RAS1 is Involved in the Antifungal Resistance of Candida Albicans and the Inhibition of Farnesol to the Resistance of Biofilms
Article Information
Lulu Zuo1,2,3, Zhenzhen Zhang1,2,3, Wei Duan1,2,3, Yun Huang1,2,3, Zheng Xu1,2,3, Qinqin Zhang1,2,3, Ying Lv1,2,3, Xin Wei1,2,3*
1Jiangsu Province key Laboratory of Oral Diseases, College of Stomatology, Nanjing Medical University, China.
2Department of Operative Dentistry and Endodontics, Affiliated Stomatological Hospital of Nanjing Medical University, Nanjing, China.
3Jiangsu Province Engineering Research Center of Stomatological Translational Medicine, Nanjing, China.
*Corresponding author: Xin Wei, Department of Operative Dentistry and Endodontics, Affiliated Stomatological Hospital of Nanjing Medical University, Nanjing, China.
Received: 08 August 2022; Accepted: 18 August 2022; Published: 29 August 2022
Citation: Lulu Zuo, Zhenzhen Zhang, Wei Duan, Yun Huang, Zheng Xu, Qinqin Zhang, Ying Lv, Xin Wei. RAS1 is Involved in the Antifungal Resistance of Candida Albicans and the Inhibition of Farnesol to the Resistance of Biofilms. Archives of Clinical and Biomedical Research 6 (2022): 694-706.
View / Download Pdf Share at FacebookAbstract
Background: Farnesol enhances the susceptibility of Candida albicans biofilms to antifungals; however, its molecular mechanisms are poorly understood. RAS1 is required for the hyphal growth. This study hypothesized that RAS1 is involved in the antifungal resistance of C.albicans and the inhibition of farnesol against biofilms.
Methods: The antifungal resistances of C.albicans biofilms formed by RAS1 mutant and control strains were examined using spot assays and XTT reduction assays with and without farnesol, and morphological changes were observed using CLSM and SEM. Meanwhile, RAS1 gene and RAS1 protein in biofilms exposed to farnesol were analyzed using RT-qPCR and Western blot.
Results: Compare with control strain, RAS1 overexpressing strain (RAS1OE) was more resistant to some of the antifungal drugs, while RAS1 deleted strain (RAS1Δ/Δ) was less resistant to all antifungal drugs. RAS1 was down-regulated in the C.albicans biofilms exposed to farnesol. The increased resistance of RAS1OE to antifungals was significantly decreased by farnesol, and farnesol decreased the resistance of RAS1OE more obviously than wild-type and RAS1Δ/Δ strains (P<0.05). Further, farnesol reduced the expression of RAS1 in RAS1OE biofilm more obviously than that in wild-type strain biofilm. In addition, morphological observation showed that RAS1OE increased the hyphal growth of biofilm, while RAS1Δ/Δ reduced that, and the inhibition of hyphal growth by farnesol was more obviously in RAS1OE biofilm, while that was less obvious in RAS1Δ/Δ biofilm.
Conclusions:
Keywords
Antifungal; C.albicans Biofilms; Resistance; Farnesol; RAS1
Antifungal articles; C.albicans Biofilms articles; Resistance articles; Farnesol articles; RAS1 articles
Article Details
List of Abbreviations:
ANOVA- One-Way Analysis of Variance; CLSM- Confocal Laser Scanning Microscope; RAS1OE- RAS1 Overexpressing Strain; SDA- Sabourd’s Dextrose Agar; SEM- Scanning Electron Microscopy; YPD- Yeast Peptone Dextrose.
1. Introduction
The fungus Candida albicans (C. albicans), an opportunistic human pathogen, is commonly found in the biofilms of oral cavity and gastrointestinal tract. The biofilms of C.albicans are significantly less susceptible to antimicrobial agents than that of planktonic cells, which seriously weakens the effectiveness of antifungals [1]. Indeed, 65~80% of C.albicans infections were associated with biofilms [2, 3]. The extensive use of antifungals has led to the increased resistance of biofilms to the few existing antifungals. Thus, it is crucial to explore the mechanisms of therapeutic or preventive strategies targeting biofilm-related infections.
Previous studies have demonstrated that farnesol prevented the germination of yeast cells into mycelia, repressing hyphal growth and biofilm formation [4, 5]. Further studies showed that farnesol enhanced the microbial susceptibility to antibiotics of Staphylococcus aureus [6], Paracoccidioides brasiliensis [7], Streptococcus mutans [8], and Fusarium graminearum [9]. Another in vitro study showed that farnesol inhibited the development of C.albicans biofilms formed from the resistant strains, and farnesol in combination with fluconazole, itraconazole, and 5-flurocytosine had synergistic effects against C.albicans biofilms [10]. Farnesol possibly enhance the susceptibility of planktonic C. dubliniensis and C. albicans to fluconazole [6] via regulation of the ergosterol biosynthesis pathway [11]. However, the exact mechanism underlying the farnesol-mediated increase in susceptibility to various antifungals is poorly understood in C. albicans biofilms.
albicans RAS proteins are small GTPases that can trigger the MAPK and cAMP signaling pathway, and involve in the morphogenesis [4, 12], stress resistance [13,14], and maintenance of hyphal state [15]. RAS1 is a highly conserved signaling protein that plays central roles in key physiological processes for both commensal and pathogenic lifestyles within the host [16], in this process RAS1 gene might regulate a diverse array of phenotypes that are critical to the antifungal resistance of C. albicans. However, the role of RAS1 in the resistance of C. albicans biofilms was not discussed in the previous studies, and the molecular mechanism of the RAS1 in regulating the farnesol-relevant antifungal capacity to C. albicans biofilms is still unknown. In the present study, we hypothesized that RAS1 is associated to the antifungal resistance of C. albicans biofilms, and also regulated the inhibition of farnesol on the antifungal resistance of C. albicans biofilms. To test this hypothesis, the antifungal resistance of C. albicans biofilms was examined in the RAS1 mutant strains (including RAS1 overexpressing strain (RAS1OE) and deletion strain (RAS1Δ/Δ)) with the presence or absence of farnesol.
2. Materials and Methods
2.1 Strains, Plasmids and Media
The wild-type clinical isolate C. albicans SC5314 (American Type Culture Collection, MD, America) [10] and the resistant strain were used to analyze the effects of farnesol on the RAS1 expression in C. albicans. The resistant strain derived from SC5314 induced via the serial fluconazole concentration gradient method until the MIC reached or exceeded 64 µg/ml. RAS1 overexpressing strain (RAS1OE) was generated from CAI4 (William A. Fonzi, Department of Microbiology and Immunology, Georgetown University, Washington, USA) using RAS1-pCaEXP plasmids [10,17], and the PCaEXP-CAI4 strain with wild-type phenotype was generated from CAI4 using the plasmid pCaEXP (BioVector NTCC Inc., China). The transformants then grew on the plates of SD-ura-met-cys selective culture medium, and the positive clone of PCaEXP-RAS1-CAI4 was verified using RT-PCR to detect the level of mRNA of RAS1. RAS1 deletion strain (RAS1Δ/Δ) was generated from SN152 (School of Pharmacy, The Second Military Medical University) using HIS-LEU-ARG knocking-out strategy [18]. Fusion PCR was performed to create disruption fragments of C. albicans HIS1 and LEU2 coding sequences, and the HIS1 and LEU2 disruption fragments were transformed into SN152 to obtain RAS1Δ/Δ [19]. The homologous recombination transformants grew positively on selective medium (SD-his, SD-leu, and SD-his-leu). The positive clone was verified using colony PCR to detect the presence of correct knockout junctions with the designed primers. The strains and plasmids used in the study are listed in Table 1.
|
Strains |
Genotype and Descriptiona |
Reference |
|
C. albicans strains |
||
|
SC5314 |
Wild-type clinical isolate |
American Type Culture Collection, America |
|
Resistant strain |
derived from SC5314 induced via the serial fluconazole concentration gradient method until the MIC reached or exceeded 64 µg/ml |
Yu LH et al. 2011 |
|
Xia J et al. 2017 |
||
|
CAI4 |
ura3:: λimm434/ura3:: λimm434 |
Fonzi and Irwin. 1993 |
|
URA3 auxotrophic strain |
||
|
PCaEXP-CAI4 |
ura3:: λimm434/ura3:: λimm434-(pCaEXP URA3) |
In this study |
|
strain transformed with pCaEXP, used as a control of overexpression experiments |
||
|
RAS1OE |
ura3:: λimm434/ura3:: λimm434-(RAS1-pCaEXP RAS1, URA3) |
In this study |
|
RAS1-overexpressing strain |
||
|
SN152 |
arg4/arg4 leu2/leu2 his1/his1 URA3/ura3::imm434 IRO1/iro1::imm434 |
Noble SM et al. 2005 |
|
Arg4, Leu2, His1 auxotrophic strain |
||
|
RAS1Δ/Δ |
arg4/arg4 leu2/leu2 his1/his1 URA3/ura3::imm434 IRO1/iro1::imm434 |
In this study |
|
RAS1::C.d.HIS1/RAS1::C.m.LEU2 |
||
|
RAS1-deleted strain |
||
|
Plasmids |
||
|
pCaEXP |
URA3 and MET3 promoter integrating plasmid |
R. S. Care et al. 1999 |
|
RAS1-pCaEXP |
Constructed by integration of RAS1 |
In this study |
|
pSN52 |
With HIS1 marker |
Noble SM et al. 2005 |
|
pSN40 |
With LEU2 marker |
Noble SM et al. 2005 |
Table 1: Strains and Plasmids used in this Study.
aC.m., C. maltose: the DNA fragments LEU2 was generated from C. m. ; C.d., C. dubliniensis: the DNA fragments HIS1 was generated from C.d. The DNA fragments LEU2 and HIS3 from C. dublinensis and C. maltosa have been homologous recombinated to replace the target genes RAS1 in the C. albicans.
2.2 Biofilm Formation and Farnesol Treatment
Freshly grown yeast cells from Sabourd’s Dextrose Agar (SDA) plates were incubated in yeast peptone dextrose (YPD) medium and grown overnight in an orbital shaker at 30 °C. The cells were collected and resuspended in RPMI 1640 (Gibco Ltd., Paisley, U.K.). The solution was adjusted to a cell density of 1×106 cells/ml for the experiments. Standardized suspensions of the strains were dispensed into flat-bottom microtiter dishes (Corning, Inc., N.Y., USA). The cells were incubated at 37 °C in a moist chamber with 5% CO2. After 2 h of incubation, non-adherent cells were removed by thoroughly washing the biofilms three times with PBS. Then the biofilms formed depending on the selected time periods (6, 12, 24, and 36 h). For the farnesol treatment, equal volumes of farnesol (100-300 μM) or sterile water (with an equal concentration of methanol) were added to the farnesol-treated and untreated control groups, respectively. Stock solutions (100 mM) of farnesol (E, E farnesol; Sigma Chemical Co., St. Louis, Mo) were dissolved in 100% (vol/vol) methanol and frozen at -70 °C until use. The effect of methanol on the growth of C. albicans was evaluated by testing different methanol concentrations without antifungals, ranging from 0.05–1% (v/v) [20]. The highest methanol concentration used in the microdilution plates was 0.1% (v/v), which presented no antifungal activity.
2.3 Susceptibility Tests
The antifungals used in the tests were fluconazole (Sigma-Aldrich, St Louis, MO, USA), itraconazole (Selleckchem, Houston, TX, USA), amphotericin B (Sigma-Aldrich, St Louis, MO, USA), caspofungin (Sigma-Aldrich, St Louis, MO, USA), terbinafine (Selleckchem, Houston, TX, USA), 5-flucytosine (Selleckchem, Houston, TX, USA), and nystatin (Selleckchem, Houston, TX, USA). The susceptibility of RAS1 mutant strains in planktonic form to antifungals was determined using a spot assay [21]. The strains were incubated in the mediums and grown for 16 h in an orbital shaker at 30 °C. The yeast cells were harvested during the logarithmic growth phase, washed twice with PBS and suspended in fresh medium to an OD600nm of 1.0. A 5 μl of tenfold serial dilution of the suspension was spotted onto plates in the presence or absence of antifungals with serial concentration gradients. Antifungals were added to the medium at a concentration of 4 μg/ml for fluconazole, 0.5 μg/ml for itraconazole, 2 μg/ml for amphotericin B, 0.5 μg/ml for caspofungin, 15 μg/ml for terbinafine, 8 μg/ml for 5-fluorocytosine, and 2 μg/ml for nystatin. Growth differences were measured after incubation at 30 °C for 48 h. All experiments were performed in thrice on three separated occasions. The susceptibility of RAS1 mutant strains in biofilms to antifungals was determined using the XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide]-menadione (0.5 mg/ml XTT, 1 μM menadione) reduction assay (Sigma-Aldrich, St Louis, MO, USA) [22]. Colorimetric changes were analyzed in a microtiter plate reader (BioTek Instruments, Inc., VT, USA) at a wavelength of 490 nm, which measured the changes in the metabolic activity of the biofilms [23]. The biofilms were incubated as described above. A series of antifungal agent-free wells and biofilm-free wells were also included to serve as the positive and negative controls. Drugs were prepared in serial 2-fold dilutions, and their final concentrations ranged from 4 to 1024 µg/ml for fluconazole, 1 to 256 µg/ml for itraconazole, 0.25 to 64 µg/ml for amphotericin B, caspofungin, and nystatin, and 2 to 512 µg/ml for terbinafine and 5-flucytosine. Antifungals were added to the biofilms and incubated for an additional 24 h at 37 °C. The value of the background OD measurement, obtained from biofilm-free wells processed in the same manner as the inoculated wells, was subtracted from the OD measurement of each well. After this subtraction, the percentage of growth in each well was calculated as the OD of each well /OD of the drug-free well. The lowest drug concentration that inhibited biofilms growth by 50% was considered the sessile MIC50 (SMIC50) of this antifungal. Each concentration was tested in five wells and the average value from the five wells was used as the SMIC50. All experiments were performed in thrice on three separated occasions.
2.4 The Morphological Observations using CLSM and SEM
Biofilms from mutant, control, and farnesol-treated mutant and control strains were formed on the glass bottom of cell culture dishes for morphological observation. For confocal laser scanning electron microscopy (CLSM) observation, the formed biofilms were fixed with 4% paraformaldehyde overnight, washed with PBS and stained with 500 μl calcofluor white stain (0.0025 g/ml; Sigma Chemical Co., St. Louis, MO) [24] for 30 min at 37 °C in the dark. The biofilms were observed using a Zeiss LSM700 microscope with a video capture system, automatic camera, image analysis hardware and software (Carl Zeiss, Inc., Oberkochen, Germany), and a 488 nm argon ion laser. For scanning electron microscopy (SEM) observation, the formed biofilms were fixed with 2.5% glutaraldehyde solution overnight. Biofilms were subsequently washed twice with distilled water, dehydrated in ethanol series (70% for 10 min, 95% for 10 min, and 100% for 20 min), and air-dried overnight in a desiccator. The biofilms were then metalized by gold sputtering for 45 s in a High Vacuum Evaporator, followed by bonding to carbon double-side tape and processing for SEM (FEI Inc., Hillsboro, USA).
2.5 RT-qPCR and Western Blot Analysis
RNA samples of the farnesol-treated and untreated biofilms were purified using a modified hot phenol method [25]. The cDNA was synthesized using the reverse transcription system (TaKaRa, Bio Co., Ltd., Dalian, China) and performed on an ABI 7300 Fast real-time PCR machine (Applied Biosystems, Rotkreuz, Switzerland) using qPCR SYBR Green Mix (Thermo Scientific, Waltham, MA, USA). The PCR program used was as follows: activation at 95 °C for 10 min; 40 cycles of amplification (95 °C for 15 s, 55 °C for 1 min); 70 °C for 20 s; cooling at 4 °C. After amplification, a melting curve analysis was performed to ensure the absence of primer dimers. The expression of genes was calculated using the 2-ΔΔCt method [11]. ACT1 was used as a reference gene. Primers were all designed by Shanghai Generay Bio-Tech Co., Ltd (Shanghai, China). (Table 2).
|
Gene |
Sequences |
|
RAS1 |
F: GGTAAATCCGCTTTAACCATTC |
|
R: GCCAGATATTCTTCTTGTCCAG |
|
|
ACTIN |
F: GCCGGTGACGACGCTCCAAGAGCTG |
|
R: CCGTGTTCAATTGGGTATCTCAAGGTC |
Table 2: Primer Sequences in Identifying RAS1 Expression Strains.
Total protein extracts were prepared as described for C. albicans biofilms using an immunoprecipitation protocol [26]. Protein concentrations were determined using a BCA Protein Assay Kit (Sigma-Aldrich, St. Louis, MO, USA). A total of 15 μg protein diluted in 6× loading buffer was separated on a 10% SDS-PAGE gel and blotted onto PVDF membranes (Millipore, MA, USA). Membranes were blocked in 0.01 M PBS containing 5% skim milk and 0.1% Tween-20 at room temperature, followed by incubation of membranes overnight with the primary antibodies (RAS1, 1:1000 dilution; GAPDH, 1:8000 dilution; Bioworld, MN, USA) at 4 °C. Membranes were then rinsed with PBST (0.01 M PBS plus 0.1% Tween-20) and incubated with secondary antibodies (1:10000 dilution, Bioworld, MN, USA) at room temperature for 1 h. Membranes were visualized with Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific Inc., Rockford, IL, USA) and exposed to Kodak X-ray films. GAPDH was used as an internal control in the experiments.
2.6 Statistical Analysis
All quantitative experiments were performed in thrice for statistical analyses. Data were analyzed using SPSS19.0 software (SPSS Inc., Chicago, IL, USA). One-way analysis of variance (ANOVA) was used to analyze the differences between the groups, while a paired t-test was performed for intragroup comparisons. The rank sum test was used to analyze ranked data. Comparisons resulting in P values of less than 0.05 were considered statistically significant.
3. Results
3.1 RAS1 is Involved in the Resistance of C. albicans to Antifungals
For the spot assay to analyze the resistance of planktonic C. albicans to antifungals, RAS1OE was more resistant to fluconazole (4 µg/ml) and itraconazole (0.5 µg/ml) than control strain (pCaEXP-CAI4 with wild-type phenotype) (Fig. 1). Moreover, RAS1Δ/Δ was more susceptible to fluconazole (4 µg/ml), itraconazole (0.5 µg/ml), amphotericin B (2 µg/ml), nystatin (2 µg/ml), caspofungin (0.5 µg/ml), terbinafine (15 µg/ml), and 5-fluroctosine than wild-type strain (SN152) (Figure 1).
Yellow spot: represents resistance of planktonic C. albicans to antifungal drugs under serial concentration gradients.
For the XTT-reduction assay to analyze the resistance of C. albicans biofilms to antifungal drugs, the biofilms of RAS1OE strain had higher SMIC50 values for fluconazole (at 12 and 24 h biofilms), 5-flurocytosine (at 6 and 12 h biofilms), nystatin (at 6, 12, and 24 h biofilms), itraconazole, amphotericin B, caspofungin, and terbinafine (at 6, 12, 24, and 36 h biofilms) than the biofilms of control strain with wild-type phenotype (Table 3-1, Part A). Furthermore, biofilms of RAS1Δ/Δ had lower SMIC50 values for itraconazole (at 6, 12 and 24 h biofilms), amphotericin B (at 12 and 24 h biofilms), nystatin (at 6 h biofilm), fluconazole, caspofungin, terbinafine, and 5-flurocytosine (at the 6 and 12 h biofilms), than the biofilms of control strain (SN152) (Table 3-1, Part A). On the other hand, the SMIC50 of the antifungal drugs were increased with the mature of biofilms of all the studied strains.
|
Part A |
SMIC50 of antifungals(µg/ml) with 0 µM farnesol |
||||||
|
6 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Terb |
5-Fc |
Cas |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
>16 |
1 |
>256 |
2 |
0.125 |
2 |
|
RAS1OE |
>1024 |
64 |
4 |
512 |
512 |
1 |
4 |
|
SN152 |
128 |
16 |
8 |
64 |
>512 |
16 |
4 |
|
RAS1Δ/Δ |
4 |
4 |
8 |
16 |
256 |
4 |
2 |
|
12 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Terb |
5-Fc |
Cas |
Nys |
|
|
pCaEXP-CAI4 |
512 |
>16 |
0.5 |
>256 |
256 |
>8 |
4 |
|
RAS1OE |
>1024 |
128 |
64 |
>512 |
512 |
32 |
8 |
|
SN152 |
1024 |
>256 |
8 |
512 |
>512 |
32 |
4 |
|
RAS1Δ/Δ |
>1024 |
4 |
4 |
64 |
128 |
4 |
4 |
|
24 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Terb |
5-Fc |
Cas |
Nys |
|
|
pCaEXP-CAI4 |
1024 |
>16 |
1 |
>256 |
512 |
>8 |
4 |
|
RAS1OE |
>1024 |
256 |
64 |
>512 |
512 |
32 |
8 |
|
SN152 |
1024 |
>256 |
32 |
>512 |
>512 |
64 |
4 |
|
RAS1Δ/Δ |
1024 |
>64 |
16 |
>512 |
>512 |
64 |
4 |
|
36 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Terb |
5-Fc |
Cas |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
>16 |
0.25 |
>256 |
512 |
>8 |
8 |
|
RAS1OE |
>1024 |
256 |
64 |
>512 |
512 |
32 |
8 |
|
SN152 |
1024 |
>256 |
32 |
>512 |
>512 |
>64 |
4 |
|
RAS1Δ/Δ |
1024 |
>256 |
32 |
>512 |
>512 |
>64 |
4 |
Table 3-1: The SMIC Values of Antifungals in C. albicans Biofilms of Mutant Strains.
* “Flu” is an acronym for Fluconazole; “Itr” is an acronym for Itraconazole; “AmB” is an acronym for Amphotericin B; “Cas” is an acronym for Caspofungin; “Terb” is an acronym for Terbinafine; “5-Fc” is an acronym for 5-Flucytosine; “Nys” is an acronym for Nystatin. *Part A: SMIC50 of antifungals (µg/ml) to C. albicans biofilms without farnesol (0 µM farnesol).
3.2 RAS1 of the C. albicans Biofilms Regulated by Farnesol
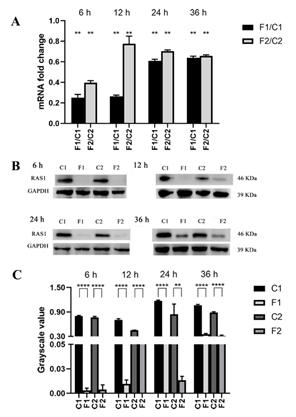
When the biofilms exposed to farnesol, compare with the farnesol-untreated control, RAS1 expression and RAS1 level in the biofilms of SC5314 and resistant strain (formed from SC5314) were all down-regulated at four growth phases (6, 12, 24, and 36 h), respectively (P<0.01) (Figure 2).
A: Comparisons of gene expression between the farnesol-treated and untreated groups of C. albicans biofilms at all studied phases; B: The levels of RAS1 in the strains with or without farnesol-treated were assessed using western blot at all studied phases; C: The bands in B were quantified and analyzed using grayscale values. C1: farnesol-untreated SC5314; F1: farnesol-treated SC5314; C2: farnesol-untreated resistant strain; F2: farnesol-treated resistant strain. Farnesol: 200 µM. *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001.
3.3 Effects of Farnesol on the Susceptibility of Biofilms of RAS1 Mutant Strains to Antifungals
Significant SMIC reductions were observed when farnesol was added to the biofilms of RAS1OE strains. Compared with the untreated biofilms, the resistance of RAS1OE biofilms to fluconazole (at 6 and 12 h biofilms), itraconazole (at 6, 12, 24, and 36 h biofilms), amphotericin B (at 12, 24 and 36 h biofilms), caspofungin (at 6, 12, 24, and 36 h biofilms), terbinafine (at 6, 12, and 36 h biofilms), 5-flurocytosine (at 6, 12, 24, and 36 h biofilms), and nystatin (at 12 h biofilm) were decreased after farnesol treatment. Moreover, the inhibitory effects of farnesol on the antifungal (including fluconazole, itraconazole, amphotericin B, nystatin, caspofungin, and 5-flucytosine) resistance of RAS1OE were more obvious than that of control strain (PCaEXP-CAI4) (P<0.05) (Table 3-2 Part B, C, and D). Further, compared with the untreated control, farnesol did not change the resistance of RAS1Δ/Δ biofilms to fluconazole, itraconazole, and 5-flurocytosine; however, farnesol decreased the resistance of RAS1Δ/Δ biofilms (at 6, 12, 24, and 36 h biofilms) to amphotericin B and caspofungin and increased that to nystatin (at 6, 12, 24, and 36 h biofilms) and terbinafine (at 6 and 12 h biofilms) (Table 3-2 Part B, C, and D).
|
Part B |
SMIC50 of antifungals (µg/ml) with 100 µM farnesol |
||||||
|
6 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
>16 |
0.5 |
0.0625 |
>256 |
1 |
4 |
|
RAS1OE |
256 |
16 |
4 |
0.0625 |
256 |
4 |
4 |
|
SN152 |
128 |
16 |
8 |
0.25 |
>512 |
512 |
8 |
|
RAS1Δ/Δ |
8 |
4 |
1 |
0.25 |
>512 |
128 |
8 |
|
12 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
512 |
>16 |
0.5 |
>8 |
>256 |
1 |
4 |
|
RAS1OE |
512 |
64 |
8 |
0.125 |
256 |
1 |
4 |
|
SN152 |
1024 |
256 |
4 |
0.5 |
>512 |
512 |
8 |
|
RAS1Δ/Δ |
128 |
4 |
4 |
0.5 |
>512 |
512 |
8 |
|
24 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
1024 |
>16 |
1 |
>8 |
>256 |
2 |
2 |
|
RAS1OE |
>1024 |
>64 |
8 |
16 |
512 |
4 |
8 |
|
SN152 |
1024 |
>256 |
16 |
0.5 |
>512 |
512 |
4 |
|
RAS1Δ/Δ |
1024 |
64 |
16 |
0.5 |
>512 |
512 |
8 |
|
36 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
>16 |
0.25 |
0.25 |
>256 |
0.5 |
4 |
|
RAS1OE |
>1024 |
>64 |
2 |
0.125 |
64 |
0.5 |
8 |
|
SN152 |
1024 |
>256 |
2 |
1 |
>512 |
512 |
8 |
|
RAS1Δ/Δ |
1024 |
>256 |
2 |
0.5 |
>512 |
512 |
8 |
|
Part C |
SMIC50 of antifungals (µg/ml) with 200 µM farnesol |
||||||
|
6 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
>16 |
1 |
0.0625 |
>256 |
2 |
2 |
|
RAS1OE |
128 |
1 |
4 |
0.25 |
256 |
2 |
2 |
|
SN152 |
64 |
8 |
4 |
0.25 |
>512 |
512 |
4 |
|
RAS1Δ/Δ |
4 |
4 |
4 |
0.25 |
>512 |
256 |
8 |
|
12 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
512 |
>16 |
0.5 |
0.25 |
>256 |
4 |
2 |
|
RAS1OE |
256 |
1 |
4 |
0.25 |
512 |
4 |
2 |
|
SN152 |
1024 |
>256 |
4 |
0.5 |
>512 |
16 |
4 |
|
RAS1Δ/Δ |
128 |
4 |
4 |
1 |
>512 |
64 |
8 |
|
24 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
512 |
>16 |
0.5 |
>8 |
>256 |
2 |
2 |
|
RAS1OE |
512 |
16 |
8 |
0.25 |
>512 |
2 |
2 |
|
SN152 |
1024 |
>256 |
8 |
0.25 |
>512 |
512 |
4 |
|
RAS1Δ/Δ |
1024 |
16 |
8 |
0.5 |
>512 |
512 |
8 |
|
36 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
0.125 |
0.125 |
0.125 |
>256 |
16 |
2 |
|
RAS1OE |
1024 |
32 |
16 |
0.5 |
512 |
2 |
2 |
|
SN152 |
1024 |
>256 |
2 |
0.5 |
>512 |
512 |
4 |
|
RAS1Δ/Δ |
1024 |
>256 |
2 |
0.5 |
>512 |
512 |
8 |
|
Part D |
SMIC50 of antifungals (µg/ml) with 300 µM farnesol |
||||||
|
6 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
>16 |
1 |
0.0625 |
>256 |
16 |
4 |
|
RAS1OE |
128 |
16 |
2 |
0.0625 |
256 |
16 |
4 |
|
SN152 |
64 |
4 |
2 |
0.25 |
>512 |
512 |
8 |
|
RAS1Δ/Δ |
4 |
4 |
8 |
0.25 |
>512 |
128 |
16 |
|
12 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
512 |
>16 |
0.5 |
0.25 |
>256 |
16 |
4 |
|
RAS1OE |
256 |
64 |
16 |
0.0625 |
512 |
0.5 |
4 |
|
SN152 |
1024 |
4 |
2 |
0.25 |
>512 |
512 |
16 |
|
RAS1Δ/Δ |
1024 |
4 |
2 |
0.5 |
>512 |
512 |
32 |
|
24 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
512 |
>16 |
1 |
>8 |
>256 |
64 |
4 |
|
RAS1OE |
512 |
64 |
16 |
2 |
512 |
0.5 |
4 |
|
SN152 |
1024 |
>256 |
8 |
0.5 |
>512 |
512 |
16 |
|
RAS1Δ/Δ |
1024 |
128 |
8 |
0.5 |
>512 |
512 |
16 |
|
36 h biofilms |
|||||||
|
Flu |
Itr |
AmB |
Cas |
Terb |
5-Fc |
Nys |
|
|
pCaEXP-CAI4 |
>1024 |
0.5 |
0.125 |
0.0625 |
>256 |
64 |
0.5 |
|
RAS1OE |
>1024 |
64 |
16 |
16 |
512 |
64 |
1 |
|
SN152 |
1024 |
>256 |
2 |
0.25 |
>512 |
512 |
16 |
|
RAS1Δ/Δ |
1024 |
>256 |
2 |
0.5 |
>512 |
512 |
16 |
Table 3-2: The SMIC Values of Antifungals in C. albicans Biofilms of Mutant Strains with Farnesol.
* “Flu” is an acronym for Fluconazole; “Itr” is an acronym for Itraconazole; “AmB” is an acronym for Amphotericin B; “Cas” is an acronym for Caspofungin; “Terb” is an acronym for Terbinafine; “5-Fc” is an acronym for 5-Flucytosine; “Nys” is an acronym for Nystatin. *Part B: SMIC50 of antifungals (µg/ml) to C. albicans biofilms with 100 µM farnesol; Part C: SMIC50 of antifungals (µg/ml) to C. albicans biofilms with 200 µM farnesol; Part D: SMIC50 of antifungals (µg/ml) to C. albicans biofilms with 300 µM farnesol.
3.4 The Morphologic Changes of Biofilms
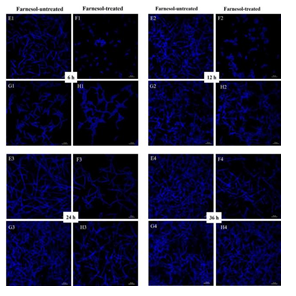
Both CLSM and SEM observations showed that the inhibitory effects of farnesol on hyphal growth of RAS1OE were more obvious than that of the control strain (pCaEXP-CAI4) (Figure 3 and 4), while the inhibitory effects of farnesol on hyphal growth of RAS1Δ/Δ were less obvious than that of the control biofilm (SN152) (Figure 3 and 4). The CLSM showed that the biofilms of RAS1OE and RAS1Δ/Δ exposed to farnesol had fewer hyphae but more pseudohyphae and blastospores than the control biofilms without farnesol (Figure 3 C1D1, C2D2, C3D3, C4D4; G1H1, G2H2; G3H3; G4H4), respectively. In addition, the biofilms formed from RAS1OE had more extensively grown hyphae and pseudohyphae with few blastospores than the biofilms formed from the control strain (pCaEXP-CAI4) (Figure 3 A1C1, A2C2, A3C3, A4C4). Moreover, the biofilms formed from RAS1Δ/Δ had fewer hyphae but more pseudohyphae and blastospores than the biofilms formed from the control strain (SN152) (Figure 3 E2G2, E3G3).
A: farnesol-untreated pCaEXP-CAI4; B: farnesol-treated pCaEXP-CAI4; C: farnesol-untreated RAS1OE; D: farnesol-treated RAS1OE. E: farnesol-untreated SN152; F: farnesol-treated SN152; G: farnesol-untreated RAS1Δ/Δ; H: farnesol-treated RAS1Δ/Δ. A1, B1, C1, D1 E1, F1, G1, and H1: 6 h biofilms; A2, B2, C2, D2, E2, F2, G2, and H2: 12 h biofilms; A3, B3, C3, D3, E3, F3, G3, and H3: 24 h biofilms; A4, B4, C4, D4, E4, F4, G4, and H4: 36 h biofilms. The biofilms of C1C2 had fewer hyphae but more pseudohyphae and blastospores than that of D1 and D2. The inhibitory effects of farnesol were more obvious in D1, D2, D3, and D4 than that in B1, B2, B3, and B4. The biofilms of G1 and G2 had fewer hyphae but more pseudohyphae and blastospores than that of H1and H2. The inhibitory effects of farnesol in H1, H2, H3, and H4 were less obvious than that in F1, F2, F3, and F4. Magnification: 400×; farnesol: 200 µΜ
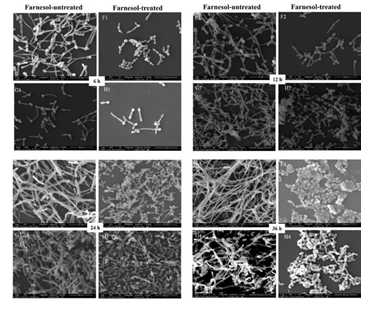
SEM analysis showed that the cells in farnesol-treated biofilms of RAS1OE and RAS1Δ/Δ appeared in the short pseudohyphae and had a rough cell surface than those in the biofilms without farnesol (Fig. 4 C1D1, C2D2, C3D3, C4D4, G1H1, G2H2, G3H3, G4H4). In addition, the biofilms formed from RAS1OE had more hyphae than those formed from the control strain (pCaEXP-CAI4) (Fig. 4 A1C1, A2C2, A3C3, A4C4), while the biofilms formed from RAS1Δ/Δ had fewer hyphae and more pseudohyphae than those formed from the control strain (SN152) (Fig. 4 E1G1, E2G2, E3G3, E4G4). The surfaces of cells in RAS1OE biofilms were similar to the biofilms of RAS1Δ/Δ.
A: farnesol-untreated pCaEXP-CAI4; B: farnesol-treated pCaEXP-CAI4; C: farnesol-untreated RAS1OE; D: farnesol-treated RAS1OE. E: farnesol-untreated SN152; F: farnesol-treated SN152; G: farnesol-untreated RAS1Δ/Δ; H: farnesol-treated RAS1Δ/Δ. A1, B1, C1, D1, E1, F1, G1, and H1: 6 h biofilms; A2, B2, C2, D2, E2, F2, G2, and H2: 12 h biofilms; A3, B3, C3, D3 E3, F3, G3, and H3: 24 h biofilms; A4, B4, C4, D4, E4, F4, G4, and H4: 36 h biofilms. The biofilms of D1 to D4 appeared in short pseudohyphae and had a rough cell surface, while C1 to C4 appeared in long hyphae and had smooth surfaces. The inhibitory effects of farnesol in D1, D2, D3, and D4 were more obvious than that in B1, B2, B3, and B4. The biofilms of F1 to F4 and H1 to H4 appeared in short pseudohyphae and had a rough cell surface, while E1 to E4 and G1 to G4 appeared in long hyphae and had smooth surfaces. The inhibitory effects of farnesol in H1, H2, H3 and H4 were less obvious than that in F1 F2, F3, and F4. Magnification: 2000×; farnesol: 200 µΜ.
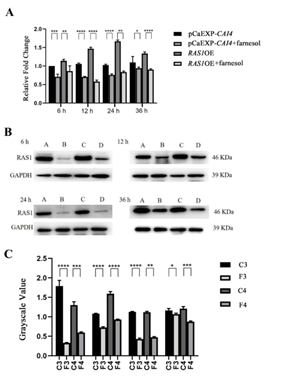
3.5 Farnesol Decreased the Expression of RAS1 in the Biofilms formed from the RAS1OE Strains
RT-qPCR and western blotting showed that farnesol significantly reduced the expression of RAS1 and the level of RAS1 in the biofilms formed by RAS1OE and the control strains (PCaEXP-CAI4), as compared with that of the farnesol-untreated biofilms at all studied phases (at 6, 12, 24, and 36 h biofilms) (P<0.05) (Figure 5), respectively. Moreover, farnesol reduced the expression of RAS1 and the level of RAS1 more obviously in the biofilms of RAS1OE than that of control strains (PCaEXP-CAI4) (P<0.05). Meanwhile, there were no expression of RAS1 in the biofilms formed by RAS1Δ/Δ, with or without farnesol.
A: Comparison of gene expression between farnesol-treated and untreated groups of C. albicans biofilms at all studied phases (6, 12, 24, and 36 h); B: The level of RAS1 in the strains with or without farnesol-treated was assessed using western blot at all studied phases; C: The bands in B was quantified and analyzed using grayscale value. C1: farnesol-untreated pCaEXP-CAI4; F1: farnesol-treated pCaEXP-CAI4; C2: farnesol-untreated RAS1OE; F2: farnesol-treated RAS1OE. Farnesol: 200 µM; *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001.
4. Discussion
Previous investigations have clearly showed that RAS1 gene is required for the induction of hyphal growth and the maintenance of hyphal state [12, 14], and also has a close relationship with pathogenic processes [27], such as cell adhesion and biofilm formation. RAS1 is a highly conserved signaling protein that might play the roles in the antifungal resistance of C. albicans and the relevant farnesol regulation mechanisms of the biofilm resistance, which are discussed in the present study. The study found that RAS1 is involved in the antifungal resistance of C. albicans. For the C. albicans in planktonic form, RAS1-overexpressing strain increased the resistance of C. albicans to fluconazole and itraconazole, and RAS1 deletion strain reduced that to all the tested antifungals. For the C. albicans in biofilm form, RAS1-overexpressing strain increased the resistance of biofilms to all the tested antifungal drugs, and RAS1 deletion strain reduced that. The findings indicates that RAS1 is associated with the antifungal resistance of C. albicans in planktonic and biofilm forms, which has not been reported in the previous studies.
Currently, azoles, polyenes, echinocandins and miscellaneous antifungals are the main antifungal families used in clinical practices. In the present study, the antifungal drugs for susceptibility test were fluconazole, itraconazole, amphotericin B, nystatin, caspofungin, terbinafine, and 5-flurocytosine. Fluconazole/itraconazole impairs ergosterol synthesis and leads to a cascade of membrane abnormalities in the fungus [28]. Amphotericin B/nystatin induces the ergosterol sequestration after binding to ergosterol and acts through pore formation at the cell membrane [29]. Caspofungin blocks the synthesis of β-(1,3)-D-glucan which is an essential component of the fungal cell wall [30]. Terbinafine prevents the conversion of squalene to lanosterol by inhibiting squalene epoxidase and inhibits the ergosterol synthesis [31]. 5-flurocytosine works by being converted into 5-fluorouracil inside the fungal, and blocks its ability to make protein [32]. Each of the antifungal drugs has its own mechanism of action, thus, the effects of antifungals to the mutant strains might be complex and different. The results that RAS1 overexpression increased the antifungal susceptibility to the drugs, and RAS1 deletion decreased that, regarding multiple antifungals with different mechanisms, implies that RAS1 has an ability in antifungal resistance and might be involved in a non-specific mechanism for the resistant regulation by C. albicans.
In the present study, farnesol decreased the expression of RAS1 gene and the level of RAS1 protein in C. albicans biofilms of RAS1-overexpressing strain and the strain with wild-type phenotype. Meanwhile, the inhibitory effects by farnesol on the resistance of biofilms of RAS1-overexpressing strain were more obvious than that of the wild-type phenotype (PCaEXP-CAI4) and RAS1-deletion strains. The results suggested that RAS1 is involved in the antifungal resistance regulated by farnesol. On the other hand, farnesol decreased the resistance of biofilms from RAS1-deletion strain to amphotericin B and caspofungin, while increased that to nystatin and terbinafine. The possible reason for this might be the complexity of multiple mechanisms of antifungals, which leads to the variations of antifungal resistance as the RAS1-deletion strain exposed to farnesol.
A previous study showed that RAS1-deletion strain was impaired in hyphal development under many different induction conditions [33]. In the present study, morphological observations showed that RAS1-deletion decreased the hyphal growth of the C. albicans biofilms and RAS1-overexpression increased the hyphal growth of C. albicans biofilms, which was in accordance with the previous study[33]. Furthermore, both CLSM and SEM observations showed that the inhibitory effects of farnesol on RAS1-overexpression were more obvious than that on the control strain with wild-type phenotype, and the inhibitory effects of farnesol on RAS1 deletion were less obvious than that on the control strain with wild-type phenotype. The morphological results suggest that farnesol is involved in the antifungal resistance of C. albicans biofilms via decreasing the hyphal growth caused by RAS1 regulation.
The formation of mature biofilms was composed of a series developed steps. C. albicans biofilms started with the initial adherence of yeast cells (0-2 h), followed by germination and micro-colony formation (2-4 h), filamentation (4-6 h), monolayer development (6-8 h), proliferation (8-24 h), and maturation (24-48 h) [34]. The main stages of biofilms were investigated here to clarify the effects of farnesol on the RAS1 regulation in C. albicans biofilms. A phase-specific mechanism might be involved in biofilm resistance, which appeared of the increased resistance of C. albicans with the mature biofilms (24 and 36 h). Because of this phase-specific resistance, the inhibitory effects of farnesol were reduced following with the mature of biofilms. In addition, the study showed that the inhibitory effects of farnesol on RAS1 level and the antifungal resistance of biofilms were consistent with the growth phases of RAS1-overexpressing strain, which further confirmed that RAS1 is involved in the antifungal resistance caused by farnesol. On the other hand, the effects of farnesol on the resistance of biofilms from the RAS1-deletion strain are not accordance with the mature of biofilms, other complicated and multiple antifungal mechanisms might affect the resistance of biofilms fromed from the RAS1-deletion strain, which need to be discussed in the future.
5. Conclusion
The extensive use of antifungals has led to the increased resistance of C. albicans biofilms to the few existing antifungals. Farnesol enhances the susceptibility of Candida albicans biofilms to antifungals, while the mechanisms of this behavior are poorly understood. Thus, it is crucial to explore the mechanisms of preventive strategies for the farnesol-mediated increase in susceptibility to various antifungals targeting biofilm-related infections. For the first time, this study demonstrated that RAS1 is involved in the antifungal resistance of C. albicans and the inhibition regulated by farnsol on the resistance of biofilms. Additional research regarding the specific molecular mechanisms should be further pursued to elucidate the antifungal resistance mechanisms of C. albicans biofilms.
Declarations
Ethics Approval
Not applicable
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Acknowledgments
Not applicable
Competing Interests
Lulu Zuo, Zhenzhen Zhang, Wei Duan, Yun Huang, Zheng Xu, Qinqin Zhang, Ying Lv, and Xin Wei declare that they have no conflict of interest.
Availability of Data and Materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' Contributions
XW conceived the study, participated in study design, and data analysis, and was responsible for writing and submitting the final manuscript. LLZ structured the RAS1 overexpressing and deletion strains and performed statistical analysis and drafted the manuscript. ZZZ carried out the XTT reduction assay and spot assay. WD and QQZ participated in statistical analysis. YL participated in RT-qPCR and western blot analysis. YH and ZX participated in morphological observations by CLSM and SEM. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Sciences Foundation of China under Grant Nos. 81970945 and 81371156 and the Foundation of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2018-87). Xin Wei was the recipient of these funds.
References
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence 4 (2013): 119-128.
- Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes and infection 5 (2016): 310-321.
- Nobile CJ, Johnson A. Candida albicans Biofilms and Human Disease. Annual review of microbiology 69 (2015): 71-92.
- Chen S, Xia J, Li C, et al. The possible molecular mechanisms of farnesol on the antifungal resistance of albicans biofilms: the regulation of CYR1 and PDE2. BMC Microbiology 18 (2018).
- Hall R, Turner KJ, Chaloupka J, et al. The Quorum-Sensing Molecules Farnesol/Homoserine Lactone and Dodecanol Operate via Distinct Modes of Action in Candida albicans. Eukaryotic Cell 10 (2011): 1034-1042.
- Jabra-Rizk MA, Meiller TF, James CE, et al. Effect of farnesol on staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrobial Agents and Chemotherapy 50 (2006): 1463-1469.
- Polke M, Leonhardt I, Kurzai O, et al. Farnesol signalling in Candida albicans – more than just communication. Critical Revies in Microbiology 44 (2018): 230-243.
- Koo H, Hayacibara MF, Schobel BD, et al. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. Journal of Antimicrobial Chemotherapy 52 (2003): 782-789.
- Luo K, Rocheleau H, Qi P, et al. Indole-3-acetic acid in Fusarium graminearum: Identification of biosynthetic pathways and characterization of physiological effects. Fungal biology 120 (2016): 1135-1145.
- Xia J, Qian F, Xu W, et al. In vitro inhibitory effects of farnesol and interactions between farnesol and antifungals against biofilms of Candida albicans resistant strains. Biofouling 33 (2017): 283-293.
- Yu L, Wei X, Ma M, et al. Possible Inhibitory Molecular Mechanism of Farnesol on the Development of Fluconazole Resistance in Candida albicans Antimicrobial Agents and Chemotherapy 56 (2011): 770-775.
- Davis-Hanna A, Piispanen AE, Stateva LI, et al. Farnesol and dodecanol effects on the Candida albicans RAS1-cAMP signalling pathway and the regulation of morphogenesis. Molecular Microbiology 67 (2008): 47-62.
- Martin R, Walther A, Wendland J. RAS1-Induced Hyphal Development in Candida albicans requires the Formin Bni1. Eukaryotic Cell 4 (2005): 1712-1724.
- Lindsay AK, Deveau A, Piispanen AE, et al. Farnesol and Cyclic AMP Signaling Effects on the Hypha-to-Yeast Transition in Candida albicans. Eukaryotic Cell 11 (2012): 1219-1225.
- Piispanen AE, Bonnefoi O, Carden S, et al. Roles of RAS1 Membrane Localization during Candida albicans Hyphal Growth and Farnesol Response. Eukaryotic Cell 10 (2011): 1473-1484.
- Chen S, Xia J, Li C, et al. The possible molecular mechanisms of farnesol on the antifungal resistance of albicans biofilms: the regulation of CYR1 and PDE2. BMC microbiology 18 (2018): 203.
- Noble S, Johnson A. Strains and Strategies for Large-Scale Gene Deletion Studies of the Diploid Human Fungal Pathogen Candida albicans. Eukaryotic Cell 4 (2005): 298-309.
- Zuo L, Chengxi LI, Zheng L, et al. Comparison of knocking-out the RAS1 gene in candida albicans using different auxotrophic markers. Stomatology (2018).
- Xia J, Zhang Z, Xu S, et al. A study of modified lithium acetate transformation for plasmid transformation in candida albicans. Journal of Nanjing Medical University (Natural Sciences) (2017).
- Ramage G, Saville S, Wickes B, et al. Inhibition of Candida albicans Biofilm Formation by Farnesol, a Quorum-Sensing Molecule. Applied and Envirnmental Microbiology 68 (2002): 5459-5463.
- Xiao J, Moon Y, Li L, et al. Candida albicans Carriage in Children with Severe Early Childhood Caries (S-EC) and Maternal Relatedness. PLoS ONE 11 (2016).
- Ku TS, Palanisamy SK, Lee S. Susceptibility of Candida albicans biofilms to azithromycin, tigecycline and vancomycin and the interaction between tigecycline and antifungals. International journal of antimicrobial agents 5 (2010): 441-446.
- Manoharan RK, Lee J, Kim Y. Alizarin and Chrysazin Inhibit Biofilm and Hyphal Formation by Candida albicans. Fronters in Cellular and Infection Microbiology 7 (2017).
- Richard M, Nobile CJ, Bruno V, et al. Candida albicans Biofilm-Defectiv Mutants. Eukaryotic Cell 4(2005): 1493-1502.
- Barbier FF, Chabikwa TG, Ahsan M, et al. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods 15 (2019).
- Ma S, Li H, Yan C, et al. Antagonistic effect of protein extracts from Streptococcus sanguinis on pathogenic bacteria and fungi of the oral cavity. Experimental and Therapeutic Medicine 7 (2014): 1486-1494.
- Katragkou A, McCarthy M, Alexander EL, et al. In vitro interactions between farnesol and fluconazole, amphotericin B or micafungin against Candida albicans The Journal of antimicrobial chemotherapy 70 (2015): 470-478.
- Martel CM, Parker J, Bader O, et al. Identification and Characterization of Four Azole-Resistant erg3 Mutants of Candida albicans. Antimicrobial Agents and Chemotherapy 54 (2010): 4527-4533.
- Gray KC, Palacios DS, Dailey I, et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proceedings of the National Academy of Sciences 109 (2012): 2234-2239.
- Hao B, Cheng S, Clancy C, et al. Caspofungin Kills Candida albicans by Causing both Cellular Apoptosis and Necrosis. Antimicrobial Agents and Chemotherapy 57 (2012): 326-332.
- Koutsoulas C, Pippa N, Demetzos C, et al. Preparation of liposomal nanoparticles incorporating terbinafine in vitro drug release studies. Journal of nanoscience and nanotechnology 14 (2014): 4529-4533.
- Gopinathan S, Janagond AB, Agatha D, et al. Detection of FUR1 Gene in 5-Flucytosine Resistant Candida Isolates in Vaginal Candidiasis Patients. Journal of clinical and diagnostic research: JCDR 7 (2013): 2452-2455.
- Martin R, Walther A, Wendland J. RAS1-Induced Hyphal Development in Candida albicans requires the Formin Bni1. Eukaryotic Cell 4 (2005): 1712-1724.
- Mathé L, Dijck PV. Recent insights into Candida albicans biofilm resistance mechanisms. Current Genetics 59 (2013): 251-264.
Supplemental Materials
The primers used to obtain the RAS1 deletion strain are listed in Table 1.
|
Primer |
Sequences (5’-3’) |
|
Universal F |
ccgctgctaggcgcgccgtgAGCTCGGATCCACTAGTAACG |
|
Universal R |
gcagggatgcggccgctgacGCCAGTGTGATGGATATCTGC |
|
RAS1-F |
GATGAATATGACCCAACT |
|
RAS1-R |
AAGAAACACCTCCATTAC |
|
RAS UP-F |
CTCCCTAACCCCAGTAAA |
|
RAS UP-R |
cacggcgcgcctagcagcggTCAAGGTCAATGTCCAAT |
|
RAS DOWN-F |
gtcagcggccgcatccctgcTGATTGTTTCCAAGTTAC |
|
RAS DOWN-R |
AAGGTATGAAGAGGATGT |
|
RAS UP check |
ATGACAATCCCTCCCTAA |
|
RAS HIS left |
TGCATAAACGGTGGCACATT |
|
RAS LEU left |
AAACCTCTTTCTTGACCC |
|
RAS HIS right |
TCCAATTCAACGACGGCTGA |
|
RAS LEU right |
GATTCTGATTGGCTCTTT |
|
RAS DOWN check |
GAGACAAAGGTATGAAGAGG |
Table 1: Primer Sequences in Constructing Knockout Strain RAS1Δ/Δ.
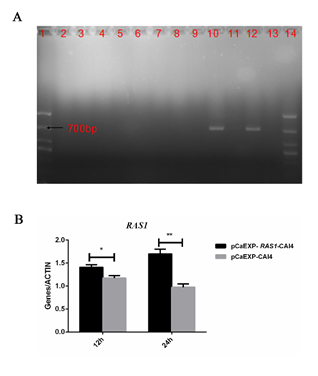
Identification of RAS1 Overexpressing Strain (RAS1OE)
The results of sequencing showed that results of overexpression strain (pCaEXP-RAS1) were the same as NCBI C. albican RAS1 sequence (Fig. 1A). RT-PCR showed that the RAS1 expression (at 12 and 24 h) increased significantly in the RAS1OE (pCaEXP- RAS1-CAI4) compared to the control strain (pCaEXP-CAI4) (Figure 1B).
Figure 1: Identification of RAS1 Gene Overexpressing Strains.
A: Agarose gel electrophoresis; 1: maker; 2-7: CaEXP-CAI4: 8-13: transformants; 10 and 12: postive transformations of PCaEXP-RAS1-CAI4; 14: maker; B: RT-PCR showed that the expression of RAS1 (at 12 and 24 h) significantly increased compared to the control strain. *: P<0.05; **: P<0.01.
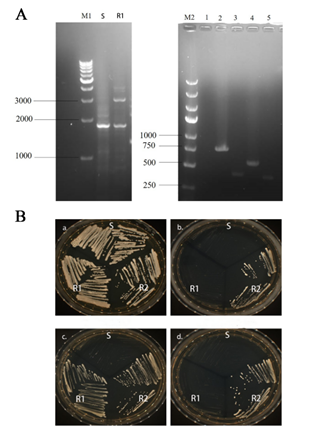
Identification of RAS1 Deletion Strain (RAS1Δ/Δ)
The results of agarose gel electrophoresis showed that C. albicans RAS1Δ/Δ (RAS1 double allelic deletion strain) was successfully constructed using HIS-LEU-ARG knocking-out strategy (Fig. 2A). As shown in Fig. 2B, only C. albicans RAS1Δ/Δ (RAS1 double allelic deletion strain) could grow on selective medium (SD-Leu-His).
Figure 2: Identification of RAS1 Gene Knockout Strains.
A: Agarose gel electrophoresis; M1: 1 kb DNA marker; S: SN152 strain; R1: RAS1Δ; M2: DL5000 DNA marker; 1: RAS1 gene; 2: HIS1 upstream fragment; 3: HIS1 downstream fragment; 4: LEU2 upstream fragment; 5: LEU2 downstream fragment; B: S: SN152 strain; R1: RAS1Δ strain; R2: RAS1Δ/Δ strain. The growth of the control strains and mutant strains in the following media are shown: a: YPD agar; b: SD-Leu agar, c: SD-His agar; d: SD-Leu-His agar; The schematic drawing of a plate indicates the location of the plated strains (S, R1 and R2).