PIK3CA Mutational Profiling in a Patient Cohort with HR+/HER2- Advanced Metastatic Breast Cancer at a Tertiary Cancer Center
Article Information
Osama Alsmadi*, Hikmat Abdel-Razeq, Yazan Talab, Hazem Abdulelah, Zeena Shaheen and Abdelghani Tbakhi
King Hussein Cancer Center, Queen Rania St 202, Amman, Jordan
*Corresponding Author: Osama Alsmadi, King Hussein Cancer Center, Queen Rania St 202, Amman, Jordan.
Received: 13 February 2024; Accepted: 07 March 2024; Published: 27 March 2024
Citation: Osama Alsmadi, Hikmat Abdel-Razeq, Yazan Talab, Hazem Abdulelah, Zeena Shaheen and Abdelghani Tbakhi. PIK3CA Mutational Profiling in a Patient Cohort with HR+/HER2- Advanced Metastatic Breast Cancer at a Tertiary Cancer Center. Fortune Journal of Health Sciences. 7 (2024): 158-163.
View / Download Pdf Share at FacebookAbstract
Common PIK3CA gene activating mutations can be found in 20-30% of all breast cancer cases, and regarded as predictive markers for therapeutic response to PI3K inhibitors. The therascreen PIK3CA mutation companion assay and the alpha-specific PI3K inhibitor, Alpelisib, are FDA-approved for selecting and treating patients with advanced PIK3CA-mutated metastatic breast cancer. The main objective behind this report was to investigate the composition and proportion of PIK3CA mutations using a PIK3CA mutation Therascreen RT-PCR assay, in a patient cohort with receptor-positive/HER2-negative (HR+/HER2) metastatic breast cancer, who were diagnosed and treated at King Hussein Cancer Center (KHCC). Patients with PIK3CA-mutated tumors represented 39.4% (91/231) of all patients. Four PIK3CA mutations comprised 86.8% of all PIK3CA mutations; mainly H1047R (33.3%), E545K (20.9%), E542K (24.2%), and H1047L (8.8%). The four main mutations map to the helical and kinase domains of the PIK3CA encoded protein. C420R was found in only one patient, and E545A was found in two patients. Nine of the 91 mutated patients had shown double PIK3CA mutations (9.9%). In conclusion, PIK3CA is frequently mutated in multiple types of cancers at known ‘Hot-spots’, mainly in the kinase and helical modular domains, which we found consistent with our findings. PIK3CA mutational signature in our metastatic breast cancer cohort varied with a 39.4% (91/231) positivity rate. The PIK3CA mutational screening panel did not capture mutations in the remaining 140 (60.6%) cases; these patients may be mutated in other genes related to breast cancer, or in PIK3CA loci not covered by the Therascreen assay. Survival and clinical outcomes in association with PIK3CA mutational profiles shall be addressed in a follow-up investigation for this patients’ cohort.
Keywords
PIK3CA, AKT, KHCC, FDA, SAFIR02
Article Details
Introduction
PIK3CA activating mutations occur in 20–30% of all cancer cases and considered as predictive markers for response to PI3K inhibitors. PIK3CA belongs to the lipid phosphoinositide 3-kinases (PI3Ks), also known as phosphatidylinositol 3-kinases, that are a family of enzymes composed of four classes who drive a range of physiological functions and cellular processes including cell proliferation, growth, survival, motility and metabolism [1]. Class I PI3Ks are composed of a regulatory and a catalytic subunits that heterodimerize to form a functional biomolecule. PIKs are further divided into IA & IB subsets with the former structured by dimerization between the catalytic p110 (alpha, beta or delta) and p85 regulatory subunits [2]. The p85 subunit has two Src-homology 2 (SH2) domains, and one inter-SH2 domain that constitutively binds to the catalytic p110α subunit, keeping it in an inactive state [3].
Under normal biological conditions, p110 activation occurs when the cell surface receptor tyrosine kinase (RTKs) is autophosphorylated upon its specific ligand binding (e.g. growth factor such as insulin), which then recruits and binds to the cytoplasmic p85/p110 heterodimer complex. This molecular interaction between the phosphorylated receptor and the p85/p110 complex frees the p110 catalytic subunit, enabling it to convert the cellular phosphatidylinositol (3,4)-bisphosphate (PIP2) to phosphatidylinositol (3,4,5)-triphosphate (PIP3) [4]. PIP3, also known as the second messenger, subsequently phosphorylates PDK1 (3-phosphoinositide-dependent protein kinase-1) which in turn activates AKT (also known as protein kinase B; PKB) [5, 6]. The overall phosphorylation-based signaling cascade promotes cancer cells proliferation and survival through the mTOR pathway [7–9].
Activating alterations in the PI3K-Akt signaling pathway are frequently seen in many human cancers, particularly the common types such as breast, colorectal, endometrial and prostate. Activation by RTKs and somatic driver mutations in specific components of the pathway are the two main underlying oncogenic mechanisms. Female breast cancer is the most commonly diagnosed cancer at the global level, accounting for 11.7% of the total cases [10]. This malignancy and other tumor types has been the focus of many investigations that led to the discovery and design of anti-endocrine and targeted therapies that improved cancer overall survival and clinical outcomes [11]. Many therapies have been approved for the treatment of various malignancies by targeting the PI3K pathway using inhibitory drugs such as BYL719 (alpelisib), CAL101 (idelalisib), BAY 80-6946 (copanlisib), the mTOR inhibitors RAD001 (everolimus), and CCI-779 (temsirolimus). Nevertheless, several resistance mechanisms facilitated by tumor related intrinsic adaptive responses through the reactivation of the PI3K pathway had emerged. These mechanisms are underlined by limited treatment responses and tolerance to therapeutic agents, plus other changes in cellular plasticity, which unfortunately resulted in an increased cancer-associated prevalence and mortalities [12].
More than 70% of breast cancers are hormone receptor (HR) positive and human epidermal growth factor receptor 2 (HER2) negative [13]. Approximately 40% of patients with HR-positive, HER2-negative breast cancer, have activating mutations in the gene PIK3CA (gene encoding p110α catalytic subunit) leading to a hyperactivated and dysregulated PI3K pathway [14]. PIK3CA gene is 34 kb in length and located within the chromosome 3q26.3 region. It encodes 1,068 amino acids spanning five modular protein functional domains: the amino-terminal p85 binding domain, Ras binding domain, C2 domain, helical domain, and the kinase domain [15]. As exon 1 is entirely UTR, many studies have accordingly numbered the exons according to the coding exons (1-20). Subsequently, these studies refer to E545 mutations as being in exon 9 and H1047 mutations in exon 20 [16]. In general, patients found with mutated PIK3CA and hormone receptor-positive, HER2-negative advanced breast cancer, have a worse prognosis compared to those patients with a wild-type gene [17].
The recent p110α-specific FDA approved PI3K pathway inhibitor and degrader, alpelisib, has captured much of interest for its efficacy against the invasive and metastatic breast cancer in patients harboring PIK3CA mutations. These mutations mainly occur in one of the two hotspot gene domains, specifically p.E542K and p.E545K in exon 9 (α-helicase domain), and p.H1047R/L in exon 20 (kinase domain) [18]. This group of patients in our study were eligible for receiving the second line therapy, alpelisib alone, or as a combination therapy protocol. More recent data by others, revealed alpelisib therapy had enhanced estrogen signaling in HR+ breast cancer, raising a flag for its use as the sole therapeutic agent [19]. Alpelisib when used in combination with fulvestrant endocrine treatment agent according to the SOLAR-1 study, had shown synergistic effects and promising results in patients with advanced metastatic breast cancer [20]. Here, we report on the mutational profiling of PIK3CA in a patient cohort diagnosed with metastatic breast cancer with ER+/HER2- profile, who were considered for treatment based on their genetic profiles.
Methods
Patient samples and characteristics
In this single center retrospective study, FFPE samples from 231 patients with metastatic breast cancer were received by the molecular diagnostic laboratory, to investigate the presence of PIK3CA actionable mutations using the therascreen Qiagen real-time PCR assay kit. All samples were of breast tissue origin, and ER+/HER-. Histopathological and immunohistochemical examination by qualified pathologists had confirmed the primary diagnosis and tumor percentile, in each case. This study (22 KHCC 041) was approved by KHCC Research Council, and the Institutional Review Board (IRB), in accordance with the international scientific and ethical guidelines. Participants’ written informed consent form (ICF) waiver was approved by KHCC IRB for this retrospective report.
PIK3CA mutational analysis
PIK3CA mutational analysis was performed using the FDA-approved Qiagen therascreen PIK3CA RGQ real-time qualitative PCR assay Kit, according to the manufacturer instructions. The kit detects 11 mutations in PIK3CA (exon 7: C420R; exon 9: E542K, E545A, E545D, E545G, E545K, Q546E, and Q546R; and exon 20: H1047L, H1047R, and H1047Y) using genomic DNA derived from formalin-fixed, paraffin-embedded (FFPE) breast tumor tissue with ≥20% tumor percentile, or ctDNA extracted from plasma taken from patients with breast cancer.
Results:
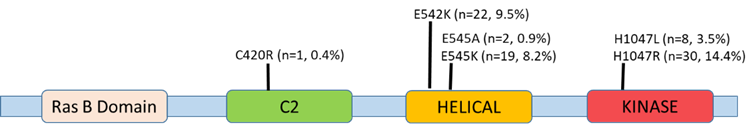
Tumor contents of the 231 studies samples range was at 20-95%. Participants’ age ranged between 24-86 years, mean age 53.4 years, median age at 53 years, and the mode age was 46 years. All 231 samples originating from metastatic breast cancer FFPE tissues were analyzed for the presence of PIK3CA mutations. Eleven targeted mutations were screened for in this analysis spanning exons 7 (C420R, E542K, E545A, E545D), 9 (E545G, E545K, and Q546E, Q546R, H1047L), and 20 (H1047R and H1047Y). Of the 11 assay-targeted mutations, E542K (22/231) and E545K (19/231) who map to helicase domain, in addition to H1047R (30/231) and H1047L (8/231) that both map to the kinase domain, were the most prevalent mutations amongst the overall 231 assessed samples (Figure 1), which is consistent with the global hotspot mutational rates in PIK3CA [21]. C420R located within the surface loops of C2 domain, that is proposed to be part of the membrane binding region of p110α [22], was seen just once in one patient, while E545A was seen in two patients. Interestingly, E545K/H1047L and E545K/H1047R double mutations were detected simultaneously in 2 and 7 patients, respectively. Of the 11 mutations covered in the assay, five (E545D, E545G, Q546E, Q546R, and H1047Y) had not been detected in any of the study participants’ samples.
Of all participants, 91/231 (39.4%) had at least one mutation detected, while the remaining 140 cases (60.6%) were wild-type for the tested mutations (Table 1). Tumor percent (20-95%) was neither age related, nor mutation presence related. Furthermore, PIK3CA wild-type samples were unexpectedly associated with higher percentile tumors at ≥50%, while presence of mutation was associated with tumors of lower tumor contents, which we find intriguing and needing further explorations.
Table 1: PIK3CA mutations count and percentiles of the positive samples.
|
Coding Exon # |
Amino Acid Change |
Base Change |
P110α Functional domain |
PIK3CA mutated (mutation percent this study, n=91) |
PIK3CA mutated |
% mutated in all patients |
% mutated in global patients |
|
7 |
C420R |
1258 T>C |
C2 |
1 (1.1) |
22 (2.7) |
0.4 |
1.1 |
|
7 |
E542K |
1624 G>A |
Helicase |
22 (24.2) |
94 (11.6) |
9.5 |
4.7 |
|
7 |
E545A |
1634 A>C |
Helicase |
2 (2.2) |
21 (2.6) |
0.9 |
1.1 |
|
7 |
E545D |
1635 G>T |
Helicase |
0 |
9 (1.1) |
0 |
0.5 |
|
9 |
E545G |
1634 A>G |
Helicase |
0 |
14 (1.7) |
0 |
0.7 |
|
9 |
E545K |
1633 G>A |
Helicase |
19 (20.9) |
163 (20.2) |
8.2 |
8.2 |
|
9 |
Q546E |
1636 C>G |
Helicase |
0 |
2 (0.2) |
0 |
0.1 |
|
9 |
Q546R |
1637 A>G |
Helicase |
0 |
7 (0.9) |
0 |
0.4 |
|
20 |
H1047Y |
3139 C>T |
Kinase |
0 |
11 (1.4) |
0 |
0.6 |
|
20 |
H1047L |
3140 A>T |
Kinase |
8 (8.8) |
52 (6.4) |
3.5 |
2.6 |
|
20 |
H1047R |
3140 A>G |
Kinase |
30 (33) |
288 (35.6) |
13 |
14.4 |
|
9&20 |
E545K and H1047L |
2 (2.2) |
0 |
0.9 |
0 |
||
|
9&20 |
E545K and H1047R |
7 (7.7) |
0 |
3 |
0 |
||
|
Total Mutated Samples and percentile |
91/231 (39.4) this study; 808/1995 (40.5) global study |
||||||
|
Total Wild-Type samples and percentile |
140/231 (60.6) this study; 1187/1995 (59.5) global study |
||||||
Discussion
PIK3CA is frequently mutated in multiple types of cancers at known ‘Hot-spots’, mainly in the kinase and helical modular domains. These are thought to occur early and possibly initiate events leading to malignancies like breast cancer [23]. Of notice, there is a noteworthy absence of mutations in both the Ras Binding Domain (RBD) and within the conserved catalytic site of the kinase domain of PIK3CA [24].
Preclinical evidence in model systems suggests PIK3CA mutations in general have activating (gain of function) properties, however domain-specific mutations may be associated with different AKT (and other pathways) phenotypic consequences, that may be cancer type-specific, or in general common in malignancies. PIK3CA somatic mutations rate varies between 0-50% between the different cancer types, and at 35% with respect to breast cancer [25, 26]. Several studies reported PIK3CA mutations were strongly associated with AKT pathway activation, growth factor-independent cell proliferation, resistance to apoptosis, and increased invasion and cell migration [18, 24, 27, 28].
Kinase domain mutations appear to be linked more strongly with pathway features enabling cell proliferation [25, 29], while helical domain mutations are linked more with features enabling metastatic cell migration and dissemination (e.g. RHO GTPases) [29, 30]. In our patients’ samples, mutations were detected in both domains, suggesting a direct role for the observed phenotypic features underlying breast cancer development and subsequent invasiveness in these patients. Nevertheless, with respect to possible pathogenic mechanisms, clinical outcomes, and survival in our patient cohort, an in-depth analysis is warranted, and this limitation shall be revisited in a further follow-up investigation.
Scarce ethnical/population PIK3CA mutational prevalence currently exists at the global level, mainly limited by the clinical trials data at various geographic areas. The largest data set had been recently presented in a poster session at the San Antonio Breast Cancer Symposium 2021 [21]. The authors presented data that is part of the SOLAR-1 clinical trial, where a total of 1995 patients were enrolled from across 4 regions covering Asia, Europe, Latin America, and Middle East, which included 633, 1025, 136, and 200 participants, respectively. In our study, PIK3CA mutational rate of 39.4% was close but distinct from other regional and global studies from Asia (38.2%), Europe (43.3%), Latin America (38.2%), and Middle East (35.0%). The representative Middle Eastern data in particular was on participants from Egypt and Lebanon. Apart from the European data, our Jordanian participants’ mutational prevalence rate is essentially similar to that reported from populations of Asian, Latin America and the Middle East. When combining all 4 continental geographical areas covered by this SOLAR-1 trial study, the overall prevalence (40.5%) was essentially very comparable to our study (39.4%). The double somatic mutations (E545K/H1047L and E545K/H1047R) seen in 9 (9.9%) patients amongst the overall 91 positive cases could potentially exist and act in cis, leading to augmented PIK3CA gain of function and increased PI3K3CA signaling that promote further cellular proliferation and tumor growth [31]. These nine patients are predicted to have an improved sensitivity to PI3Ka inhibitors, compared with patients harboring just a single-hotspot mutation. To this regard, a met-analysis in silico study [32] combining 10 studies with 6338 patients, showed doubly mutated patients were at 4% of all cases, which is essentially similar to our study (3.9%), suggesting our observations are in line with the global data.
In another clinical trial (SAFIR02) that enrolled 364 HR+, HER2– patients with metastatic breast cancer, PIK3CA mutations were detected in 104 (28.6%) of all these patients, who poorly responded to chemotherapy, and a shorter overall survival [17]. Altogether, our described patients cohort seem to have an overall PIK3CA mutational rates falling within the regional and global rates, including when individual mutations are considered, suggests the pathogenic mechanisms and therapeutic outcomes for our patient are predicated to be in line with regional and global data. Nevertheless, it remains to be seen how these projections will pan out in our planed follow-up clinical investigations.
Acknowledgment
We are grateful to the Science Health Education (SHE) Center and Dana-Farber Cancer Institute for their guidance in editing and review of this manuscript. Special thank you to Editor, Elizabeth Hamlin, for her contribution to the finalization of this manuscript. We also wish to express our gratitude to KHCC and KHCF for their support of our research and scientific advancements. This work was approved by the KHCC IRB.
Author Contribution
OA and AT conceived the study scope. OA analyzed data and wrote the manuscript. HR, YT and HA clinically characterized the patients and reviewed the manuscript. ZS performed genotyping and organized the genetic data. All authors read and approved the manuscript.
Data Availability
All data generated or analyzed during this study are included in this article (Table 1).
References
- Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol 13 (2012): 195-203.
- Carpenter CL, Duckworth BC, Auger KR, Cohen B, Schaffhausen BS, Cantley LC. Purification and characterization of phosphoinositide 3-kinase from rat liver. J Biol Chem 265 (1990): 19704-19711.
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 15 (2015): 7-24.
- Dannemann N, Hart JR, Ueno L, Vogt PK. Phosphatidylinositol 4,5-bisphosphate-specific AKT1 is oncogenic. Int J cancer 127 (2010): 239-244.
- Hiles ID, Otsu M, Volinia S, et al. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell 70 (1992): 419-429.
- Stephens L, Anderson K, Stokoe D, et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279 (1998): 710-714.
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 4 (2002): 658-665.
- Ebner M, Lucic I, Leonard TA, Yudushkin I. PI (3,4,5) P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell 65 (2017): 416-431.
- Hung C-M, Garcia-Haro L, Sparks CA, Guertin DA. mTOR-dependent cell survival mechanisms. Cold Spring Harb Perspect Biol 4 (2012).
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71 (2021): 209-249.
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9 (2009): 550-562.
- Wright SCE, Vasilevski N, Serra V, Rodon J, Eichhorn PJA. Mechanisms of Resistance to PI3K Inhibitors in Cancer: Adaptive Responses, Drug Tolerance and Cellular Plasticity. Cancers (Basel) 13 (2021).
- Lim E, Metzger-Filho O, Winer EP. The natural history of hormone receptor-positive breast cancer. Oncology (Williston Park) 26 (2012): 688-694, 696.
- Hempel D, Ebner F, Garg A, et al. Real-world data analysis of next generation sequencing and protein expression in metastatic breast cancer patients. Sci Rep 10 (2020): 10459.
- Alqahtani A, Ayesh HSK, Halawani H. PIK3CA Gene Mutations in Solid Malignancies: Association with Clinicopathological Parameters and Prognosis. Cancers (Basel) 12 (2019).
- Moon A, Chin S, Kim HK, et al. EGFR, COX2, p-AKT expression and PIK3CA mutation in distal extrahepatic bile duct carcinoma. Pathology 48 (2016): 35-40.
- Mosele F, Stefanovska B, Lusque A, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol Off J Eur Soc Med Oncol 31 (2020): 377-386.
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304 (2004): 554.
- André F, Ciruelos E, Rubovszky G, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med 380 (2019): 1929-1940.
- Rugo HS, Lerebours F, Ciruelos E, et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol 22 (2021): 489-498.
- Pathmanathan Rajadurai, Tatiana Semiglazova, Alinta Hegmane E al. San Antonio Breast Cancer Symposium 2021. In: PIK3CARegistry: A Noninterventional, Descriptive, Retrospective Cohort Study of PIK3CAMutations in Patients with Hormone Receptor-Positive (HR+), Human Epidermal Growth Factor Receptor 2-Negative (HER2–) Advanced Breast Cancer (ABC) (2021): P5-13-25.
- Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402 (1999): 313-320.
- Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol 347 (2010): 21-41.
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA 105 (2008): 2652-2657.
- Zardavas D, Phillips WA, Loi S. PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data. Breast Cancer Res 16 (2014): 201.
- Mitchell CB, Phillips WA. Mouse Models for Exploring the Biological Consequences and Clinical Significance of PIK3CA Mutations. Biomolecules 9 (2019).
- Wu X, Renuse S, Sahasrabuddhe NA, et al. Activation of diverse signalling pathways by oncogenic PIK3CA mutations. Nat Commun 2014 51 5 (2014): 1-13.
- Janku F, Wheler JJ, Naing A, et al. PIK3CA mutations in advanced cancers: characteristics and outcomes. Oncotarget 3 (2012): 1566-1575.
- López-Knowles E, Segal C V, Gao Q, et al. Relationship of PIK3CA mutation and pathway activity with antiproliferative response to aromatase inhibition. Breast Cancer Res 16 (2014): R68.
- Krygowska AA, Castellano E. PI3K: A Crucial Piece in the RAS Signaling Puzzle. Cold Spring Harb Perspect Med 8 (2018).
- Vasan N, Razavi P, Johnson JL, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 366 (2019): 714-723.
- Martínez-Sáez, O., Chic, N., Pascual, T. et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res 22 45 (2020).