Prenatal findings of 2q13 Duplication and Deletion: Further Evidence for Lack of Phenotypic-Genotype Correlation
Article Information
Lu Li1,2, Xiuzhu Huang1,2, Mei Ye 2, Jieping Chen2, Zhipeng Zeng2, Hui Guo2, Qiuyan Liao2, Wenlong Hu*2, Donge Tang*2, Yong Dai*2
1MOE Key Laboratory of Tumor Molecular Biology and Key Laboratory of Functional Protein Research of Guangdong Higher Education Institutes, Institute of Life and Health Engineering, College of Life Science and Technology, Jinan University, Guangzhou, Guangdong 510632, P.R. China
2Clinical Medical Research Center, The Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen People’s Hospital, Shenzhen, Guangdong 518020, P.R. China
*Corresponding author:
- Yong Dai, MD, PhD. Clinical Medical Research Center. The Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen People’s Hospital, Shenzhen, Guangdong 518020, P.R. China.
- Donge Tang, PhD, Clinical Medical Research Center, The Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen People’s Hospital, Shenzhen, Guangdong 518020, P.R. China.
- Wenlong Hu, Clinical Medical Research Center, The Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen People’s Hospital, Shenzhen, Guangdong 518020, P.R. China.
Received: 26 December 2022; Accepted: 2 January 2023; Published: 31 March 2023;
Citation:
Yong Dai, Donge Tang, and Wenlong Hu. Prenatal findings of 2q13 Duplication and Deletion: Further Evidence for Lack of Phenotypic-Genotype Correlation. Journal of Bioinformatics and Systems Biology. 6 (2023): 90-96.
View / Download Pdf Share at FacebookAbstract
Objective: In previous studies, 2q13 CNV was associated with various diseases, with a lack of consensus. This study aimed to analyze the prenatal diagnosis and clinical presentation of fetuses with different deletions or duplications of 2q13.
Materials and methods: Detailed prenatal screening and laboratory examinations, including prenatal ultrasound diagnosis and amniocentesis, were performed, and genetic analysis was performed using multiplex ligation-dependent probe amplification (MLPA) and chromosome microarray analysis (CMA).
Results: CMA analysis showed that four fetuses had deletion in the proximal region of 2q13, one had duplication, and one had duplication in the distal region of 2q13. Four fetuses had inherited copy number variation (CNV) from their parents; however, they had variable outcomes.
Conclusion: Individuals with the same CNV of 2q13 may have different phenotypes or are unaffected; multiple individuals with the same deletion or duplication need to be evaluated to capture feature sets associated with that CNV. Genetic counseling and follow-up to the fetus’s mother and family are essential. Genomic diseases’ characteristics should be explained in detail when providing prenatal genetic counseling to mothers and their families.
Keywords
chromosome 2q13, copy number variation, prenatal diagnosis, chromosomal microarray analysis
chromosome 2q13 articles; copy number variation articles; prenatal diagnosis articles; chromosomal microarray analysis articles
chromosome 2q13 articles chromosome 2q13 Research articles chromosome 2q13 review articles chromosome 2q13 PubMed articles chromosome 2q13 PubMed Central articles chromosome 2q13 2023 articles chromosome 2q13 2024 articles chromosome 2q13 Scopus articles chromosome 2q13 impact factor journals chromosome 2q13 Scopus journals chromosome 2q13 PubMed journals chromosome 2q13 medical journals chromosome 2q13 free journals chromosome 2q13 best journals chromosome 2q13 top journals chromosome 2q13 free medical journals chromosome 2q13 famous journals chromosome 2q13 Google Scholar indexed journals copy number variation articles copy number variation Research articles copy number variation review articles copy number variation PubMed articles copy number variation PubMed Central articles copy number variation 2023 articles copy number variation 2024 articles copy number variation Scopus articles copy number variation impact factor journals copy number variation Scopus journals copy number variation PubMed journals copy number variation medical journals copy number variation free journals copy number variation best journals copy number variation top journals copy number variation free medical journals copy number variation famous journals copy number variation Google Scholar indexed journals prenatal diagnosis articles prenatal diagnosis Research articles prenatal diagnosis review articles prenatal diagnosis PubMed articles prenatal diagnosis PubMed Central articles prenatal diagnosis 2023 articles prenatal diagnosis 2024 articles prenatal diagnosis Scopus articles prenatal diagnosis impact factor journals prenatal diagnosis Scopus journals prenatal diagnosis PubMed journals prenatal diagnosis medical journals prenatal diagnosis free journals prenatal diagnosis best journals prenatal diagnosis top journals prenatal diagnosis free medical journals prenatal diagnosis famous journals prenatal diagnosis Google Scholar indexed journals chromosomal microarray analysis articles chromosomal microarray analysis Research articles chromosomal microarray analysis review articles chromosomal microarray analysis PubMed articles chromosomal microarray analysis PubMed Central articles chromosomal microarray analysis 2023 articles chromosomal microarray analysis 2024 articles chromosomal microarray analysis Scopus articles chromosomal microarray analysis impact factor journals chromosomal microarray analysis Scopus journals chromosomal microarray analysis PubMed journals chromosomal microarray analysis medical journals chromosomal microarray analysis free journals chromosomal microarray analysis best journals chromosomal microarray analysis top journals chromosomal microarray analysis free medical journals chromosomal microarray analysis famous journals chromosomal microarray analysis Google Scholar indexed journals ultrasound diagnosis articles ultrasound diagnosis Research articles ultrasound diagnosis review articles ultrasound diagnosis PubMed articles ultrasound diagnosis PubMed Central articles ultrasound diagnosis 2023 articles ultrasound diagnosis 2024 articles ultrasound diagnosis Scopus articles ultrasound diagnosis impact factor journals ultrasound diagnosis Scopus journals ultrasound diagnosis PubMed journals ultrasound diagnosis medical journals ultrasound diagnosis free journals ultrasound diagnosis best journals ultrasound diagnosis top journals ultrasound diagnosis free medical journals ultrasound diagnosis famous journals ultrasound diagnosis Google Scholar indexed journals MLPA articles MLPA Research articles MLPA review articles MLPA PubMed articles MLPA PubMed Central articles MLPA 2023 articles MLPA 2024 articles MLPA Scopus articles MLPA impact factor journals MLPA Scopus journals MLPA PubMed journals MLPA medical journals MLPA free journals MLPA best journals MLPA top journals MLPA free medical journals MLPA famous journals MLPA Google Scholar indexed journals DNA articles DNA Research articles DNA review articles DNA PubMed articles DNA PubMed Central articles DNA 2023 articles DNA 2024 articles DNA Scopus articles DNA impact factor journals DNA Scopus journals DNA PubMed journals DNA medical journals DNA free journals DNA best journals DNA top journals DNA free medical journals DNA famous journals DNA Google Scholar indexed journals CMA articles CMA Research articles CMA review articles CMA PubMed articles CMA PubMed Central articles CMA 2023 articles CMA 2024 articles CMA Scopus articles CMA impact factor journals CMA Scopus journals CMA PubMed journals CMA medical journals CMA free journals CMA best journals CMA top journals CMA free medical journals CMA famous journals CMA Google Scholar indexed journals genetic diseases articles genetic diseases Research articles genetic diseases review articles genetic diseases PubMed articles genetic diseases PubMed Central articles genetic diseases 2023 articles genetic diseases 2024 articles genetic diseases Scopus articles genetic diseases impact factor journals genetic diseases Scopus journals genetic diseases PubMed journals genetic diseases medical journals genetic diseases free journals genetic diseases best journals genetic diseases top journals genetic diseases free medical journals genetic diseases famous journals genetic diseases Google Scholar indexed journals
Article Details
1. Introduction
Copy number variation (CNV) is the gain or loss of genomic material greater than 1 KB in the genome [1]. Although CNV is common in normal people, in some cases, due to chromosomal rearrangement, it can affect certain genes' expression, leading to disease development [2, 3]. Deletion or duplication of the long arm of chromosome 2 have been reported to be associated with a variety of phenotypes, including orofacial clefting, developmental delay (DD), failure to thrive and dysmorphism [4, 5]. Not all CNVs have a pathogenic phenotype, and 2q13 CNVs have unknown clinical significance.
Compared with karyotype analysis, the chromosome microarray analysis (CMA) technique can detect small deletions and duplicates, effectively improving the detection rate of chromosomal abnormalities [6]. It has rapidly replaced the standard G-band karyotype and has become an important diagnostic method for detecting chromosomal abnormalities in pregnancy products [7]. Here, six new cases have been identified through the CMA with a 2q13 genomic imbalance and each with similar or different outcomes at prenatal diagnosis. Nevertheless, it may help identify these features early in the prenatal period for reference by parents and healthcare providers and monitor the condition or intervene promptly.
2. Materials and Methodology:
The six probands with 2q13 duplication or deletion were seen in the Antenatal Diagnosis Center of Shenzhen People's Hospital. We collected clinical information on these cases. The following data were collected through medical record reviews: age, sex, gestational age and growth parameters at birth, parameters of obstetric examination during pregnancy, the risk of multiple genetic diseases and diagnostic CMA analysis, and multiplex ligation-dependent probe amplification (MLPA) assays were performed as part of their clinical evaluations.
Samples were taken from fetal exfoliated cells in amniotic fluid and lymphocytes of cord blood or parent peripheral blood. CMA was performed for each sample using a Cytoscan HD array (Affymetrix Inc., Santa Clara, CA, USA), and DNA was extracted according to the manufacturer's instructions. The annotation of the results was conducted in accordance with the Human Feb.2009 (GRCh37/hg19) Assembly.
3. Results
Table 1 summarizes detailed clinical information for each fetus. The mothers ranged in age from 29 to 43, and the average gestational age at the time of testing was about 21 weeks. Three fetuses were at high risk for Down syndrome. Three women had a history of spontaneous abortion, and two had a history of induced abortion. The mother of fetus 1 had two sons, one who died of neuroblastoma at age three and the other who died of intracranial hemorrhage at age 2. The maternal and fetal outcomes of pregnancy were followed up (Table 2). All six cases were delivered successfully, with four fetuses delivered to term and one premature. All the fetuses were in the normal range of height and weight. Among them, the baby boy in case 4 was born with congenital hydrocephalus and congenital pulmonary cystadenomatous.
Table 1: Clinical data
|
Fetus |
1 |
2 |
3 |
4 |
5 |
6 |
|
Mother’s age |
29 |
29 |
31 |
29 |
43 |
30 |
|
Sample type |
Cord blood |
Cord blood |
Cord blood |
Amniotic fluid |
Cord blood |
Amniotic fluid |
|
Gestational weeks |
18+ |
22+ |
25+ |
19+ |
22+ |
20+ |
|
MLPA |
duplication of exon 8 of the SMN1; heterozygous deletions of exon 8 of the SMN2 |
N |
heterozygous deletions of exon 7 and exon 8 of the SMN2; duplication of terminal subtelomeres of PAR region of Xp22 |
N |
N |
duplication of exon 7 and exon 8 of the SMN1; heterozygous deletions of exon 7 and exon 8 of the SMN2 |
|
AFP(MoM) |
2.01 |
1.23 |
0.77 |
0.58 |
0.89 |
0.67 |
|
Free β-hCG(MoM) |
7.16 |
2.46 |
0.9 |
2.63 |
2.177 |
2.87 |
|
uE3(MoM) |
0.47 |
1.14 |
0.732 |
0.64 |
1.443 |
0.26 |
|
Ds risk |
1/10000 |
Jan-68 |
1/473 |
1/250 |
Jan-65 |
1/204 |
|
NTD |
Low risk |
Low risk |
Low risk |
U |
U |
Low risk |
|
Clinical diagnosis |
FGR |
bowel strong echo and left ventricular strong echo |
N |
left ventricle punctate strong echo |
Elderly couple, two renal pelvis dissection in the fetus |
N |
|
Abnormal pregnancy-labor history of mother |
gave birth to two boys who died in infancy |
one spontaneous abortion |
one induced labor due to "cleft lip and cleft palate" and one spontaneous abortion due to fetal termination |
N |
two induced abortions |
N |
Abbreviations: MPLA, multiplex ligation-dependent probe amplification; AFP, alpha-fetoprotein (Normal range: 0.61-2.49); Free β-hCG, free beta-human chorionic gonadotropin (0.41-2.39); uE3, unconjugated estriol (>0.73); MoM, multiple of median; DS, Down syndrome (Normal range: < 1/1000); NTD, neural tube defects; FGR, fetal growth restriction; U, unknown; N, normal.
Table 2: Pregnancy outcome
|
Case |
1 |
2 |
3 |
4 |
5 |
6 |
|
Gestational weeks |
40 |
39 |
36 |
39 |
38 |
38 |
|
Delivery situation |
Caesarean section |
Normal delivery |
Caesarean section |
Caesarean section |
Caesarean section |
Normal delivery |
|
Sex of the baby |
Girl |
Girl |
Boy |
Boy |
Girl |
Boy |
|
Weight (g) |
3000 |
3130 |
2800 |
3450 |
3500 |
3000 |
|
Height (cm) |
50 |
49 |
49 |
50 |
50 |
50 |
The UCSC database shows duplication or deletion regions and their involved genes in six fetuses (Figure1).
Five fetuses had deletions or duplications in the proximal or distal regions of 2q13. Among them, four were deletions in the proximal region of 2q13, and one was duplication. A search of the Decipher database (https://decipher.sanger.ac.uk/) revealed that this region contained ten genes, of which 4 were OMIM genes: NPHP1 (OMIM: 607100), RGPD5 (OMIM: 612708), MALL (OMIM: 602022), RGPD6 (OMIM: 612709), and 1 was Morbid genes: NPHP1. Another fetus had a duplication of the distal region of 2q13, which the Decipher database showed contained 27 genes, including 8 OMIM genes: BUB1 (OMIM: 602452), ANAPC1 (OMIM: 608473), MERTK (OMIM: 604705), MIR4435-2HG (OMIM: 617144), BCL2L11 (OMIM: 603827), TMEM87B (OMIM: 617203), FBLN7 (OMIM: 611551), RGPD8 (OMIM: 602752), and 3 Morbid genes: BUB1, ANAPC1, MERTK. Figure 2 shows the chromosomal locations of duplication and deletion in six fetuses.
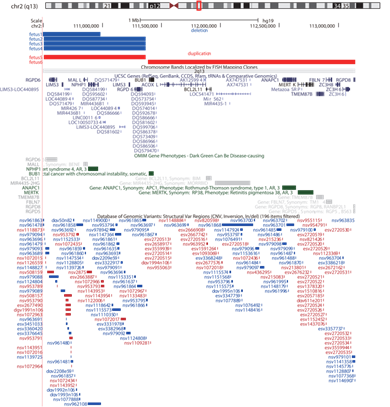
Figure 1: UCSC Genome Browser (http://genome.ucsc.edu/) view of 2q13. The top panel shows the deletion or duplication reported here. UCSC genes, OMIM genes and Database of Genomic Variants (DGV) cases are shown below the custom track.
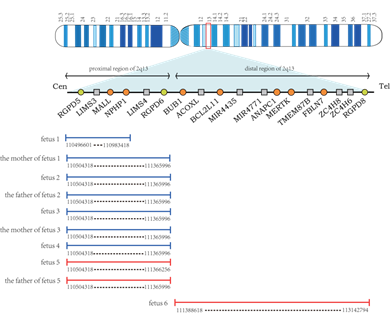
Figure 2: Schematic maps of human chromosome 2q13. Circles are OMIM genes. The proximal and distal region of 2q13 is shown with locations of the deletion and duplication reported here. Deletion of the fetus and the parents is depicted in blue, and duplication is shown in red.
4. Discussion
The deletion of the proximal region of 2q13.
One of the genes located in 2q13, NPHP1, has been reported to be closely associated with human disease. Nephronophthisis (NPHP) is a tubulo-interstitial, autosomal recessive cystic kidney disease, which is one of the most frequent genetic diseases causing end-stage renal disease (ESRD) in children and adolescents [8]. At first, Hildebrandt et al. found homozygous deletion of Mall and NPHP1 genes in 16 out of 22 families with nephropathy, suggesting a relationship between nephronophthisis and 2q13 [9]. This could be explained by interchromosomal or intrachromosomal mispairing of the genome inverted repeat, followed by an interchromosomal unequal crossover event [10]. In addition to NPHP, Cogan-type congenital ocular motor apraxia (COMA), retinitis pigmentosa (Senior-Loken syndrome), and Joubert syndrome were also found in patients with a deletion of the NPHP1 gene [11-13].
However, deletion of the proximal region of 2q13 involving RGPD5, RGPD6, and LIMS3 genes has not been reported. Ciccarelli et al. found that genomic rearrangement of RANBP2 and GCC2 genes on chromosome 2 of primates produced 8 RGPD genes, including RGPD5, RGPD6, and RGPD8 [14]. LIMS3 encodes a conserved protein containing the LIM domain, which plays a role in cell-cell and cell-matrix adhesion in the formation of multi-protein complexes. MALL is a member of the MyD88 adapter-like (Mal) family, which functions in a variety of tumors [15].
The duplication of the proximal region of 2q13
Previous studies have shown that duplication of the 2q13 proximal region is commonly associated with autism spectrum disorder (ASD). Baris et al. reported a single-copy gain in the NPHP1 gene in patients with speech delay, global developmental delay, attention hyperactivity disorder (AHDH), and veformities of varying degrees. [16]. Previous reports also found some cases of 2q13 duplication associated with ASD, DD, and mental retardation (ID)[17, 18]. Similarly, two brothers with ASD were reported. Both brothers had 2q13 duplication, including MALL, NPHP1, RGPD6, and BUB1 genes, and both suffer from ID and liver disorder [19]. It is clear that duplication of the 2q13 proximal region increases the risk of ASD.
The duplication of the distal region of 2q13
Although the penetrance of duplication is lower than that of deletion [4]. Duplications at the distal region of 2q13 are increasingly accepted as risk factors for developmental delay, autism, adult neuropsychiatric expression, and deformities [20-22].
Rudd et al. reported that two patients with patrilineal 2q13 had dental crowding [22], while FBLN7 plays a crucial role in odontoblast differentiation and maintenance, as well as in dentine formation [23]. Another gene in the region, BUB1, belongs to a family of genes that encodes proteins that bind to the kinetochore, and BUB1 is a vital component of the spindle checkpoint. Tang et al. also demonstrated the role of hBUB1 in centromeric cohesion during mitosis in mammalian cells [24]. There are three isoforms of BCL2L11, which sense apoptotic stimuli and initiate apoptosis by activating BAK, BAX, and other multi-domain pro-apoptotic proteins [25, 26]. ANAPC1 is the largest subunit of anaphase-promoting complex/cyclosome (APC/C) and ANAPC1 deficiency is the cause of Rothmund-Thomson syndrome type 1 [27]. Homozygous mutations in MERTK, a tyrosine kinase, are associated with retinal dystrophy and retinitis pigmentosa [28].
Cases
Six fetuses in our study showed deletion or duplication at a different region of 2q13 on the CMA results. Fetus 1-5 was a deletion or duplication of the proximal region of 2q13, and fetus 6 was a duplication of the distal region of 2q13. Among them, four fetuses had the same breakpoint, which could be considered a 2q13 recurrent breakpoint. The deletions of fetuses 1 and 3 were inherited from their mother. The deletion of fetus 3 was identical to that of his mother, while the deletion of fetus 1 was smaller than that of her mother. Both the deletion of fetus 2 and the duplication of fetus 5 are inherited from the father. The deletion of fetus 2 is identical to her father, and the termination site of fetus 5 is slightly different from her father.
Although, almost all pathogenic CNVs have two characteristics: variable expression and incomplete penetrance, which implies a series of phenotypic outcomes and unaffected family members carrying the same CNV [29, 30]. To date, many 2q13 deletions and duplications have been hereditary, but parental phenotypes have been poorly described. Half of the six fetuses were at high risk for Down syndrome, and four mothers in this study had a history of abnormal pregnancy-labor. Fetus 1 was accompanied by fetal growth restriction (FGR), and the mother had given birth to two boys who both died of neuroblastoma or intracranial hemorrhage. The mother of fetus 2 had a spontaneous abortion due to the termination of the embryo. Fetus 2 was found to have strong echoes in the left ventricle and bowel. LISM3 is involved in cell-cell and cell-matrix adhesion processes and is involved in cytoskeleton regulation. However, molecules that regulate cell migration and cell adhesion are thought to be involved in congenital heart defect (CHD) [31]. The deletion of fetus 3 is inherited from the mother. The mother of fetus 3 had three pregnancies, including one induced labor due to "cleft lip and cleft palate" and one spontaneous abortion due to fetal termination, while fetus 3 was accompanied by Xp22 subtelomere microduplication. It is widely accepted that subtelomere chromosome rearrangement may be a common cause of idiopathic mental retardation (MR) [32]. The genotypes of aborted fetuses cannot be determined, but chromosomal abnormalities may play a role in early pregnancy loss. Deletion of 2q13 also occurs in fetus 4; congenital hydrocephalus and congenital pulmonary cystadenomatous were found after birth. Fetus 5 was born to elderly parents. It is believed that elderly parturient women have a higher incidence of chromosomal abnormalities than younger mothers [33]. Furthermore, MLPA results showed an abnormal copy number of the SMN gene in fetal 1, 3, and 6, which is the primary pathogenic gene of spinal muscular atrophy (SMA) [34].
5. Conclusion:
The presence of a chromosomal-specific low-copy repeat sequence (LCR) increases the risk of chromosomal rearrangement and makes chromosome 2 susceptible to many unbalanced structural variants, including deletions and duplications. These CNVs can be developed de novo or inherited from parents with similar deletions or duplications. However, almost all CNVs have two characteristics of variable expression and incomplete penetrance, and patients with the same CNV may have different phenotypes or be unaffected [29, 30], and the penetrance of duplication is lower than that of deletion [4]. Therefore, it is necessary to evaluate multiple individuals with the same CNV to capture characteristic sets associated with genomic diseases.
Of the six fetuses reported here, four had deletions or duplicates inherited from their parents, and they had different phenotypes. The characteristics of variable expression and incomplete penetrance of genomic diseases pose challenges for prenatal diagnosis and genetic counseling. These two characteristics should be explained in detail when providing prenatal genetic counseling to mothers and their families.
Authorship:
The corresponding authors are responsible for the study design and revision of the paper. Lu Li and Xiuzhu Huang worked together on analyzing the genetic data and drafted the present manuscript. Mei Ye, Jieping Chen and Zhipeng Zeng collected clinical information and provided genetic counseling. Hui Guo and Qiuyan Liao contributed to the data analysis and interpretation. All authors have read and approved the final manuscript.
Funding:
This work was supported by Science and Technology Planning Project of Guangdong Province, China (No. 2017B020209001), the science and technology plan of Shenzhen (No. JCYJ20180305163846927).
Acknowledgements:
We sincerely thank the six families for supporting our research.
Declaration of competing interest:
The authors declare no conflicts of interest.
References
- Feuk L, Carson AR, and Scherer SW. Structural variation in the human genome. Nat Rev Genet 7(2) (2006): 85-97.
- Shaw CJ and Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13(1) (2004): 57-64.
- Giglio S, Broman K, Matsumoto N, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. American journal of human genetics 68(4) (2001): 874-83.
- Yu HE, Hawash K, Picker J, et al. A recurrent 1.71 Mb genomic imbalance at 2q13 increases the risk of developmental delay and dysmorphism. Clin Genet 81(3) (2012): 257-64.
- Wenger S, Bleigh O, and Hummel M. Cleft palate in a newborn with duplication 2(q13q23). The Cleft palate-craniofacial journal: official publication of the American Cleft Palate-Craniofacial Association 41(5)(2004): 568-70.
- Dhillon RK, Hillman SC, Morris RK, et al. Additional information from chromosomal microarray analysis (CMA) over conventional karyotyping when diagnosing chromosomal abnormalities in miscarriage: a systematic review and meta-analysis. BJOG 121(1) (2014): 11-21.
- Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86(5) (2010): 749-64.
- Hildebrandt F and Otto E, Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nature reviews. Genetics 6(12) (2005): 928-40.
- Hildebrandt F, Otto E, Rensing C, et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet 17(2) (1997): 149-53.
- Saunier S, Calado J, Benessy F, et al. Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. American journal of human genetics 66(3) (2000): 778-89.
- Betz R, Rensing C, Otto E, et al. Children with ocular motor apraxia type Cogan carry deletions in the gene (NPHP1) for juvenile nephronophthisis. The Journal of pediatrics 136(6) (2000): 828-31.
- Parisi M, Bennett C, Eckert M, et al. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. American journal of human genetics 75(1) (2004): 82-91.
- Caridi G, Murer L, Bellantuono R, et al. Renal-retinal syndromes: association of retinal anomalies and recessive nephronophthisis in patients with homozygous deletion of the NPH1 locus. American journal of kidney diseases : the official journal of the National Kidney Foundation 32(6) (1998): 1059-62.
- Ciccarelli FD, von Mering C, Suyama M, et al. Complex genomic rearrangements lead to novel primate gene function. Genome Res 15(3) (2005): 343-51.
- Marazuela M and Alonso M. Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue. Histology and histopathology 19(3)(2004): 925-33.
- Baris H, Bejjani B, Tan W, et al. Identification of a novel polymorphism--the duplication of the NPHP1 (nephronophthisis 1) gene. American journal of medical genetics. Part A 17(2006): 1876-9.
- Kaminsky E, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in medicine: official journal of the American College of Medical Genetics 13(9)(2011): 777-84.
- Yasuda Y, Hashimoto R, Fukai R, et al. Duplication of the NPHP1 gene in patients with autism spectrum disorder and normal intellectual ability: a case series. Annals of general psychiatry 13(2014): 22.
- Chen C, Lin S, Lee C, et al. Recurrent 2q13 microduplication encompassing MALL, NPHP1, RGPD6, and BUB1 associated with autism spectrum disorder, intellectual disability, and liver disorder. Taiwanese journal of obstetrics & gynecology 56(1) (2017): 98-101.
- Costain G, Lionel A, Fu F, et al. Adult neuropsychiatric expression and familial segregation of 2q13 duplications. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics 4(2014): 337-44.
- Yu H, Hawash K, Picker J, et al. A recurrent 1.71 Mb genomic imbalance at 2q13 increases the risk of developmental delay and dysmorphism. Clinical genetics 81(3) (2012): 257-64.
- Rudd M, Keene J, Bunke B, et al. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Human molecular genetics 18(16) (2009): 2957-62.
- de Vega S, Iwamoto T, Nakamura T, et al. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. The Journal of biological chemistry 282(2007) (42): 30878-88.
- Tang Z, Sun Y, Harley S, et al. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proceedings of the National Academy of Sciences of the United States of America 101(52) (2004): 18012-7.
- O'Connor L, Strasser A, O'Reilly L, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. The EMBO journal 17(2) (1998): 384-95.
- Willis S, Chen L, Dewson G, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes & development 19(11) (2005): 1294-305.
- Ajeawung NF, Nguyen TTM, Lu L, et al. Mutations in ANAPC1, Encoding a Scaffold Subunit of the Anaphase-Promoting Complex, Cause Rothmund-Thomson Syndrome Type 1. Am J Hum Genet 105(3) (2019): 625-630.
- Thompson D, McHenry C, Li Y, et al. Retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. American journal of human genetics 70(1)(2002): 224-9.
- Rosenfeld J, Coe B, Eichler E, et al. Estimates of penetrance for recurrent pathogenic copy-number variations. Genetics in medicine: official journal of the American College of Medical Genetics 15(6) (2013): 478-81.
- Deak K, Horn S, and Rehder C. The evolving picture of microdeletion/microduplication syndromes in the age of microarray analysis: variable expressivity and genomic complexity. Clinics in laboratory medicine 31(4) (2011): 543-64.
- Sailani M, Makrythanasis P, Valsesia A, et al. The complex SNP and CNV genetic architecture of the increased risk of congenital heart defects in Down syndrome. Genome research 23(9) (2013): 1410-21.
- Hélias-Rodzewicz Z, Bocian E, Stankiewicz P, et al. Subtelomeric rearrangements detected by FISH in three of 33 families with idiopathic mental retardation and minor physical anomalies. Journal of medical genetics 39(9) (2002): 53.
- Gou L, Liu T, Wang Y, et al. Clinical utilization of chromosomal microarray analysis for the genetic analysis in subgroups of pregnancy loss. J Matern Fetal Neonatal Med 7(2020): 1-8.
- Lefebvre S, Bürglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80(1) (1995): 155-65.
