Rapid Diagnosis of Invasive Fungal Infections Caused by Candida and Aspergillus Species in Patients Admitted to Intensive Care Unit of a Tertiary Care Hospital
Article Information
Shaila Akhtar, Shaheda Anwar, Ahmed Abu Saleh
Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University, Dhaka, Bangladesh
*Corresponding author: Shaila Akhtar, Department of Microbiology and Immunology, Bangabandhu Sheikh Mujib Medical University, Dhaka, Bangladesh.
Received: 01 October 2023; Accepted: 10 October 2023; Published: 19 October 2023
Citation: Shaila Akhtar, Shaheda Anwar, Ahmed Abu Saleh. Rapid Diagnosis of Invasive Fungal Infections Caused by Candida and Aspergillus Species in Patients Admitted to Intensive Care Unit of a Tertiary Care Hospital. Fortune Journal of Health Sciences. 6 (2023): 378-385.
View / Download Pdf Share at FacebookAbstract
Introduction: Invasive fungal infections (IFI) are common nosocomial infections in immunosuppressed individuals. Rapid diagnosis of IFIs is important to support the growing number of at-risk patients and standardize the treatment guidelines. The study aimed to assess the role of serum 1,3-β-D-Glucan (BDG) and galactomannan (GM) biomarkers and real-time PCR in the rapid diagnosis of IFIs.
Method: It was a cross-sectional and observational study. A total of 60 peripheral venous blood samples were collected from clinically suspected IFI patients from the intensive care unit (ICU) of Bangabandhu Sheikh Mujib Medical University (BSMMU). The study was conducted from December 2022 to August 2023.
Result: Out of 60 clinically suspected IFI patients, 12 (20%) were positive for fungus in blood culture, of which Candida species accounted for 11 (18.33%) and Aspergillus species accounted for 1 (1.67%). Using serum biomarkers (GM and BDG), 46.7% of patients were positive for the BDG assay, and 20.0% of patients were positive for the GM detection assay. Using real-time PCR, Candida species (C. albicans, C. parapsilosis, C. glabrata, C. krusei) were detected in 24 (40.0%) cases and Aspergillus fumigatus were detected in 14 (23.33%) cases. The agreement between real-time PCR and serum biomarkers was 81.67% and the kappa value was 0.639, which was considered good. The sensitivity of GM, BDG, and real-time PCR were 67.65%, 92.00%, and 92.00% respectively. The specificity of GM, BDG, and real-time PCR were 100.00%, 80.77%, and 58.34% respectively.
Conclusion: Serum GM and BDG biomarkers and PCR are promising and highly sensitive tests for rapidly diagnosing at-risk patients suspected of having invasive fungal infections.
Keywords
invasive fungal infections, serum biomarkers, real-time PCR, 1,3-β-D-Glucan, galactomannan
invasive fungal infections articles, serum biomarkers articles, real-time PCR articles, 1,3-?-D-Glucan articles, galactomannan articles
Article Details
1. Introduction
Fungal pathogens cause at least 13 million infections and 1.5 million deaths worldwide each year, mainly in people with compromised immune function [1]. It is estimated that approximately 3 million people worldwide suffer from severe chronic fungal infections and nearly 1.9 million patients develop acute invasive fungal infection (IFI) each year [1]. In recent years, the prevalence of IFI has been increasing, mainly among hospitalized patients. The pathogens causing IFIs are mostly opportunistic, including Candida, Cryptococcus, and Aspergillus species [2]. The most common pathogenic fungi in immunocompromised patients are Candida and Aspergillus species [3].
The rate of candidemia varies significantly by country, and using a conservative 5 per 100,000 rate, a study estimated 8100 cases in Bangladesh each year [4]. Among other serious fungal infections, invasive aspergillosis (IA) is hardly recognized in Bangladesh. Among the estimated 34.5 million adults over the age of 40, there are an estimated 2,481,444 patients with COPD in Bangladesh (7.2%) [5]. Assuming that 13% of these patients are admitted to hospital each year and that 1.3% develop IA. Given the aforesaid data, invasive candidemia and aspergillosis constitute a serious burden in Bangladesh. Diagnosis of IFIs is conventionally done by microscopy and cultures which have limitations in cases of sensitivity, time to positivity (2-4 days) for Candida and Aspergillus species, and difficulty in collecting invasive specimens. The sensitivity of culture is only 2-8% in the case of Aspergillus species and 20-40% in the case of Candida species [6]. Novel diagnostic approaches based on non-culture-based methods have been developed that could enable rapid diagnosis and treatment of IFIs. The most promising techniques include the detection of fungal antigens (BDG and GM) in blood and real-time polymerase chain reaction (PCR).
GM is a carbohydrate component of the Aspergillus cell wall that is released by all Aspergillus species during growth in the tissue. GM is escaped into the blood and other body fluids even in the early steps of Aspergillus invasion. Circulating GM could be sensed in the serum before diagnosis by clinical and radiological examination in about 65% of patients [3]. BDG is a cell wall component of many fungi and can be detected in IFIs due to Aspergillus species, Candida species, Pneumocystis jirovecii, Fusarium species, Trichosporon species, and Saccharomyces species, but absent in mucormycetes and cryptococcus. However, unlike GM, which is specific to Aspergillus, BDG is found in several fungal species. Therefore it can be considered a pan-fungal marker and a predictor of systemic fungal infection when demonstrable in blood or other normally sterile body fluids [7]. Pathogen detection using real-time PCR is undoubtedly the most powerful tools for rapid detection of human pathogens. The potential value of nucleic acid-based methods for the detection and identification of fungus in immunosuppressed patients is undeniable. IFIs present a major challenge in the treatment of immune-compromised patients. Early diagnosis is required to improve survival from these infections. As numerous articles show, conventional microbiological, histological, and radiological techniques still form the basis of diagnosis but are insensitive and have little impact on clinical decision-making. There is an urgent need to develop new effective diagnostic methods. These tests must be rapid and highly sensitive.
2. Materials and Methods
2.1 Study design
This is an observational and cross-sectional study conducted in a tertiary care hospital in a time span of 9 months, from December 2022 to August 2023.
2.2 Study sites
Peripheral venous blood was collected from the patients admitted to the intensive care unit of Bangabandhu Sheikh Mujib Medical University (BSMMU). Laboratory work was performed in the Department of Microbiology & Immunology, (BSMMU).
2.3 Participants
Clinically suspected patients of IFIs were admitted to the intensive care unit of BSMMU.
2.4 Inclusion criteria
Clinically suspected patients of IFIs with any of the following factors were included in this study.
The factors are:
- Neutropenia (<0.5x 109 neutrophils/L).
- Use of corticosteroid for a prolonged time (minimum dose of 0.3 mg/kg/day of prednisone equivalent, for >3 weeks).
- Fever refractory to at least 3 days of appropriate antibiotics/fever relapsing after a period of defervescence of at least 48 hours while still receiving antibiotics.
2.5 Exclusion criteria
Participants were not be enrolled if they
- Had any bacterial/parasitic infections.
- Were receiving immunoglobins and albumin.
- Were on the regimen for anti-fungal therapy.
2.6 Data collection
Relevant data were collected from patients or their attendants or the clinical history records and investigations of the patients in a predesigned data collection sheet. Results obtained from laboratory methods were recorded in a separate data collection sheet.
Among the 60 patients’, X-ray was done in all the patients. HRCT chest was done where clinically indicated.
2.7 Procedures in the laboratory
Peripheral venous blood was collected under aseptic conditions using a sterile disposable syringe tagged with a butterfly needle after preparing the patient’s skin with, at first 70% alcohol and hereafter with 1% tincture iodine [8]. A total of 16 ml of blood was collected and divided into three parts. The first part is for blood culture which consists of 10ml. According to the procedure, this part was inoculated into an automated blood culture bottle. The second part consists of 3ml of blood, which was taken in a sterile EDTA-coated vials for DNA extraction and shaken gently for proper mixing. Another 3 ml of blood was taken into a sterile test tube without anticoagulant for serum separation.
2.8 Blood culture and identification of yielding fungi
Inoculation of 10 ml of blood into the BD-BACTECTM Plus Aerobic/F blood culture bottle after disinfection of the head of the bottle with 70% alcohol [9]. Then the blood culture bottle was inserted into the BD-BACTECTM FX40 (Becton, Dickinson and Company, Sparks, MD 21152, USA) blood culture machine for incubation at 370 C temperature for 1-5 days according to manufacturer’s instruction [10].After showing positive indicators on the machine; with all necessary aseptic precautions, isolation of microorganisms was done by sub-culturing a small amount of liquid media on Sabouraud dextrose agar (SDA) media, blood agar media, and MacConkey agar media and were incubated at 370C for 24 hours aerobically, up to 4-5 days for SDA media, as maximum time required for Candida and Aspergillus species to grow in SDA media is 4-5 days [11]. A smear was also made from growth on subculture media and Gram stain preparations were performed. Culture plates with growth of organism other than Candida and Aspergillus species were excluded. All the bottles and the culture media were discarded after 7 days according to the proper safety procedure [12]. Identification of Candida spp. and Aspergillus spp. by colony morphology, wet film, Gram staining, and microscopy was done.
2.9 Galactomannan assay
Properly stored serum from patients of clinically suspected invasive fungal infections was used for determination of Galactomannan (GM) by using sandwich ELISA according to the manufacturer’s instruction (Genobio Pharmaceutical Co., Ltd, Tianjin). After measuring the absorbance values for each control and samples, the mean cut-off control value was calculated and then sample index (SI) was calculated. SI value ?0.5, were considered as positive.
2.10 1,3-β-D-Glucan assay
Properly stored serum from patients of clinically suspected invasive fungal infections was used for determination of BDG by using the Kinetic ELISA method according to manufacturer’s instruction (Genobio Pharmaceutical Co., Ltd, Tianjin). The results were expressed in pg/mL of sample and ranged from non-detectable (<31.25 pg/mL) to >500 pg/mL and read from the standard curve. BDG values <60 pg/mL were interpreted as negative results. Values ≥80 pg/mL were interpreted as positive. Values from 60 to 79 pg/mL were considered possible fungal infections.
2.11 DNA extraction
DNA extraction from peripheral venous blood was done according to the manufacturer’s instructions (Qiagen QIAamp DNA Mini extraction kit). The volume of the clinical sample for DNA extraction was 200µl. Extracted DNA stored at -20°C until use. The concentration of DNA was measured by spectrophotometric assay performed using a Nanodrop 200 spectrophotometer (Thermo Fisher scientific, Waltham, MA, USA) according to the manufacturer’s instruction.
2.12 Detection of Candida species and Aspergillus species by real-time PCR
Molecular detection of Aspergillus species (Aspergillus fumigatus, Aspergillus flavus and Aspergillus terreus) And Candida species (Candida albicans, Candida parapsilosis, Candida glabrata and Candida krusei) was done using an Aspergillus differentiation kit (VIASURE) and Candida spp. Detection kit v2 (Anatolia Geneworks, Bosphore, Turkey) according to the manufacturer’s instructions respectively. The real-time PCR instrument was CFX Opus 96 TouchTM real-time detection system (Bio-Rad). PCR was considered as positive when for a single sample, the CT value was ≤40 for Aspergillus species detection and ≤37 for Candida species detection.
2.13 Ethical approval
This study was ethically approved by the Institutional Review Board (IRB), BSMMU on 12/12/2022.
2.14 Statistical analysis
Data analysis was done using SPSS software package version-27 (Strata Corporation, College station, Texas). P value was calculated by Chi-square test to explore the association of categorical data. P value <0.05 was considered statistically significant. Median were calculated for asymmetric quantitative data and mean with 95% confidence interval and standard deviation. Agreement between the three methods was analyzed using Kappa statistic. EORTC/MSG guideline 2020 [13] was as reference gold standard to calculate true positive and true negative cases. Proven and probable cases were considered as true positive and no IFIs cases were considered as true negative.
3. Results
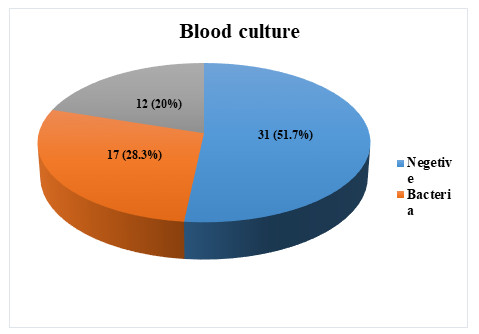
In this study, a total of 60 blood samples were collected from clinically suspected patients of invasive fungal infections were collected. Automated blood culture, measurement of serum biomarkers (BDG and GM), and real-time PCR were performed in all 60 blood samples. Out of 60 clinically suspected invasive fungal infections patients’ blood samples, 12 (20%) were positive for fungus, 17 (28.3%) were positive for bacteria and 31 (51.7%) yielded no growth, shown in a pie chart Figure 1. In Table 1, elaboration of the laboratory test results of patients with clinical suspicion of IFIs are shown. Out of 60 clinically suspected IFIs patients’, 12 (20%) were positive for fungus in automated blood culture, of which Candida species accounted for 11 (18.33%) and Aspergillus species accounted for 1 (1.67%). Using serum biomarkers, 46.7% of patients enrolled were positive for a BDG detection assay, and 20.0% of patients enrolled were positive for a GM detection assay. Using real-time PCR, Candida species were detected in 24 (40.0%) of cases and Aspergillus species were detected in 14 (23.33%) of cases. In the present study thirty-eight (63.33%) cases were positive by real-time PCR assay. Among them, 24 (40.00%) were Candida species and 14 (23.22%) were Aspergillus species. All the Aspergillus were Aspergillus fumigatus. Among the identified Candida species 8 (21.60%) were Candida albicans, 9 (23.30%) were Candida parapsilosis, 6 (15.78%) were Candida glabrata and 1 (2.70%) case was Candida krusei.
Table 1: Laboratory test results of patients with clinical suspicion of invasive fungal infections (n=60)
|
Patients of clinically suspected IFIs (n=60) |
||
|
Mycological findings |
No |
% |
|
Blood Culture |
||
|
Positive |
12 |
20.00% |
|
Candida species |
11 |
18.30% |
|
Aspergillus species |
1 |
1.67% |
|
Negative |
48 |
80.00% |
|
Serum biomarkers |
||
|
Galactomannan |
12 |
20.00% |
|
1,3-β-D-Glucan |
28 |
46.70% |
|
Real-time PCR |
||
|
Positive |
38 |
63.30% |
|
Candida species |
24 |
40.00% |
|
Candida albicans |
8 |
21.00% |
|
Candida parapsilosis |
9 |
23.30% |
|
Candida glabrata |
6 |
15.70% |
|
Candida krusei |
1 |
2.70% |
|
Aspergillus species |
14 |
23.30% |
|
Aspergillus fumigatus |
14 |
36.80% |
|
Negative |
22 |
36.60% |
Blood culture showed a 55.0% agreement with real-time PCR, and the kappa coefficient is 0.211, which was considered fair. On the other hand, blood culture exhibited a higher agreement of 73.33% with the serum biomarkers (BDG and GM) and, the kappa value is 0.444, which was statistically significant. Agreement between Real-time PCR and serum biomarkers (BDG and GM) was 81.67% and the kappa value was 0.639, which was considered good, described in Table 2. Table 3, shows clinically suspected IFI patients' classification according to EORTC/MSG criteria (Peter Donnelly et al., 2020). According to this criteria, 11 (18.33%) cases were classified as proven cases, 12 (20.00%) cases were classified as probable cases, 16 (26.67%) cases were classified as possible cases and 21 (35.00%) cases were classified as no IFI cases. In Table 4, the sensitivity, specificity, PPV, and NPV are shown. Here updated EORTC/MSG criteria,2020 was used as the gold standard to calculate true positive and true negative cases. Proven and probable cases were considered true positive, and no IFIs cases were considered true negative. The sensitivity of GM, BDG, and real-time PCR were 67.65%, 92.00%, and 92.00% respectively. The specificity of GM, BDG, and real-time PCR were 100.00%, 80.77%, and 58.34% respectively. The PPV of GM, BDG, and real-time PCR were 100.00%, 82.14%, and 91.31% respectively. The NPV of GM, BDG, and real-time PCR were 65.63%, 91.31%, and 91.31% respectively. Table 5, describes the time taken by each method to diagnose invasive fungal infections. The automated blood culture method took the most time while PCR serum biomarkers took the least time.
Table 2: Agreement between blood culture, serum biomarkers (1,3-β-D-Glucan and Galactomannan), and real-time PCR used for diagnosis of invasive fungal infections.
|
Test |
Agreement % |
κ* (Kappa) |
P value |
|
Blood culture vs real-time PCR |
55.00% |
0.211 |
0.017 |
|
Blood culture vs serum biomarkers |
73.33% |
0.444 |
<0.001 |
|
Real-time PCR vs serum biomarkers |
81.67% |
0.639 |
<0.001 |
*Value of κ
Strength of agreement <0.2: poor; 0.2-0.4: fair; 0.41-0.6: moderate; 0.61-0.8: good; 0.81-1.00: very good
Table 3: Classification of cases according to EORTC/MSG criteria
|
Clinically suspected invasive fungal infection cases (n=60) |
||
|
Class |
No |
% |
|
Proven |
11 |
18.33 |
|
Probable |
12 |
20 |
|
Possible |
16 |
26.67 |
|
No IFI |
21 |
35 |
Table 4: Sensitivity, specificity, PPV, and NPV of galactomannan, 1,3-β-D-Glucan assay, and real-time PCR in detection of invasive fungal infections (n=60)*
|
Test |
Sensitivity |
Specificity |
PPV |
NPV |
|
Galactomannan |
67.65% |
100.00% |
100.00% |
65.63% |
|
1,3-β-D-glucan assay |
92.00% |
80.77% |
82.14% |
91.31% |
|
real-time PCR |
92.00% |
58.34% |
60.53% |
91.31% |
*Considering Updated EORTC/MSG criteria, 2020 as the gold standard
Table 5: Time taken by each method to diagnose invasive fungal infections
|
Method |
Time |
|
Automated blood culture |
Up to 1 week |
|
Serum biomarkers assay |
3-4 hours |
|
Real-time PCR |
3-4 hours |
4. Discussion
Despite new antifungal drugs, mortality and life-threatening complications of invasive fungal infections are still frequently reported in critically ill patients. The diagnosis of IFI is negatively affected by suboptimal culture results. Therefore, new tools are required to rapidly diagnose IFIs in patients in the ICU. In the present study, fungi were isolated in 12 (20%) automated blood cultures. Except for 1, all were Candida species. The single one was the Aspergillus species. This is consistent with Montagna et al. in Italy in 2013 who found 87.6% of yeast and 12.4% of mold in all infections [14]. The study was conducted on 5561 patients over 18 months. The large study population and different types of samples (e.g., blood, peritoneal and cerebrospinal fluid, bronchial aspirate, sputum) may be the reason for 12.4% of mold infections in their study. Apart from this, most Candida infections are endogenous while Aspergillus infection is mainly exogenous, this could be another reason for higher Candida species infection.
Candida was isolated in 18.33% of the current study population. A similar rate of 13.5% was reported by Yang, Cheng and Lo, in 2006 in Taiwan [15]. As only one case of Aspergillus species (1.67%) was identified it is difficult to compare with other studies, where also isolation rate of mold was relatively low. The single case of Aspergillus in our study was Aspergillus fumigatus. Binder and Lass-Flörl, in 2011 reported Aspergillus fumigatus as the most common species [16]. Aspergillus fumigatus is the most common mold in the environment, this could be the reason of most common species identified in blood culture. Even Aspergillus fumigatus is the most common contaminant in culture. In our case apart from culture, the patients were also positive for biomarkers (BDG and GM) and PCR from serum, which indicates case of invasive infection not contamination. BDG detection test, 46.7% of patients were positive in the study population. BDG detection test was positive in all the blood culture-positive cases and in 26.67% blood culture-negative cases. In the case of GM, 12 (20%) enrolled patients were positive including the single Aspergillus positive automated blood culture case in our study population. These are in agreement with Azab et al. [17]. Azab et al. in 2015 reported using BDG detection test 37.9% patients positive in their study [17]. Biomarkers detection test was positive in all the blood culture positive cases and 25% of the blood culture negative cases.
In the present study thirty-eight (63.33%) cases were positive by real-time PCR assay. Among them, 24 (40.00%) were Candida species and 14 (23.22%) were Aspergillus species. All the Aspergillus were Aspergillus fumigatus. Among the identified Candida species 8 (21.60%) were Candida albicans, 9 (23.30%) were Candida parapsilosis, 6 (15.78%) were Candida glabrata and 1 (2.70%) case was Candida krusei. Majority of the Candida infections 65.2% were caused by non-albicans Candida species, whereas 34.8% were caused by Candida albicans. Similar results were reported by Montagna et al. in 2013 in Italy [18]. They reported that 59.8% of IFI were caused by non-Candida albicans species, and Candida albicans, accounted for 40.2% of yeast infections [18]. About 38 (63.33%) of enrolled patients were positive by real-time PCR assay. However, 20.0% were positive by automated blood culture. This number increases to 45% by serum biomarkers detection assay and further increases to 63.33% cases out of the 60 study populations, with a statistically significant difference. These results are in accordance with the work of El-Sayed in 2012, who detected 57.7% cases positive by PCR [22]. 50% of PCR positive cases were missed by blood culture. Another study stated that 64.5% were positive by both culture and PCR, of these samples 19.4% showed no growth of fungi but were positive by PCR [23]. The diagnosis of patients with IFIs in the present study revealed a good agreement between serum biomarkers assay of fungal antigen in serum and real-time PCR. However, there is moderate agreement between the results of blood culture and serum biomarkers assay. Additionally, there is a fair agreement between the results of blood culture and real-time PCR. By the results, Azab et al. also detected a good agreement (94.4%) between the two non-culture-based methods [17].
IFIs in this study were defined and classified using EORTC/MSG Consensus Group criteria [13], and according to these criteria 11 (18.33%) cases were classified as proven, 12 (20%) were probable, 16 (26.67%) were possible, and 21 (35.00%) cases were classified as no IFI cases. This is consistent with a study [24]. This study results of BDG are in agreement with the study conducted by Giacobbe et al. [25]. In which the sensitivity, specificity, PPV, and NPV of BDG were 92.00%, 81.00%, 79.00%, and 93.00%. A meta-analysis by Karageorgopoulos et al. showed an overall pooled sensitivity and specificity of serum BDG assay for diagnosis of invasive fungal infections 76.8% and 85.0% respectively [26]. However, marked statistical heterogeneity was noted. In this study, BDG assay gave false positive results in only 5 patients. False positive results in ICU patients may be due to many clinical variables and conditions such as surgical gauzes, renal replacement therapy, albumin transfusion, and broad-spectrum antibiotics [28,30]. Specificity and PPV can be increased by two consecutive BDG testing without significant impact on NPV [28]. The result of GM is consistent with many studies. A meta-analysis by Leeflang et al. showed the sensitivity and specificity of GM assay 49% to 77% and 89% to 97% respectively [29]. In our study, the GM assay gave no false positive result. GM showed false negativity in 11 proven cases but these cases were Candida blood culture positive, so GM obviously should be negative in these cases. Another cause of false negative results may be steroid intake [30].
In this study, the sensitivity, specificity, PPV, and NPV of real-time PCR were consistent with the study conducted by Gupta et al. [31] . This result is also consistent with the study conducted by Springer et al. where sensitivity was 85.10% and specificity was 64.50% [32]. Here, real-time PCR gave 15 false positive results. Compared to gold standard as observed by several other authors, these false positive results could be due to subclinical infection, fungal colonization, contamination by airborne fungal spores, fungal PCR product carryover, and cross-reactivity with nonfungal DNA. The risk of contamination is relatively low as the positive material (specimens and positive control) was stored separately from all other reagents. Workstations were wiped with 5% hypochlorite and 70% ethanol at every stage. The extraction of DNA, preparation of PCR products, mixing of PCR products, and amplification were carried out in biosafety hoods. All the reagents were mixed and dispensed in a pre-mix area. Jordanides et al. commented on the difficulty in determining an actual ‘false-positive’ result from an early ‘true positive’ result, reflecting the fact that PCR may be a more sensitive indicator of early invasive fungal infections [33]. In this study the specificity of real-time PCR is not good because of statistical analysis many of the PCR positive cases were considered as false positive. But we could not consider them as false positive because these patients were clinically suspected cases of invasive fungal infections. They had the signs and symptoms of invasive fungal infections and most of them had radiological and others laboratory evidences. Blood culture revealed no bacteremia also. So, we could not consider them as false positive cases.
In the study, false negative results were observed in 2 out of the 60 patients evaluated. The real-time PCR assay for Candida species detection used in this study can detect only four species of Candida. Several many species of Candida can cause invasive fungal infections in intensive care unit patients. In addition, in the study, samples for PCR were taken only once and no subsequent samples were taken. This might have led to the false negative reports in some of the cases as sequential positive PCR reports have been shown to increase the sensitivity of PCR in the studies by Landlinger et al. [34].
5. Conclusion
Real-time PCR and serum biomarkers detect more cases of IFIs than blood culture in blood from clinically suspected cases of IFIs in the ICU. BDG is positive in all the Candida and Aspergillus culture-positive cases and GM is positive in one Aspergillus culture positive case. Concerning the time consumed to diagnose IFIs, real-time PCR and serum biomarkers take the least time. The diagnosis of patients with IFIs reveals a good agreement between real-time PCR and serum biomarkers.
Conflict of interest
The authors have none to declare.
References
- Bongomin F, Gago S, Oladele R, Denning D. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. Journal of fungi 3 (2017): 57.
- Li Y, Gao Y, Niu X, Wu Y, Du Y, Yang Y, et al. A 5-Year Review of Invasive Fungal Infection at an Academic Medical Center. Frontiers in Cellular and Infection Microbiology 10 (2020): 1-10.
- Erjavec Z, Verweij PE. Recent progress in the diagnosis of fungal infections in the immunocompromised host. Drug Resistance Update 5 (2002): 3-10.
- Gugnani HC, Denning DW, Rahim R, Sadat A, Belal M, Mahbub MS. Burden of serious fungal infections in Bangladesh. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology. 36 (2017): 993-997.
- Alam DS, Chowdhury MA, Siddiquee AT, Ahmed S, Clemens JD. Prevalence and determinants of chronic obstructive pulmonary disease (COPD) in Bangladesh. HHS Public Access12 (2015): 658-667.
- Zeng W, Huang Y, Deng Y, Wen M, Han Y, Zhong W, et al. Clinical evaluation of the (1, 3) -β-D-glucan assay as an aid to diagnosis of fungal infections in severe pneumonia patients. Chinese Journal of Emergency Medicine 25 (2016): 659-662.
- Cuenca-Estrella M, Bernal-Martinez L, Buitrago MJ, Castelli MV, Gomez-Lopez A, Zaragoza O, et al. Update on the epidemiology and diagnosis of invasive fungal infection. International Journal of Antimicrobial Agents. 32 (2008): 143-147.
- Little JR, Murray PR, Traynor PS, Spitznagel E. A randomized trial of povidone-iodine compared with iodine tincture for venipuncture site disinfection: Effects on rates of blood culture contamination. American Journal of Medicine 107 (1999): 119-125.
- Doern GV, Carroll KC, Diekema DJ, Garey KW, Rupp ME, Weinstein MP, et al. A comprehensive update on the problem of blood culture contamination and a discussion of methods for addressing the problem. Clinical Microbiology Reviews 33 (2020): 1-21.
- Becton D and C. BACTEC TM FX40 Instrument User’s Manual. 8090414 (2021).
- Chander J. Textbook of medical mycology. 4th ed. Chander J, editor. New Delhi: Jaypee Brothers Medical Publishers (P) Ltd (2018).
- Madhavan P, Jamal F, Chong PP. Laboratory isolation and identification of Candida species. Journal of Applied Science 11 (2011): 2870-2877.
- Peter Donnelly J, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, et al. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clinical Infectious Diseases 71 (2020): 1367-1376.
- Montagna MT, de Giglio O, Napoli C, Lovero G, Caggiano G, Delia M, et al. Invasive fungal infections in patients with hematologic malignancies (Aurora Project): Lights and shadows during 18-months surveillance. International Journal of Molecular Sciences 13 (2012): 774-787.
- Yang YL, Cheng HH, Lo HJ. Distribution and antifungal susceptibility of Candida species isolated from different age populations in Taiwan. Medical Mycology 44 (2006): 237-242.
- Binder U, Lass-Flörl C. Epidemiology of invasive fungal infections in the Mediterranean area. Mediterranean Journal of Hematology and Infectious Diseases 3 (2011): 2035-3006.
- Azab MM, Taleb AFA, Mohamed NAE, Omran FH. Original Research Article Rapid Diagnosis of Invasive Fungal Infections. International Journal of current Microbiology and Applied Sciences 4 (2015): 470-486.
- Montagna MT, Caggiano G, Lovero G, De Giglio O, Coretti C, Cuna T, et al. Epidemiology of invasive fungal infections in the intensive care unit: Results of a multicenter Italian survey (AURORA Project). Infection 41 (2013): 645-653.
- Shabaan AE, Elbaz LM, El-Emshaty WM, Shouman B. Role of serum (1,3)-β-D-glucan assay in early diagnosis of invasive fungal infections in a neonatal intensive care unit. Jornal de Pediatria 94 (2018): 559-565.
- Kim SH, Yoon YK, Kim MJ, Sohn JW. Risk factors for and clinical implications of mixed Candida/bacterial bloodstream infections. Clinical Microbiology and Infection 19 (2013): 62-68.
- Kami M, Fukui T, Ogawa S, Kazuyama Y, Machida U, Tanaka Y, et al. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clinical Infectious Diseases 33 (2001): 1504-1512.
- El-sayed ZA, Hasan ZE, Nasr RAR. Original article Real-Time PCR in the early detection of invasive fungal infection in immunodeficient infants and children. Egyptian Journal of Paediatric Allergy and Immunology 10 (2012): 67-74.
- Ashrafi, Nabili, Shokohi, Janbabaie, Hedayati, Ali-Moghaddam. A real time PCR assay on blood for diagnosis of invasive candidiasis in immunocompromised patient. Current Medical Mycology 1 (2015): 35-41.
- Srinivas S, Kumari P, Gupta, Kumar D. Utility of Panfungal PCR in the diagnosis of invasive fungal infections in febrile neutropenia. Journal of Family Medicine and Primary Care. 10 (2021): 169-170.
- Giacobbe DR, Mikulska M, Tumbarello M, Furfaro E, Spadaro M, Losito AR, et al. Combined use of serum (1,3)-β-d-glucan and procalcitonin for the early differential diagnosis between candidaemia and bacteraemia in intensive care units. Critical Care 21 (2017): 1-9.
- Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. β-D-glucan assay for the diagnosis of invasive fungal infections: A meta-analysis. Clinical Infectious Disease 52 (2011): 750-770.
- Lamoth F, Akan H, Andes D, Cruciani M, Marchetti O, Ostrosky-Zeichner L, et al. Assessment of the Role of 1,3-β-d-Glucan Testing for the Diagnosis of Invasive Fungal Infections in Adults. Clinical Infectious Diseases 72 (2021): 102-108.
- Martínez-Jiménez MC, Muñoz P, Valerio M, Vena A, Guinea J, Bouza E. Erratum: Combination of Candida biomarkers in patients receiving empirical antifungal therapy in a Spanish tertiary hospital: A potential role in reducing the duration of treatment. Journal of Antimicrobial Chemotherapy 71 (2016): 2679.
- Leeflang MMG, Debets-Ossenkopp YJ, Wang J, Visser CE, Scholten RJPM, Hooft L, et al. Galactomannan detection for invasive aspergillosis in immunocompromised patients. Cochrane Database of Systematic Reviews 2017 (2015): 1-126.
- Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clinical Microbiology Review 27 (2014): 490-526.
- Gupta P, Ahmad A, Khare V, Kumar A, Banerjee G, Verma N, et al. Comparative evaluation of pan-fungal real-time PCR, galactomannan and (1-3)-β-D-glucan assay for invasive fungal infection in paediatric cancer patients. Mycoses 60 (2017): 234-240.
- Springer J, Morton O, Perry M, Heinz WJ, Paholcsek M, Alzheimer M, et al. Multicenter comparison of serum and whole-blood specimens for detection of aspergillus DNA in high-risk hematological patients. Journal of Clinical Microbiology 51 (2013): 1445-1450.
- Jordanides NE, Allan EK, McLintock LA, Copland M, Devaney M, Stewart K, et al. A prospective study of real-time panfungal PCR for the early diagnosis of invasive fungal infection in haemato-oncology patients. Bone Marrow Transplantation 35 (2005): 389-395.
- Landlinger C, Preuner S, Bašková L, Van Grotel M, Hartwig NG, Dworzak M, et al. Diagnosis of invasive fungal infections by a real-time panfungal PCR assay in immunocompromised pediatric patients. Leukemia 24 (2010): 2032-2038