function to help characterize the relative abundance of different constituents and follow the progression of a lung pathological condition. Specifically, a study has used in situ spatial proteomics to assess the abundance of normal constituents (alveolar type I and II, bronchial epithelia, endothelial, muscular, stromal and hematopoietic cells), and to follow lung fibrosis over an extended period [31]. Additionally, recent research using spatial proteomics approach in assessing the pathophysiological state of colorectal cancer cells show that a panel of 52 hyperplexed immunofluorescence probes could usefully predict clinical outcomes for patients [32].
Fluorescence correlation spectroscopy is perhaps the first high throughput subcellular spatial proteomics approach. Specifically, in one study, 53 nuclear proteins labelled with appropriate antibody based fluorophore were interrogated with fluorescence spectroscopy, yielding a high dimensional dataset.33 Overall, 60000 measurements were made in 10000 cells that affords new data on which insights on cell cycle dependent fluctuations in protein abundance was observed [33]. Using a similar approach, another research team also successfully imaged the co-localisation of hundreds of proteins using cycles of fluorescence tagging and imaging [34. Current multiplexed immunofluorescence imaging spatial proteomics approaches are still limited to 20 to 100 proteins [10]. Hence, spatial proteomics efforts have been limited to a relatively small subset of well-chosen protein targets important for discerning physiological and metabolic effects of the experiment system. In addition, choice of fluorophore conjugation method for imaging-based spatial proteomics is critical to experiment success. A study examining this issue for the ensemble of proteins in mammalian cells revealed 80% concordance in the subcellular localisation of proteins detected via immunohistochemistry (fluorophore conjugated to antibody), and fluorescent protein conjugation method, suggesting general applicability of both approaches in routine spatial proteomics experiments [35].
To put the numbers in perspective. Technologies exists for subcellular mapping of Saccharomyces cerevisiae spatial proteomes at the 1000s of proteins level using a combination of subcellular organelles isolation and isotope tagged mass spectrometry detection and analysis [36]. But, mass spectrometry-based profiling and imaging-based proteomics could be used in combination, for example, to gain a holistic view of protein trafficking and migration within and between vesicles and the larger trans-Golgi network or in between different compartments of a plant cell [4]. This trend is gaining momentum in the field as researchers are increasingly using both mass spectrometry, and high content imaging to address both broad scale and granular questions in the field [37]. Specifically, the large field of view of fluorescence imaging approach allows the tackling of broad questions, while the details inherent in the mass spectrometry approach affords granular insights to be discerned.
3.4 Type of microscopy methods for spatial proteomics
Super-resolution microscopy is one potential solution for the current challenge of gaining a more refined understanding of protein-protein interactions at the molecular level. One effort in this direction is the development of a super-resolution microscopy tool for mapping the proximity radii of proximity labelling platforms in spatial proteomics [38]. While the conventional notion is to improve spatial resolution in order to more accurately map out the localisation of different proteins at the nanometre level, there is also the possibility of using the newer inverse technique of expansion microscopy to map out the distribution and localisation of proteins in a spatially resolved manner. Specifically, expansion microscopy relies on the use of swellable polymer to expand the tissue sample in an isotropic manner, which pulls the different proteins and biomolecules further apart. This then allows the previously closely associated proteins to be better resolved spatially and aids in identification. In microscopy terms, the previously overlapping Airy or point spread functions of closely associated proteins are well separated and resolved after the use of swellable polymer in expansion microscopy, and this aids fluorescence imaging. For example, a 330 µm tissue sample comprising 260 cells could be expanded to 1 mm in size, and allows the detection of 655 proteins using a conventional optical microscope [39]. Thus, use of expansion microscopy techniques allow more conventional microscopes to be utilized in spatial proteomics experiments. Current implementation of expansion microscopy technique in spatial proteomics should allow the interrogation of protein-protein interaction partners, as well as the relative abundance and localisation of different proteins in the original size sample.
4. Application of machine learning in spatial proteomics
Figure 3: General concept of applying machine learning to aid large-scale data analysis in spatial proteomics experiments. Image obtained from: Minjie Mou, (2022), Application of Machine Learning in Spatial Proteomics.
Prior to the name, spatial proteomics, the effort to pinpoint the localisation of proteins through fluorescence microscopy is known as location proteomics. Back in 2004, there were already fairly advanced efforts to use artificial intelligence (i.e., neural network and machine learning) approaches to derive more understanding of wide-field fluorescence microscopy images of the subcellular milieu [40]. As another example, developing a set of membrane biomarkers for segmenting individual cells in spatial proteomics studies is important for single cell profiling as well as probing the interactions between different cell types. A recent effort in this direction sought to use convolutional neural network (CNN) to identify the optimal set of biomarkers for segmenting different cell types [41]. In general, as depicted in Figure 3, machine learning and artificial intelligence tools aids in analysing the wealth of data from spatial proteomics experiments, and help answers protein subcellular localisation questions.
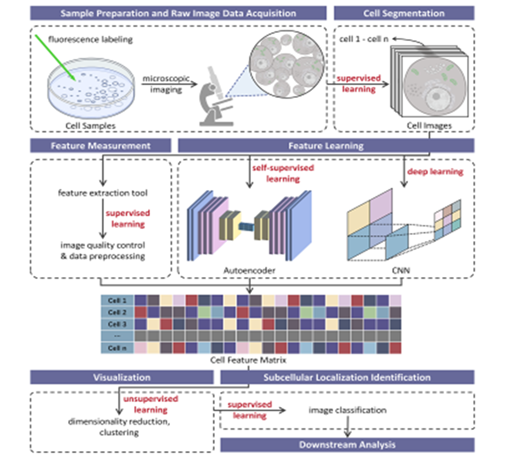
Figure 4: Schematic diagram showing the main workflow steps in applying modern machine learning concepts to analyse spatial proteomics dataset from fluorescence microscopy. Image obtained from: Minjie Mou, (2022), Application of Machine Learning in Spatial Proteomics.
Typically, the workflow from applying machine learning and neural networks approaches to analysing imaging-based spatial proteomics experimental data involves data preprocessing to normalize for effects of contrast and brightness. This is followed by image segmentation (Figure 4) using a supervised (training) approach. Deep learning and self-supervised learning via convolutional neural networks come next to process and teased the data apart in different dimensions to reveal hidden signals in the data such as protein abundance and localisation. Finally, the features of the original datasets extracted by the deep learning neural network is extracted and send to a binning and classifier for final data display as a map to reveal protein relative abundance, protein-protein interaction partners, and protein spatial localisation pattern.
Data from spatial proteomics experiment can be highly heterologous as there are efforts to use both mass spectrometry-based and immunofluorescence imaging-based methods to access the subcellular proteome. A study has shown how transfer learning models based on nearest neighbours and support vector machines can potentially help bring together disparate datasets of different data structures to help glean hitherto inaccessible insights into the spatial proteomics [42]. Method has been validated by its use in uncovering and annotating unknown proteins in pluripotent mouse embryonic stem cells [42]. What emerges from the above analysis is that the disparate datasets emanating from spatial proteomics experiments conducted in either the mass spectrometry or fluorescence imaging paradigm require reconciliation to help maximise the extraction of insights from otherwise very expensive experimental approaches [43]. Requirements for data analysis approaches as well as statistical tools needed are outlined in a recent review paper [43]. An example of this trend in increasing the robustness of data reporting in the spatial proteomics field is the use of relative weighted consistency and area under the curve in reporting the protein signatures from different organelles and micro-compartments in the cell [44].
5. Pushing the envelope to single molecule protein imaging with super-resolution microscopy
5.1 Is that possible with our understanding of optics and the methods we have for manipulating light of different wavelengths?
The goal in spatial proteomics is to identify as many proteins as possible in the cellular milieu as well as determine their localisation. To do this, it is best to achieve single molecule resolution in locating the protein’s location. Current super-resolution microscopy methods such as PALM and STORM does have the ability to achieve this resolution limit. The target resolution for observing a single hydrated protein molecule is about 5 nm, and the best resolution possible with PALM and STORM methods is about 10 to 20 nm. Hence, putting things into context, it appears that current single molecule localisation microscopy methods could achieve close to single molecule detection of many types of proteins, except for the small proteins of length less than 50 amino acid residues. Such capabilities have afforded us with unprecedented ability to interrogate the chemical information and protein concentration of a cell at the subcellular and compartmental level. Overall, the conclusion is that the current state of the art in single molecule protein imaging allows us to view a cluster of a few protein molecules (2 to 6 molecules) as a single point spread function.
5.2 What is spatial proteomics at 10 to 20 nanometre resolution? Can it be used to interrogate protein-protein interactions in liquid-liquid phase separation or membraneless organelles?
The current resolution limit for single molecule localisation microscopy via the PALM and STORM approach is about 10 to 20 nm. So, what does this means for spatial proteomics experiment? Specifically, could we use two different fluorophores of different colours to image single protein binding to another protein molecule? Answers to this question requires an understanding of the current direction of the spatial proteomics field. Reading through the contemporary literature of this young field reveals that researchers are still trying to expand the types of fluorophores that could be used in spatial proteomics, as well as understanding the fluorescence images obtained for wide field high resolution spatial proteomics imaging experiment.
At the theoretical level, the kinds of questions that could be answered with a super-resolution imaging technique capable of spatial resolution of 10 to 20 nm include: (i) the spatial distribution of proteins of the same type in a given space, (ii) the arrangement and sequence of proteins along a pathway such as the different protein complexes that constitute the electron transport chain on the Gram-negative bacterial double membrane, (iii) relative abundance or stoichiometry of binding of protein complexes with subunits labelled with different colour fluorophore, and (iv) relative ratio of different proteins involved in a liquid-liquid phase separation or biomolecular condensates. Specifically, if two proteins are involved in a biomolecular condensate with stoichiometry of 1:1, we are likely to get a blurry overlapping point spread function of mixed colour (i.e., the overall colour is a mesh of the two colours of the fluorophores used to image this biomolecular condensates). At a spatial scale of 10 to 20 nm, interaction of light remains in the classical regime, and is not quantum mechanically controlled. Thus, it can be expected that we would image an overlapping point spread function with mesh colours that is a proportional mix of the intensities of the two different colours. In this sense, we are imaging a discrete biomolecular interaction between two different proteins in the continuum regime.
It is to be remembered that, from the thought experiment above, the final image of the biomolecular condensate involving the directional interaction of two different protein is a blurred image. One possibility to improve the spatial resolution of the image to the range of 5 nm would be to use a nanometre feature size diffraction grating to perform what is essentially nanoscale structural illumination microscopy (nanoscale SIM). In this approach, the nanometre feature size diffraction grating serves as a long-pass filter for high frequency waves, which helps improve the resolution of the imaging process by about 2-fold from 10 nm to 5 nm using traditional linear fluorophores where the emission intensity scale directly with the excitation intensity.
5.3 Probe-based method may allow the imaging of many types of DNA binding proteins with relevance to expanding our understanding of DNA replication and transcription.
Spatial proteomics is currently thought about as a wide-field fluorescence microscopy method able to reveal the distribution and localisation of different types of proteins in the cell. However, the technologies that brought forth the spatial proteomics technique could also be used in a micro-level to reveal protein-protein interactions important to assembly of macromolecular complexes [45 46] or signalling complexes [47]. One example is the assembly of the DNA replication fork complexes or transcription initiation complexes. Theoretically, by labelling each subunit with a fluorophore, it is possible to visualise the assembly of the DNA replication macromolecular complexes. However, current spatial resolution of single molecule localisation microscopy (SMLM) may not afford the detection of subunits that localise in close proximity to each other. But PALM and STORM techniques could reveal where the DNA replication and transcription occur (for example, near to the nucleus centre or close to the nucleus membrane through tethering), as well as a quick partial view of the different subunits that are in the macromolecular complex. Note here that for two subunits with overlapping Airy disk of different colour, the microscopy image will reveal an enlarged Airy disk of meshed colours.
Hence, current SMLM microscopy techniques such as PALM and STORM could only highlight the location of the replication or transcription complex in the cell. In terms of protein-protein interactions between different subunits of the replication and transcription complex, we could only guess the participants of a direct interaction through meshed colour enlarged Airy disk. But, in general, we could determine the molecular players involved in the macromolecular complex through profiling the colours of the Airy disk imaged. One disadvantage of current PALM and STORM techniques is that they are relatively slow and data intensive methods, and thus, we could not use them to image the time-lapse process of progressive assembly of the replication and transcription complexes.
6. But super-resolution microscopy methods are inherently information intensive and are slower techniques; hence, calling the need for high temporal resolution methods
High spatial resolution images are also inherently high image content and information intensive images, especially for multi-colour, multi-protein spatial proteomics experiment. In particular, the computational load scales exponentially with the number of proteins targeted by antibody-fluorophore in the imaging experiment. Irrespective of whether the imaging modality is STED, PALM, STORM or SIM (structured illumination microscopy), multiple images need to be taken in order to reconstruct the final images revealing high resolution localisation of the target protein in the subcellular milieu. Note here the imaging task in spatial proteomics is predominantly preoccupied with determining the central questions of protein localisation, protein migratory patterns, and protein interaction partners in the subcellular milieu. Considering that a typical mammalian cell is of 80 micrometres in diameter, this meant that signal to noise ratio is not sufficiently high, and larger number of frames need to be taken in order to obtain sufficient photon count to reconstruct a reasonable image with scientific accuracy and value. Hence, the imaging task and computational burden is gigantic for super-resolution microscopy applied to spatial proteomics questions, and this calls for higher temporal resolution method. However, this may not be possible as temporal resolution is limited by current speed limit in scientific CMOS cameras, and the speed of taking each frame.
Typically, each frame is on the time constant of milliseconds, with upwards of 1000 frames needed at least for PALM and STORM. With such a time constant, advances in speed of photoactivable, photo switchable or photoconvertible fluorophores or fluorescent proteins in terms of faster switching speeds may not be that consequential. Overall, efforts to enhance the temporal resolution of super-resolution microscopy spatial proteomics is limited by image acquisition speed, number of frames needed to reconstruct an image, as well as signal-to-noise ratio as some fluorophores may not have high quantum yield. Low quantum yield means longer time to acquire sufficient photons to yield a reasonable image useful for the image reconstruction process such as in PALM or STORM. Structured illumination microscopy (SIM) uses the concept of Fourier transform to enhance the useable signals that can be detected by the camera in order to enhance the spatial resolution of the image.
However, it takes about 9 images to reconstruct an image with spatial resolution of 100 nm in about 1 second or about 10 frames per second (10 Hz).48 On the other hand, STED could only capture a 512 x 512 pixel image in about 10 to 30 seconds, making it unable to resolve fast moving structures in the cell [48]. In the area of single molecule localisation microscopy by PALM and STORM, it takes about 1 to 10 milliseconds to track the movement of 1 molecule, but a total of at least 1000 frames are needed to reconstruct an image, which takes the total time needed to reconstruct an image to 1 to 10 seconds.
Taken together, there is a trade-off between imaging speed and spatial resolution in super-resolution microscopy. Generally, it takes about 1 to 10 seconds to obtain an image for single molecule localisation microscopy, which makes it hard to track the migratory dynamics of proteins in the subcellular milieu. Hence, at present, single molecule localisation microscopy is not very useful in live cells imaging for tracking protein migration and trafficking. But it offers unprecedented glimpse into protein localisation at the fixed/dead cells level. On the other hand, SIM may offer a coarse-grained view of protein dynamics at the subcellular level in live cells with an imaging speed of 10 Hz or 100 milliseconds. It must be noted that such a view is coarse and pertains to movements of clusters of proteins over long distances in the cell such as its path in the Golgi apparatus or secretory pathway. Short-range movement in protein in cells remains undetectable by structured illumination microscopy. Finally, STED is too slow to capture the dynamics of fast-moving organelles in the cell. Overall, super-resolution microscopy methods are unable to image protein conformational dynamics which occurs at the microsecond level, and its best spatial resolution attainable limits the time scale of imaging to 1 to 10 seconds, which places live cell protein tracking off-limits.
Technology developments that can enable high temporal resolution imaging of proteins with less toxicity to cells in live cell imaging and are there caveats?
7.1 To track the movement of individually labelled protein would require time correlated microscopy: do we have the computational capacity to deconvolute image data from high protein concentration experiment systems?
One goal of spatial proteomics efforts is to track the movement of multiple proteins across the subcellular space at least up to 1 min timescale. Considering that we need STED, PALM and STORM to achieve single molecule localisation microscopy, such time-lapse imaging would require huge amount of memory and intensive data processing. Specifically, each reconstructed image of PALM and STORM imaging require hundreds to thousands of frames of information, and it must be remembered that the proteins are at least constantly in Brownian motion. So, the final image that can be obtained from PALM and STORM image of subcellular spatial proteomics is a blurred one. This meant that it is nearly impossible with current technologies and methodologies to achieve accurate single molecule tracking of proteins using our best available mature single molecule localisation microscopy techniques. While there may be other more advanced SMLM methods, these remain in the research phase, and would likely require expensive instrumentation and optical train.
Hence, to track the movement of proteins across the subcellular milieu would require conventional high speed sCMOS enabled widefield fluorescence microscopy or spinning disk confocal fluorescence microscopy that allows single imaging frame to reveal specks of fluorescence light indicating clusters of proteins that have been differentially labelled with fluorophores. This meant that we sacrifice single molecule localisation information in exchange for faster imaging. In the case of protein-protein interactions between two differentially labelled proteins, the detected fluorescence would be a mesh of the colours of the two fluorophores. Given that it is impossible to achieve single molecule resolution in conventional diffraction-limited widefield or confocal fluorescence microscopy, the image information obtained from such higher speed imaging may be a continuum of colours across different hues depending on the relative abundance of the different proteins (and their labelled fluorophores). Overall, the image obtained from such widefield, or confocal fluorescence microscopy imaging of spatial proteomics could inform us, in a qualitative manner, the approximate distribution of different proteins in the cell at a tens of proteins resolution.
7.2 Need for excitation by low energy laser for gentle live cell imaging experiments
Single molecular spatial proteomics experiments would require the use of the current best-in-class super-resolution PALM and STORM techniques. Such techniques require multiple frames of imaging with activation of different fluorophores distributed at different points in the specimen to produce a reconstructed image. Hence, PALM and STORM are vulnerable to photobleaching and phototoxicity effects, which may produce serious imaging artefacts due to damaged fluorophores or cytotoxicity to live cells.
In the case of photobleaching, this comes about due to damage to the fluorophore’s molecular structure from high laser excitation energy. For phototoxicity, cellular processes are significantly affected by long-term exposure to the fluorophore, whether in quiescent or excited state [48]. Since the energy of the laser excitation light is higher at the blue end of the visible light spectrum compared to the red end, it means that using low energy laser in the red and infrared region may improve the duration of time-lapse fluorescence microscopy imaging with reduced photobleaching and phototoxicity. To do this, specialized photoactivated fluorophore needs to be used. In addition, more sensitive camera detector is also needed to detect the lower number of fluorescence emission photons. Research works in this direction include the design of a synthetic silicon-rhodamine fluorophore with good photostability that responses to near-infrared excitation suitable for live cell super-resolution microscopy [49].
8. Review on the high spatial resolution imaging of cytoskeletal network, endoplasmic reticulum, Golgi apparatus to pinpoint the subcellular compartmentalisation of proteins in order to provide context to subcellular proteomics.
While it is important to obtain accurate localisation of different proteins (preferably at the single molecule level), it is also crucial to have the subcellular context of these localisation studies correct. By subcellular context, we need to have high spatial resolution imaging of subcellular compartments such as endoplasmic reticulum, Golgi apparatus, lysosomes, mitochondria, cytoskeletal network, peroxisomes, spliceosomes, etc. In particular, high-resolution imaging of the cytoskeletal network would open our eyes to possible interactions between individual proteins and members of the cytoskeletal network such as actin, intermediate filaments, and microtubule. Possibilities for such imaging rests heavily on the availability of fluorophores whether conjugated to antibodies or as standalone dyes for specific labelling individual subcellular organelles and compartments. These fluorophores should ideally bind to membrane proteins of subcellular organelles which serve as markers for delineating the outline of the organelle. An example of how carefully chosen targets for bioimaging could reveal the context of super-resolution spatial proteomics experiment is the three-colour confocal fluorescence microscopy imaging of actin in Figure 5 using infrared excitable fluorophore.
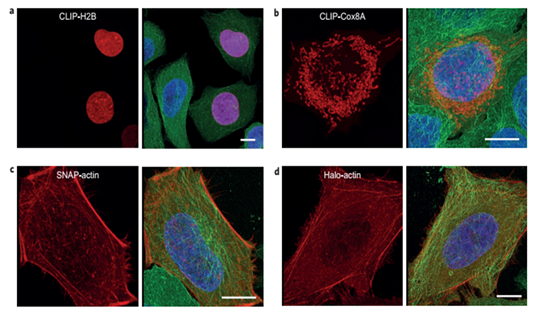
Figure 5: Three colour confocal fluorescence microscopy imaging of actin using the infrared excitable fluorophore. Image taken from Grazvydas Lukinavicius, (2013), A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins.
Overall, developing fluorophores that target membrane with high probe densities is key to achieving super-resolution imaging of subcellular organelles. Using photo switchable fluorophores, high spatial resolution of 30-60 nm and temporal resolution of 1-2 seconds was obtained for lysosomes, plasma membrane, mitochondria, and endoplasmic reticulum.50 Such spatial and temporal resolution features afford the imaging of mitochondria fission/fusion and endoplasmic reticulum remodelling [50]. Cytoskeleton is the mechanical support of the cell that provides overall architectural structure as well as providing organisation for the organelles. Comprising intermediate filament and actin bundle, imaging cytoskeletal network to high spatial resolution would offer unprecedented access to the interactions between cellular signalling and structural proteins to the architectural support of the cell. It may seem that super-resolution fluorescence microscopy methods such as STED, PALM and STORM are the natural tools of choice for imaging the cytoskeletal network. But, a lot of useful localisation and even biomolecular information could also be obtained from diffraction-limited confocal laser scanning microscopy as well as total internal reflection microscopy [51, 52]. An example is in Figure 6 which illustrates the utility of STED microscopy in improving the spatial resolution as compared to conventional confocal microscopy. Another imaging example shows how PALM improves the spatial resolution and contrast of total internal reflection microscopy (TIRF) as compared to conventional epifluorescence microscopy.
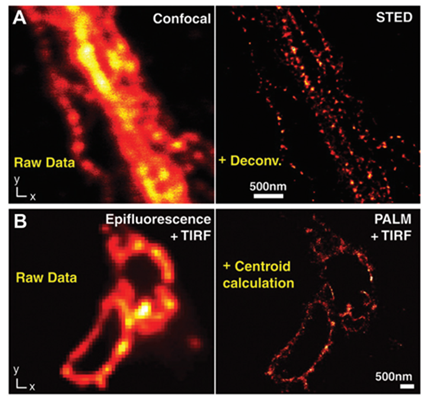
Figure 6: Comparison between conventional fluorescence microscopy and super-resolution imaging of cytoskeletal network. Image taken from Solaire A. Finkenstaedt-Quinn, (2016), Super-resolution imaging for monitoring cytoskeleton dynamics.
One example of super-resolution imaging of cytoskeleton is the use of dual objective STORM to achieve 10 nm (lateral) and 20 nm (axial) imaging of the actin cytoskeleton of mammalian cells [53]. This approach affords the visualisation of individual actin filament, and the cytoskeletal architecture of the cell (Figure 7) [53]. In another study revolving around the same theme of single molecule localisation microscopy, photoactivable fluorescent proteins were employed to image the diffusional dynamics of FtsZ in Escherichia coli, as well as mapping out the distributional network of this important cell division protein [54]. Results obtained help highlight the utility of single molecule localisation microscopy in revealing the architectural network of the cytoskeletal network at different cell states in prokaryotic systems [54]. Finally, STED microscopy was employed in the continuous wave mode to image actin filament network in both fixed and live cells using common confocal and STED microscopy dyes [55]. Such efforts revealed the continuous remodelling of the cell’s actin filament network, but the technique suffers from photobleaching and possible phototoxicity issues [55].
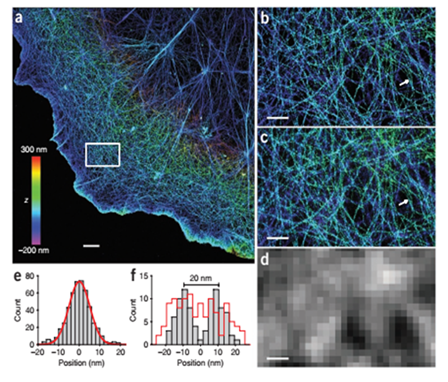
Figure 7: Comparison of single and dual objective STORM imaging of actin network. A) Dual objective STORM image of actin, B) Close up of A), C) Single objective STORM image of actin, D) Conventional widefield fluorescence image of same area, E) Cross-sectional profile of 8 actin filaments aligned by the centre filament, F) Cross sectional profile of two nearby filaments. From: Ke et al., (2012), Dual-objective storm reveals three-dimensional filament organization in the actin cytoskeleton.
Overall, cytoskeletal imaging has reached a fairly high level of sophistication with regards to obtainable spatial resolution. In particular, single molecular localisation microscopy techniques such as PALM and STORM could be reliably used to reduce the point spread function of the imaging, and thus, helps improve spatial resolution obtained. What is more gratifying is that the dyes and fluorophores used in separate studies in the literature featured above showed sufficient photostability and reduced phototoxicity and photobleaching properties to afford dynamic events in cytoskeletal network to be imaged as the cell responds to perturbations. However, the focus in cytoskeletal network high resolution imaging has been predominantly focused on actin filament, which results in the development of fairly robust tools and protocols for the imaging of this important cytoskeletal constituent. Future work may want to explore the imaging of other cytoskeletal membranes such as microtubules and intermediate filaments in spatial resolution that surpassed what is commonly attainable with conventional confocal laser scanning fluorescence microscopy.
Mitochondria is the powerhouse of the cell responsible for the synthesis of ATP. By using photo switchable fluorophores in single molecule localisation microscopy using total internal reflection microscopy as readout, it is possible to achieve a spatial resolution of 20 nm. Such resolution allows the imaging of the FoF1 ATP synthase as well as cytochrome C oxidase in the inner membrane of the mitochondria, [56] hence, allowing cytochrome C oxidase to be used as a facile marker for the mitochondria. Other approaches that afford the live cell and high spatial resolution imaging of mitochondria are FRET and STED imaging where it is possible to discern protein trafficking and mitochondria fission and fusion dynamics [57].
Endoplasmic reticulum (ER) is an organelle contiguous with the nuclear envelope that is responsible for a range of protein post-translational modifications and sorting, as well as calcium storage and release [58]. Despite such knowledge, recent research has linked the ER to new regulatory roles mediated by various bioactive species. With the help of fluorescence live cell imaging, we are beginning to unentangle the intricate and complicated roles of these regulatory actions through direct visualisation of molecular events with high spatial resolution and sensitivity, and non-invasive nature [58]. Overall, there is less emphasis on imaging the structure and protein dynamics of the ER in the scientific literature, which opens up a wide field for further research and exploration.
Golgi apparatus is the organelle that connects the endoplasmic reticulum to the plasma membrane. Important as a secretory pathway, it has mainly been imaged by electron microscopy for high resolution imagery [59]. Recently, there has been use of confocal laser scanning microscopy to obtain multi-colour high resolution images of the Golgi apparatus that reveals the release of natural cargo proteins from the ER into the Golgi apparatus for carriage to the plasma membrane [60]. Another approach uses a pH sensitive rhodamine fluorescent probe to assess the dynamic changes in pH in the Golgi apparatus, which may reveal important facets of how Golgi apparatus respond to different disease states [61]. Briefly, the super-resolution or high resolution imaging of Golgi apparatus is not well developed, and awaits new angles into addressing both basic science questions as well as medically-focussed questions concerning this important secretory apparatus. One fundamental question concerns the selection of biomarkers able to delineate the outline and contour of the Golgi apparatus as well as its dynamic response to cellular stress and external perturbations.
Figure 8: Fluorescence microscopy imaging of different organelles with different fluorophores. Image taken from: Emma Lundberg, and Georg Borner, (2019), Spatial proteomics: a powerful discovery tool for cell biology.
Overall, many of the organelles and subcellular compartments of a mammalian cell have been imaged to high spatial resolution using super-resolution techniques such as STED, PALM and STORM. In comparison, conventional fluorescence microscopy could only allow us to appreciate the subcellular context of the particular organelle in question such as the images obtained for different organelles in Figure 8. The computational burden is huge for these microscopy techniques, which meant that such high-resolution imaging is not available in most labs. More importantly, given the time needed to obtain sufficient number of imaging frames to reconstruct a high spatial resolution image, current suite of super-resolution microscopy techniques could not be used to track the dynamics of organelle movement or cytoskeletal remodelling in live cells where cellular dynamic processes are constantly occurring. Hence, these techniques need to be applied to fixed (dead) cells, which while allowing us to glean unprecedented insights into the architecture of subcellular compartments and their constituents, also meant that we cannot extrapolate our understanding to live dynamic systems. In addition, concerns over photostability and phototoxicity for dyes and fluorophores used in super-resolution microscopy also meant that possibility of imaging artefacts may limit the amount of understanding that we can extract from our imaging studies.
9. Future directions
Spatial proteomics as it is now practiced is overwhelmed with data in both types of proteins and fluorophores. Hence, it is difficult to discern more nuanced meanings from the rich set of data captured using sophisticated microscopy techniques such as STED, PALM and STORM. Information embedded in these images could potentially inform us of the different gene expression pattern in cells that underpins tumour progression, or which allows classification of different pathological states of a particular type of tissue such as pancreas. To do this would require innovations in the use of machine learning and artificial neural networks in discerning hidden signals in data-rich subcellular spatial proteomics images. One challenge is in understanding fluorescence signals along a continuum of intensity and trying to perform classification decisions based on the fluorescence intensity (and relative abundance) of a protein. Such fine shades of fluorescence intensity may not be discernible to the human eye. On the other hand, classification of cell states based on large field of view spatial proteomics experiment would require the assessment of relative fluorescence intensities of a multitude of genes, perhaps tens to twenties, which is difficult to be performed manually using visual inspection. It is hoped that machine learning and artificial intelligence tools will lend a useful hand in this exercise.
10. Conclusions
Spatial proteomics is an extension of proteomics in the era where we finally have the tools to interrogate the localisation and relative abundance of proteins at high spatial resolution. The principal enabling tools for spatial proteomics via fluorescence microscopy is the single molecule localisation microscopy technique such as PALM and STORM. Another approach that could be used is the super-resolution microscopy technique of STED. Common to these approaches is the trade-off between high spatial resolution and high temporal resolution, where current goal of high spatial resolution of 10-20 nm meant that temporal resolution is poor.
While spatial proteomics imply profiling of large ensembles of proteins, current state-of-the-art in spatial proteomics is limited to accessing a proteome of about 50 different proteins. Although limited, this nevertheless represents a rich source of information for understanding protein trafficking, protein-protein interactions, and protein localisation patterns in health and disease in different mammalian disease models at the cell line and primary cell level. True technological limitations in spatial proteomics in the contemporary era are mainly three folds: (i) the lack of large palette (>80) of photostable and low phototoxicity fluorophores available for facile conjugation to target proteins of interest, (ii) need for long imaging time per image to obtain single molecule localisation reconstructed image, and (iii) relatively large overlapping Airy disk that hinder the imaging of protein-protein interaction at the single molecule level. Efforts are being put in by the research community to address all of the above issues, but given the difficulty of these challenges, incremental advances are to be expected in the near and medium term in the spatial proteomics field.
When placed alongside the complementary mass spectrometry-based spatial proteomics field, fluorescence imaging interrogation of spatial proteomics pales significantly in comparison. This comes about due to the higher throughput of mass spectrometry imaging, as well as its amenability to label free (compared to fluorescence labelling) detection and identification of different proteins. But, at the analytical level, mass spectrometry-based spatial proteomics is not amenable to live cell imaging, and thus, could not profile dynamic cellular processes unlike the confocal imaging-like scanning structured illumination microscopy approach which could yield spatial resolution of 120 nm. Hence, taken together, both mass spectrometry-based and fluorescence-based spatial proteomics would each make their own contribution to our understanding of high spatial resolution protein migratory dynamics, protein-protein interactions, and protein localisation pattern. However, there is a slight advantage in favour of fluorescence-based approaches in that it could potentially image live cells at a lower (but still useful) resolution to yield protein (fluorescence) localisation patterns useful for cell and tissue classification decisions in clinical pathology or basic medical research.
Conflicts of interest
The author declares no conflicts of interest.
Funding
No funding was used in this work.
References
- Mao Y, Wang X, Huang P & Tian R. Spatial proteomics for understanding the tissue microenvironment. Analyst 146 (2021): 3777-3798.
- Prinz A, Reither G, Diskar M & Schultz C. Fluorescence and bioluminescence procedures for functional proteomics. PROTEOMICS 8 (2008): 1179-1196.
- Shin J J H. et al. Spatial proteomics defines the content of trafficking vesicles captured by golgin tethers. Nat. Commun. 11 (2020): 5987.
- Zhang L. et al. Spatial proteomics of vesicular trafficking: coupling mass spectrometry and imaging approaches in membrane biology. Plant Biotechnol. J 21 (2023): 250-269.
- Martinez-Val A. et al. Spatial-proteomics reveals phospho-signaling dynamics at subcellular resolution. Nat. Commun 12 (2021): 7113.
- Boisvert F-M. et al. A Quantitative Spatial Proteomics Analysis of Proteome Turnover in Human Cells *. Mol. Cell. Proteomics 11 (2012).
- Bhatia HS. et al. Spatial proteomics in three-dimensional intact specimens. Cell 185 (2022): 5040-5058.
- Christoforou A. et al. A draft map of the mouse pluripotent stem cell spatial proteome. Nat. Commun 7 (2016): 9992.
- Bottek J. et al. Spatial proteomics revealed a CX3CL1-dependent crosstalk between the urothelium and relocated macrophages through IL-6 during an acute bacterial infection in the urinary bladder. Mucosal Immunol 13 (2020): 702-714.
- Strack R. Spatial proteomics with subcellular resolution. Nat. Methods 19 (2022): 780-780.
- Measurement of Protein Dynamics from Site Directed Cu (II) Labeling - Singewald - 2023 - Analysis & Sensing - Wiley Online Library.
- Otosu T, Ishii K & Tahara T. Microsecond protein dynamics observed at the single-molecule level. Nat. Commun 6 (2015): 7685.
- Miklosi AG, Del Favero G, Marko D, Harkany T & Lubec G. Resolution Matters: Correlating Quantitative Proteomics and Nanoscale-Precision Microscopy for Reconstructing Synapse Identity. PROTEOMICS 18 (2018): 1800139.
- Zhu Y. et al. Cross-link assisted spatial proteomics to map sub-organelle proteomes and membrane protein topology. 2022.05.05.490733.
- Unnersjö-Jess D. et al. Confocal super-resolution imaging of the glomerular filtration barrier enabled by tissue expansion. Kidney Int 93 (2018): 1008-1013.
- Winter PW & Shroff H. Faster fluorescence microscopy: advances in high speed biological imaging. Curr. Opin. Chem. Biol 20 (2014): 46-53.
- Zhang Y, Park K-Y, Suazo KF. & Distefano MD. Recent progress in enzymatic protein labelling techniques and their applications. Chem. Soc. Rev 47 (2018): 9106-9136.
- Zhang G, Zheng S, Liu H & Chen PR. Illuminating biological processes through site-specific protein labeling. Chem. Soc. Rev 44 (2015): 3405-3417.
- Chen L. et al. Improved variants of SrtA for site-specific conjugation on antibodies and proteins with high efficiency. Sci. Rep 6 (2016): 31899.
- Lundberg E. & Borner GHH. Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol 20 (2019): 285-302.
- Paul I, White C, Turcinovic I. & Emili A. Imaging the future: the emerging era of single-cell spatial proteomics. FEBS J 288 (2021): 6990-7001.
- Gnann C, Cesnik AJ & Lundberg E. Illuminating Non-genetic Cellular Heterogeneity with Imaging-Based Spatial Proteomics. Trends Cancer 7 (2021): 278-282.
- Schueder F, Unterauer EM, Ganji M & Jungmann R. DNA-Barcoded Fluorescence Microscopy for Spatial Omics. PROTEOMICS 20 (2020): 1900368.
- Mou M. et al. Application of Machine Learning in Spatial Proteomics. J. Chem. Inf. Model 62 (2022): 5875-5895.
- Christopher JA. et al. Subcellular proteomics. Nat. Rev. Methods Primer 1 (2021): 1-24.
- Marx V. Mapping proteins with spatial proteomics. Nat. Methods 12 (2015): 815-819.
- Yan Y & Marriott G. Analysis of protein interactions using fluorescence technologies. Curr. Opin. Chem. Biol 7 (2003): 635-640.
- Liu X. et al. Computer-aided design of reversible hybridization chain reaction (CAD-HCR) enables multiplexed single-cell spatial proteomics imaging. Sci. Adv 8 (2022): eabk0133.
- Wang N, Li X, Wang R & Ding Z. Spatial transcriptomics and proteomics technologies for deconvoluting the tumor microenvironment. Biotechnol J 16 (2021): 2100041.
- Natter K. et al. The Spatial Organization of Lipid Synthesis in the Yeast Saccharomyces cerevisiae Derived from Large Scale Green Fluorescent Protein Tagging and High Resolution Microscopy * S. Mol. Cell. Proteomics 4 (2005): 662-672.
- Ciccimarra R. et al. The normal and fibrotic mouse lung classified by spatial proteomic analysis. Sci. Rep. 12 (2022): 8742.
- Uttam S. et al. Abstract 1642: Spatial proteomics with hyperplexed fluorescence imaging predicts risk of colorectal cancer recurrence and infers recurrence-specific protein-protein networks. Cancer Res 79 (2019): 1642.
- Wachsmuth M. et al. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol 33 (2015): 384-389.
- Schubert W. et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol 24 (2006): 1270-1278.
- Stadler C. et al. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods 10 (2013): 315-323.
- Nightingale DJH, Oliver SG & Lilley KS. Mapping the Saccharomyces cerevisiae Spatial Proteome with High Resolution Using hyperLOPIT. in Yeast Systems Biology: Methods and Protocols (eds. Oliver, S. G. & Castrillo, J. I.) (2019): 165-190.
- Mund A, Brunner A-D & Mann M. Unbiased spatial proteomics with single-cell resolution in tissues. Mol. Cell 82 (2022): 2335-2349.
- Radius measurement via super-resolution microscopy enables the development of a variable radii proximity labeling platform | PNAS.
- Toward High Spatially Resolved Proteomics Using Expansion Microscopy | Analytical Chemistry.
- Huang, K. & Murphy, R. F. Boosting accuracy of automated classification of fluorescence microscope images for location proteomics. BMC Bioinformatics 5 (2004): 78.
- Dayao MT, Brusko M, Wasserfall C & Bar-Joseph Z. Membrane marker selection for segmenting single cell spatial proteomics data. Nat. Commun 13 (2022): 1999.
- Breckels L M et al. Learning from Heterogeneous Data Sources: An Application in Spatial Proteomics. PLOS Comput. Biol 12 (2016): e1004920.
- Gatto L. et al. A Foundation for Reliable Spatial Proteomics Data Analysis *. Mol. Cell. Proteomics 13 (2014): 1937-1952.
- Zhou Y. et al. SISPRO: Signature Identification for Spatial Proteomics. J. Mol. Biol (2023): 167944
- Dietz MS. & Heilemann, M. Optical super-resolution microscopy unravels the molecular composition of functional protein complexes. Nanoscale 11 (2019): 17981-17991.
- Mito M, Kawaguchi T, Hirose T & Nakagawa S. Simultaneous multicolor detection of RNA and proteins using super-resolution microscopy. Methods 98 (2016): 158-165.
- Farrell M V. et al. Protein-PAINT: Superresolution microscopy with signaling proteins. Sci. Signal 15 (2022): eabg9782.
- Godin AG, Lounis B & Cognet L. Super-resolution Microscopy Approaches for Live Cell Imaging. Biophys. J. 107 (2014): 1777-1784.
- Lukinavicius G. et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem 5 (2013): 132-139.
- Shim S-H. et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci 109 (2012): 13978-13983.
- Finkenstaedt-Quinn SA, Qiu TA, Shin K & Haynes CL. Super-resolution imaging for monitoring cytoskeleton dynamics. Analyst 141 (2016): 5674-5688.
- McKayed KK & Simpson JC. Actin in Action: Imaging Approaches to Study Cytoskeleton Structure and Function. Cells 2 (2013): 715-731.
- Xu K, Babcock HP & Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods 9 (2012): 185-188.
- Niu L & Yu J. Investigating Intracellular Dynamics of FtsZ Cytoskeleton with Photoactivation Single-Molecule Tracking. Biophys J 95 (2008): 2009-2016.
- Neupane B. et al. Continuous-Wave Stimulated Emission Depletion Microscope for Imaging Actin Cytoskeleton in Fixed and Live Cells. Sensors 15 (2015): 24178-24190.
- van de Linde S, Sauer M & Heilemann M. Subdiffraction-resolution fluorescence imaging of proteins in the mitochondrial inner membrane with photos witchable fluorophores. J. Struct. Biol 164 (2008): 250-254.
- Jakobs S. High resolution imaging of live mitochondria. Biochim. Biophys. Acta BBA - Mol. Cell Res 1763 (2006): 561-575.
- Tang F. et al. Recent progress in small-molecule fluorescent probes for endoplasmic reticulum imaging in biological systems. Analyst 147 (2022): 987-1005.
- Koga D, Ushiki T & Watanabe T. Novel scanning electron microscopy methods for analyzing the 3D structure of the Golgi apparatus. Anat. Sci. Int 92 (2017): 37-49.
- Tojima T, Miyashiro D, Kosugi Y & Nakano A. Super-Resolution Live Imaging of Cargo Traffic Through the Golgi Apparatus in Mammalian Cells. in Golgi: Methods and Protocols (2023): 127-140.
- Feng Z. et al. A rhodamine derivative-based fluorescent probe for visual monitoring of pH changes in the Golgi apparatus. Sens. Actuators B Chem. 366 (2022): 131963.
