Prevalence and Antimicrobial Susceptibility Pattern of Extended Spectrum Beta Lactamase Producers in Gram-negative Urine Isolates at MBN Clinical Laboratories, Kampala Uganda
Article Information
Simon Dembe Kasango1, 2, 3*, Stephen Lutoti4, Izale Wewedru1, Emmanuel Aboce2 and Denish Calmax Angol2
1Mbarara University of Science and Technology, Department of Medical Laboratory science, P.O. Box 1410, Mbarara Uganda.
2MBN Clinical Laboratories Limited, Department of Microbiology, P.O. Box 35135 Kampala, Uganda.
3Ministry of health- Natural Chemotherapeutics Research Institute, P.O. Box 4864 Kampala, Uganda
4Department of Pharmacology and Therapeutics, Faculty of Medicine, Gulu University, P.O Box 166, Gulu, Uganda, East Africa
*Corresponding Author: Simon Dembe Kasango, Mbarara University of Science and Technology, Department of Medical Laboratory science, P.O. Box 1410, Mbarara Uganda, Tel: +256-703513645;
Received: 20 August 2018; Accepted: 17 September 2018; Published: 24 September 2018
Citation: Simon Dembe Kasango, Stephen Lutoti, Izale Wewedru, Emmanuel Aboce, Denish Calmax Angol. Prevalence and Antimicrobial Susceptibility Pattern of Extended Spectrum Beta Lactamase Producers in Gram-negative Urine Isolates at MBN Clinical Laboratories, Kampala Uganda. Archives of Microbiology & Immunology 2 (2018): 042-052.
View / Download Pdf Share at FacebookAbstract
Introduction: Occurrence of Extended Spectrum beta-lactamase (ESBLs) producing bacteria have presented impediment in treatment choices for urinary tract infections. ESBLs embody a major cluster of lactamases accountable for resistance to novel generations of ß-lactam drugs worldwide. The study determined prevalence of ESBL organisms in urine isolates and susceptibility patterns to 13 antibacterial agents.
Materials and methods: Two hundred samples were cultured on blood agar, MacConkey agar and incubated at 37°C utmost 48 hours. Isolates identified based on standard bacteriological culture and biochemical characteristics. Drug susceptibility centered on Clinical Laboratory Standard Institute recommended and WHO modified Kirby- Bauer disc diffusion methods. Isolates with reduced susceptibility to Ceftazidime were considered to be possible ESBL producers. Phenotypically confirmed ESBL required use of Ceftazidime in combination with Clavulanic acid. A five milimeter increase zone diameter for Ceftazidime in combination with Clavulanic acid versus its zone tested alone was considered as ESBL.
Results: Out of 200 samples, 45 (22.5%) had significant growth, majority Escherichia coli 28 (62.2%), Klebsiella pneumonae 11 (24.4%) followed and Citrobacter fruendii 2 (4.4%). Enterobacter species, Morganella morganii, Proteus mirabilis and Seratia marcescens each 1 (2.2%). Prevalence of ESBLs was 56%. Out of 25 (56%) ESBLs, highest prevalence was among Escherichia coli (15/25; 60%) followed by Klebsiella pneumonae (5/25; 24%) while Citrobacter fruendii, Enterobacter species, Morganella morganii and Proteus mirabilis each had (1/25; 4%). Susceptibility was highest to Imipenem 22 (88%) and least to Ampicillin, Amoxicillin-Clavulanic acid and Tetracycline each 1 (4%). No susceptibility t
Keywords
Extended Spectrum Beta-lactamases; Prevalence; Gram-negative isolates; Antibiotics susceptibility
Article Details
1. Introduction
Urinary tract infections (UTIs) distress up to 150 million people per annum globally and antibiotics resistance in urinary tract bacteria are growing globally, although it differs according to physical sites and in a straight line to use and misuse of antibiotics [1]. Resilient UTIs if not appropriately treated can lead to impediments such as Pelvic inflammatory Diseases (PID), ectopic pregnancy, abscess formation, Fallopian tube obstruction and associated kidneys triggering nephritis [2]. Impervious organisms have advanced a big task to infectious disease management. It is increasingly being reported in bacteria and is facilitated by genetic movable features like plasmids, transposons, and integrons [3]. Regionally, Africa in contrast with the rest of the world, there is largely a nonexistence of comprehensive data about Extended Spectrum beta-lactamase (ESBLs) manufacturing Enterobacteriaceae. Nevertheless, there is enough evidence to highlight the prevalence of ESBLs in Africa [4]. Hospital acquired infections are complex by structural or functional alterations in the urinary tract, weakened renal function or diseases which weaken the immunity, in disparity to community-acquired UTIs [5]. Urinary Tract Infections form the largest single group of hospital acquired infections and account for about 40-50% of the total hospital acquired infections.
Extended Spectrum Beta Lactamases are beta-lactamases capable of hydrolyzing oxyiminocephalosporins and are inhibited by beta-lactamase inhibitors. The occurrence of ESBL manufacturing strains among clinical isolates has been progressively increasing over the past years, causing a constraint of therapeutic choices. These enzymes are plasmid borne and confer multiple drug resistance making UTIs hard to treat. The ESBL manufacturing strains have variable vulnerability rates for aminoglycosides, Fluoroquinolones, and fourth-generation cephalosporins [6-7]. ESBLs were first recognized in 1980s and have progressively blown out throughout the world by hospital acquired routes in hospitals [8-9] and nursing homes as significant reservoirs. Patients admitted to hospitals are more probable reservoirs for resistant bacteria and ultimately, the patients in the community acquire ESBL-manufacturing [10]. ESBL-producing genes are protected on plasmids 80 kb in magnitude or greater, and most often carry resistance determinants for aminoglycosides, Fluoroquinolones, tetracycline, Cotrimoxazole and even Chloramphenicol, making the bacteria resist a wide diversity of drugs [11]. Mechanisms of drug resistance in gram-negative rods include manufacture of beta-lactamases [12], Amp C lactamases [13], efflux mechanisms and porin deficiency [14]. ESBL manufacturers may show more than one resistance mechanism, further complicating the situation.
The ESBL phenomenon is a new emerging problem that threatens hospitals and other health care centers all over the world. Occurrence and spread of these strains has resulted in treatment failure and disease complications. Recently, new cases have been reported as community-acquired ESBL infections [15]. The existence of an ESBL-manufacturing bacteria in severe infections can end up in treatment disappointment if one of these classes of drugs is used [16]. The cumulative prevalence of infections produced by antimicrobial-resistant bacteria makes empirical therapy of these infections problematic. Furthermore, these infections cause severe complications, particularly in patients with functional or structural abnormalities of the urogenital, patients who have experienced a kidney transplant, patients with polycystic kidneys, and patients with diabetes [17].
Data on epidemiology of UTIs caused by ESBL- manufacturing bacteria are inadequate in published reports in Uganda, yet there is need to monitor and check ESBLs manufacturers among urine isolates especially those associated with hospital acquired infections. Detecting ESBL producer urine isolates helps to improve care of patients mostly the immunocompromised since early detection initiates effective treatment and control measures.
Therefore, the aim of the study was to determine prevalence and antibiotics patterns of ESBL producers among Gram-negative urine isolates and their susceptibility at MBN clinical laboratories limited.
2. Materials and Methods
2.1 Study design, period and site
A cross-sectional study was conducted from February to May, 2014, at the department of microbiology-MBN Clinical Laboratories limited, located on plot 28 Nakasero road in Kampala the capital city of Uganda. MBN clinical laboratories limited is a privately owned level three laboratory offering clinical diagnostics and research services to hospitals and research institutions and walk-in patients within and around Kampala city. On a daily basis, a significant number of people from the surrounding zones and nearby districts visits the laboratory for different diagnostic services.
2.2 Study population, sample size, sampling technique, and data collection
All urine samples received for culture at the department of microbiology, MBN clinical laboratories during the study period were included in the study. A total of 200 study samples during the study period were consecutively enrolled by convenient sampling technique.
2.3 Sample collection, processing, and culture method
Only mid-stream urine collected by clean catch method in sterile containers were considered. After collection, each sample was transported to the microbiology laboratory at MBN clinical laboratories. The collected samples were inoculated on blood agar and MacConkey agar (Oxoid, Ltd.) and incubated at 350C. All the plates were incubated aerobically and initially examined for growth after 24 hours, and the ones without growth were further incubated for up to 48 hours.
2.4 Isolation, identification, and drug susceptibility testing of bacteria
After obtaining pure colonies, further identifications were done by using the standard microbiological technique, which includes Gram stain, colony morphology, and biochemical tests (Oxoid, Ltd.). Species identification of Bacterial identification was done based on standard bacteriological culture and biochemical tests [18]. The following tests were carried out to confirm the identity of the isolates; Sulphur utilization, Indole production and motility, Urea production, Citrate utilization, Triple sugar iron agar test for sugar fermentation and gas production. Antibiotic susceptibility was tested by the Disc diffusion method according to WHO modified Kirby Bauer disc diffusion method. The inoculum was adjusted to 0.5 Mac Farland standard and then swabbed on Muller Hinton agar plates (MHA). The commercial antibiotics used for gram negative isolates were ; (µg/disc) Ampicillin (10), Gentamycin (10), Ciprofloxacin (5), Co-trimoxazole/Trimethoprime-Sulfamethoxazole, (1.2/23.8), Tetracycline (30), Cefuroxime (30), Cefotaxime (30) or Ceftazidime (30) and Ceftriaxone (30), Imipenem (10), Nitrofurantoin (300), Amoxicillin-Clavulanic acid (20/10).
2.6 Extended Spectrum Beta-lactamase detection phenotypic method
Isolates were tested against third generation Cephalosporins (either Cefotaxime or Ceftazidime) using Clinical Laboratory Standard Institute [19] recommended and WHO modified Kirby Bauer disc diffusion method. Zone diameters were interpreted using the revised Clinical Laboratory Standard Institute [19]. Phenotypic confirmatory test for ESBL required use of either Cefotaxime or Ceftazidime alone and in combination with Clavulanic acid. A five milimeter increase in a zone diameter for Ceftazidime tested in combination with Clavulanic acid versus its zone when tested alone was considered an ESBL (e.g. Ceftazidime zone = 16; Ceftazidime/Clavulanic acid zone =21).
2.6 Quality control
2.6.1 Quality control the medium:
Sterility of the medium, plates were labeled QC and placed in the incubator for a maximum of three days without inoculating any organism on it and monitored every 24 hours. No growth meant it was sterile and growth implied contamination. For sterile plates, performance testing was done while for those which were not sterile, they were disposed of following the procedures of MBN Clinical laboratory waste management policy. The reliability of the findings was guaranteed by implementing quality control measures throughout the whole processes of the laboratory work. The reference strains used as control were K. pneumoniae ATCC 700603, Escherichia coli (ATCC 25922) were inoculated onto the plate and incubated for 24 hours at (35 ? 37°C) [18]. Results expected for good quality medium, were considered.
2.7 Data analysis
Data were entered and analyzed using SPSS version 18. Simple frequency was applied to determine proportions.
Ethical approval was obtained from the Mbarara University of Science and Technology, Faculty of Medicine Ethics and Research Committee. Permission was sought from MBN Clinical Laboratory management to carry out the research.
3. Results
A total of 200 urine samples were systematically sampled and received from walk-in patients at MBN clinical laboratory, samples referred from other health facilities in and outside of Kampala and these samples were cultured.
3.1 Gram negative urinary pathogens isolated
Among the 200 study samples cultured, 45 (22.5%) grew bacteria of clinical significance. Out of the 45 bacterial isolates of clinical relevancy, majority were Escherichia coli 28 (62.2%) followed by Klebsiella pneumonae 11 (24.4%) and C. fruendii 2 (4.4%) while Enterobacter species, Morganella morganii, Proteus mirabilis and Seratia marcescens were 1 (2.2%), isolated each (Table 1).
|
Pathogen |
Frequency |
Percent |
Cumulative Percent |
|
E. coli |
28 |
62.2 |
62.2 |
|
K. Pneumonae |
11 |
24.4 |
86.7 |
|
C. fruendii |
2 |
4.4 |
91.1 |
|
Enterobacter species |
1 |
2.2 |
93.3 |
|
M. Morganii |
1 |
2.2 |
95.6 |
|
Proteus mirabilis |
1 |
2.2 |
97.8 |
|
S. Marcescens |
1 |
2.2 |
100.0 |
|
Total |
45 |
100.0 |
- |
Table 1: Gram negative urinary pathogens isolated and identified.
3.2. Prevalence of ESBLs in urinary pathogens isolated
Out of the 45 Gram negative urinary bacterial pathogens 25 (56%) produced ESBLs and 20 (44%) did not produce ESBLs. Therefore, the prevalence of ESBLs among the Gram negative urinary bacterial pathogens was 56%, (Figure 1).
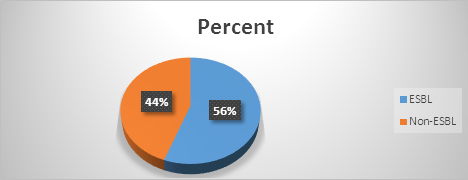
Figure 1: Prevalence of ESBL in urinary pathogens isolated.
3.3. Distribution of ESBLs among gram negative urinary pathogens isolated
Out of the 25 ESBLs producers, majority of the isolates that produced ESBLs were Escherichia coli isolates (15/25; 60%) followed by Klebsiella pneumonae (5/25; 24%) while Citrobacter fruendii, Enterobacter species, Morganella morganii and Proteus mirabilis each had (1/25; 4%) isolate that produced ESBLs ( Figure 2).
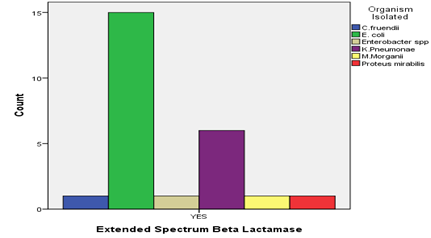
Figure 2: Distribution of ESBLs among the Gram negative urinary pathogens isolated.
3.4 Antibiotic susceptibility patterns of ESBL producers in urine bacteria isolated
The bacterial isolates showed varied antibiotics susceptibility test results to the selected antibiotics. The isolates were most susceptible to Imipenem 22 (88%), followed by Nitrofurantoin 13 (52%); Ceftazidime-Clavulanic acid 10 (40%) and Gentamycin 10 (40%). However some few isolates were less susceptible to Ofloxacin 5 (20%), Ciprofloxacin 4 (16%), Trimethoprime-Sulfamethoxazole 3 (12%), Ceftriaxone 3 (12%), Ampicillin 1 (4%), Amoxicillin-Clavulanic acid 1 (4%) and Tetracycline 1 (4%). None of the isolates showed susceptibility to Cefuroxime and Ceftazidime (Table 2).
|
Drug |
Susceptibility pattern |
||
|
Resistant |
Intermediate |
Susceptible |
|
|
Ampicillin |
24 (96) |
Nil |
1 (4) |
|
Amoxicillin-Clavulanic acid |
24 (96) |
Nil |
1 (4) |
|
Tetracycline |
24 (96) |
Nil |
1 (4) |
|
Cefuroxime |
23 (93) |
2 (8) |
Nil |
|
Ceftazidime |
23 (92) |
2 (8) |
Nil |
|
Trimethoprime-Sulfamethoxazole |
21 (84) |
1 (4) |
3 (12) |
|
Ceftriaxone |
20 (80) |
2 (8) |
3 (12) |
|
Ofloxacin |
20 (80) |
Nil |
5 (20) |
|
Ciprofloxacin |
20 (80) |
1 (4) |
4 (16) |
|
Gentamycin |
14 (56) |
1 (4) |
10 (40) |
|
Nitrofurantoin |
10 (40) |
2 (8) |
13 (52) |
|
Ceftazidime-Clavulanic acid |
7 (28) |
8 (32) |
10 (40) |
|
Imipenem |
Nil |
3 (12) |
22 (88) |
Table 2: Susceptibility pattern of ESBL producing isolates to selected antibiotics n (%).
4. Discussion
The study determined the prevalence and antibiotics susceptibility patterns of ESBL producers among Gram-negative urinary isolates. High prevalence of ESBLs was recorded in this study compared with the prevalence reported by other studies like in India [20], ESBL production was 41% in Escherichia coli and 40% in Klebsiella pneumoniae. Another study in Nigeria by Olanitola and colleagues in 2007 reported a prevalence of 30%. This study indicated that drug resistant ESBLs producers exist among urine isolated pathogens in our locality. The study showed that ESBL producers were present among urinary gram negative pathogens and they occur at an alarming rate in some species like Escherichia coli and Klebsiella pneumoniae, prevalence and incidence of ESBL producing strains among urinary isolates have been steadily increasing over the past few years resulting in limitation of therapeutic options; with outbreaks involving ESBL strains which have been reported all over the world, making them emerging pathogens as reported by Ananthkrishnan [21].
Resistant urinary tract infections, if not properly treated, can lead to complications that may result into permanent or temporary infertility, Pelvic Inflammatory Diseases (PID), ectopic pregnancy, abscess formation, Fallopian tube obstruction, epididymitis, orchitis, and the involvement of kidneys causing nephritis [2]. Highest prevalence was found among the Escherichia coli isolates (15/25; 60%) followed by Klebsiella pneumonae (5/25; 24%), while Citrobacter fruendii, Enterobacter species, Morganella morganii and Proteus mirabilis each had(1/25; 4%). This is due to the fact that ESBL producing isolates are most commonly Klebsiella species, and Escherichia coli [22]. Other isolates of family Enterobacteriaceae, such as Salmonella species and Proteus mirabilis, and isolates of Pseudomonas aeruginosa produce ESBLs but not as common as in Klebsiella species and Escherichia coli [22].
The urinary Gram negative ESBL producing isolates were most susceptible to Imipenem 22 (88%), followed by Nitrofurantoin 13 (52%); Ceftazidime-Clavulanic acid 10(40%) and Gentamycin 10(40%), due to the fact that carbapenemases and Nitrofurantoin exhibit more stability to hydrolysis by ESBLs than the 3rd generation Cephalosporins. A few isolates demonstrated susceptibility to Ofloxacin 5 (20%) and Ciprofloxacin 4 (16%), which is not the case as it was reported by Iroha and others [23-25]. Considering the susceptibility pattern of the Fluoroquinolones (Ciprofloxacin, Ofloxacin) above, newer Fluoroquinolones are unlikely to confer added benefits due to the increased Fluoroquinolones resistance among ESBLs producing isolates in our locality.
The genes encoding the ESBLs production especially CTX-M type Escherichia coli are often found to be associated with genetic structures that are genetically linked to class 1 integrons which integrates antibiotic resistance gene cassettes responsible for resistance to β-lactams, aminoglycosides, chloramphenicol, sulphonamides, tetracycline, and ciprofloxacin [26-27]. This could explain the reason of multidrug resistance pattern of the urinary Gram negative ESBL producing isolates investigated in this study.
5. Conclusion
The study showed that resistant ESBLs generating bacteria were present among urine isolates. We recommend that ESBLs bacteria isolated in urine be treated grounded on antibiotics susceptibility, continuous surveillance to guide correct treatment for urinary tract infection and to prevent the occurrence of multi drug resistant bacteria. Finally, Ministry of Health develop policies on rational antibiotics use in Uganda.
6. Limitation of the Study
We were unable to perform Minimum inhibitory concentration (MIC) and PCR to detect and establish the genes encoding for ESBLs in the isolates.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgements
The authors would like to thank MBN Clinical Laboratories for allowing us to use the laboratory facilities. The authors are very grateful to all the Department of microbiology MBN Clinical Laboratories staff for their cooperation during data collection. We also extend our sincere gratitude to staff of the Department of Medical Laboratory Science, Mbarara University of Science and Technology.
References
- Stamm W. The epidemiology of urinary tract infections: Risks factors reconsidered. Inter Sci Conf Antimicrob Agents Chemother (1999).
- Tijjani J, Arzai A, Sadiq N. Antimicrobial susceptibility pattern of extended spectrum betalactamase producers in gram-negative urogenital isolates in Kano, Nigeria. Bayero Journal of Pure and Applied Sciences 5 (2012): 20-25.
- Dessen A, Di Guilmi A, Vernet T, et al. Molecular mechanisms of antibiotic resistance in gram-positive pathogens. Current Drug Targets-Infectious Disorders 1 (2001): 63-77.
- Borg M, Scicluna E, De Kraker M, et al. Antibiotic resistance in the southeastern Mediterranean--preliminary results from the ARMed project. Euro surveillance: bulletin Europeen sur les maladies transmissibles. European communicable disease bulletin 11 (2006): 164-167.
- Mazulli T. Resistance Trends in Urinary Tract Pathogens and Impact on Management. The journal of urology (2002).
- Lautenbach E, Patel JB, Bilker WB, et al. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clinical Infectious Diseases 32 (2001): 1162-1171.
- Mazzulli T. Resistance trends in urinary tract pathogens and impact on management. The Journal of urology 168 (2002): 1720-1722.
- Kariuki S, Revathi G, Corkill J, et al. Escherichia coli from community-acquired urinary tract infections resistant to fluoroquinolones and extended-spectrum beta-lactams. The Journal of Infection in Developing Countries 1 (2007): 257-262.
- Bouchillon S, Johnson B, Hoban D, et al. Determining incidence of extended spectrum β-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: the PEARLS study 2001?2002. International journal of antimicrobial agents 24 (2004): 119-124.
- Kassis-Chikhani N, Saliba F, Carbonne A, et al. Extended measures for controlling an outbreak of VIM-1 producing imipenem-resistant Klebsiella pneumoniae in a liver transplant centre in France, 2003-2004. Euro surveillance: bulletin Europeen sur les maladies transmissibles= European communicable disease bulletin 15 (2010): 10-17.
- Arpin C, Dubois V, Coulange L, et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in community and private health care centers. Antimicrobial agents and chemotherapy 47 (2003): 3506-3514.
- Chaudhary U, Aggarwal R. Extended spectrum-lactamases (ESBL)-An emerging threat to clinical therapeutics. Indian Journal of Medical Microbiology 22 (2004): 75.
- Essack SY, Hall LM, Pillay DG, et al. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrobial agents and chemotherapy 45 (2001): 88-95.
- Jarlier V, Nicolas MH, Fournier G, et al. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Review of Infectious Diseases 10 (1988): 867-878.
- Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type β-lactamases. Antimicrobial agents and chemotherapy 46 (2002): 1-11.
- Ananthan S, Subha A. Cefoxitin resistance mediated by loss of a porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian journal of medical microbiology 23 (2005): 20.
- Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clinical microbiology reviews 18 (2005): 657-686.
- Paterson DL, Ko WC, Von Gottberg A, et al. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamases: implications for the clinical microbiology laboratory. Journal of clinical microbiology 39 (2001): 2206-2212.
- Al Otaibi FE, Bukhari EE. Clinical and laboratory profiles of urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in a tertiary care center in central Saudi Arabia. Saudi medical journal 34 (2013): 171-176.
- College of Physicians and Surgeons of Saskatchewan Laboratory Quality Assurance Program, Procedures/Guidelines for the Microbiology Laboratory, College of Physicians & Surgeons of Saskatchewan Laboratory Quality Assurance Program, Saskatchewan, Canada (2010).
- Wayne P, CLSI performance standard of antimicrobial susceptibility testing: twenty-fourth international supplement. CLSI Document M100-S24, Clinical and Laboratory Standard Institute (2011).
- Babypadmini S, Appalaraju B. Extended spectrum -lactamases in urinary isolates of Escherichia coli and Klebsiella pneumoniae - Prevalence and susceptibility pattern in a tertiary care hospital. Indian Journal of medical microbiology (2004).
- Ananthakrishnan A, Shanmuganathan C, Jayakeerthi SR, et al. Learning from an outbreak: ESBL- the essential points. Indian Journal of medical microbiology (2000).
- Elizabeth B, Hirsch Vincent H. Tam. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. Journal of Antimicrobial Chemotherapy 65 (2010): 1119-1125.
- Iroha IR, Oji AE, Esimone CO. Antimicrobial resistance pattern of plasmid-mediated extended-spectrum b-lactamase producing strains of Escherichia coli. Scientific Research (2008).
- Sorlozano A, Gutierrez J, Jimenez A, et al. Contribution of a New Mutation in parE to Quinolone Resistance in Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates. Journal of clinical microbiology (2007).
- Gunseren F, Mam?ko?lu L, Ozturk S, et al. A surveillance study of antimicrobial resistance of Gram-negative bacteria isolated from intensive care units in eight hospitals in Turkey. Journal of Antimicrobial Chemotherapy 43 (1999): 373-378.
- Bonnet R. Growing Group of Extended-Spectrum β-Lactamases: the CTX-M Enzymes” Antimicrobial agents and chemotherapy, Journal of American Society of Microbiology (2004).
- Pitout1 JDD, Hossain A, Hanson ND. Phenotypic and Molecular Detection of CTX-M-β-Lactamases Produced by Escherichia coli and Klebsiella spp. American society of microbiology. Journal of clinical microbiology (2004).
