Panel Sequencing Identified a Novel Splice Site Mutation in Hermansky-Pudlak Syndrome Type 1 Patients
Article Information
Doris Boeckelmann*#, Felix Sobotta#, Felicia Andresen, Amina Szvetnik, Salome Fels, Barbara Zieger
#Authors contributed equally
Department of Pediatrics and Adolescent Medicine, Division of Pediatric Hematology and Oncology, Medical Center – University of Freiburg, Freiburg, Germany
*Corresponding Author: Doris Boeckelmann, Department of Pediatrics and Adolescent Medicine, Division of Pediatric Hematology and Oncology, University Medical Center Freiburg, Mathildenstr. 1, 79106 Freiburg, Germany
Received: 02 October 2020; Accepted: 11 November 2020; Published: 02 December 2020
Citation:
Doris Boeckelmann, Felix Sobotta, Felicia Andresen, Amina Szvetnik, Salome Fels, Barbara Zieger. Panel Sequencing Identified a Novel Splice Site Mutation in Hermansky-Pudlak Syndrome Type 1 Patients. Archives of Clinical and Medical Case Reports 4 (2020): 1172-1181.
View / Download Pdf Share at FacebookAbstract
Introduction: Hermansky-Pudlak syndrome (HPS) is a rare autosomal recessive disorder. Patients with HPS characteristically present with oculocutaneous albinism, nystagmus and increased bleeding tendency. The symptoms are caused by malfunction of lysosome related organelles (i.e. melanosomes, platelets). Patients with HPS typically show impaired platelet delta granule secretion. We report three patients (two brothers and another unrelated patient) who presented with these typical symptoms.
Methods and Results: Platelet aggregometry and flow cytometry analyses revealed pathological platelet function and decreased platelet delta granule secretion. Using NGS panel analysis comprising all 10 HPS genes a defect in the HPS1 gene was identified. The brothers share compound heterozygous a novel splice variant (c.987+1[G>A]) and an already reported mutation (c.1189[delC]). Although the third patient is not related with the brothers, he presented with the same one base pair deletion c.1189[delC] compound heterozygous with another already reported base pair deletion (c.355[delC]).
Conclusions: Since HPS1 patients are at risk to develop pulmonary fibrosis at middle age, early genetic diagnosis of the HPS type is important for prognosis and treatment. In this study we identified a novel splice site variant (c.987+1[G>A]) in the HPS1 gene, diagnosed the patients as HPS1 type and therefore, enabled adequate follow-up and therapy.
Keywords
Hermansky-Pudlak syndrome (HPS); Next Generation Sequencing (NGS); platelet granule secretion; HPS1; Albinism
Hermansky-Pudlak syndrome (HPS) articles; Next Generation Sequencing (NGS) articles; platelet granule secretion articles; HPS1 articles; Albinism articles
Hermansky-Pudlak syndrome articles Hermansky-Pudlak syndrome Research articles Hermansky-Pudlak syndrome review articles Hermansky-Pudlak syndrome PubMed articles Hermansky-Pudlak syndrome PubMed Central articles Hermansky-Pudlak syndrome 2023 articles Hermansky-Pudlak syndrome 2024 articles Hermansky-Pudlak syndrome Scopus articles Hermansky-Pudlak syndrome impact factor journals Hermansky-Pudlak syndrome Scopus journals Hermansky-Pudlak syndrome PubMed journals Hermansky-Pudlak syndrome medical journals Hermansky-Pudlak syndrome free journals Hermansky-Pudlak syndrome best journals Hermansky-Pudlak syndrome top journals Hermansky-Pudlak syndrome free medical journals Hermansky-Pudlak syndrome famous journals Hermansky-Pudlak syndrome Google Scholar indexed journals syndrome articles syndrome Research articles syndrome review articles syndrome PubMed articles syndrome PubMed Central articles syndrome 2023 articles syndrome 2024 articles syndrome Scopus articles syndrome impact factor journals syndrome Scopus journals syndrome PubMed journals syndrome medical journals syndrome free journals syndrome best journals syndrome top journals syndrome free medical journals syndrome famous journals syndrome Google Scholar indexed journals Next Generation Sequencing articles Next Generation Sequencing Research articles Next Generation Sequencing review articles Next Generation Sequencing PubMed articles Next Generation Sequencing PubMed Central articles Next Generation Sequencing 2023 articles Next Generation Sequencing 2024 articles Next Generation Sequencing Scopus articles Next Generation Sequencing impact factor journals Next Generation Sequencing Scopus journals Next Generation Sequencing PubMed journals Next Generation Sequencing medical journals Next Generation Sequencing free journals Next Generation Sequencing best journals Next Generation Sequencing top journals Next Generation Sequencing free medical journals Next Generation Sequencing famous journals Next Generation Sequencing Google Scholar indexed journals platelet granule secretion articles platelet granule secretion Research articles platelet granule secretion review articles platelet granule secretion PubMed articles platelet granule secretion PubMed Central articles platelet granule secretion 2023 articles platelet granule secretion 2024 articles platelet granule secretion Scopus articles platelet granule secretion impact factor journals platelet granule secretion Scopus journals platelet granule secretion PubMed journals platelet granule secretion medical journals platelet granule secretion free journals platelet granule secretion best journals platelet granule secretion top journals platelet granule secretion free medical journals platelet granule secretion famous journals platelet granule secretion Google Scholar indexed journals HPS1 articles HPS1 Research articles HPS1 review articles HPS1 PubMed articles HPS1 PubMed Central articles HPS1 2023 articles HPS1 2024 articles HPS1 Scopus articles HPS1 impact factor journals HPS1 Scopus journals HPS1 PubMed journals HPS1 medical journals HPS1 free journals HPS1 best journals HPS1 top journals HPS1 free medical journals HPS1 famous journals HPS1 Google Scholar indexed journals treatment articles treatment Research articles treatment review articles treatment PubMed articles treatment PubMed Central articles treatment 2023 articles treatment 2024 articles treatment Scopus articles treatment impact factor journals treatment Scopus journals treatment PubMed journals treatment medical journals treatment free journals treatment best journals treatment top journals treatment free medical journals treatment famous journals treatment Google Scholar indexed journals CT articles CT Research articles CT review articles CT PubMed articles CT PubMed Central articles CT 2023 articles CT 2024 articles CT Scopus articles CT impact factor journals CT Scopus journals CT PubMed journals CT medical journals CT free journals CT best journals CT top journals CT free medical journals CT famous journals CT Google Scholar indexed journals surgery articles surgery Research articles surgery review articles surgery PubMed articles surgery PubMed Central articles surgery 2023 articles surgery 2024 articles surgery Scopus articles surgery impact factor journals surgery Scopus journals surgery PubMed journals surgery medical journals surgery free journals surgery best journals surgery top journals surgery free medical journals surgery famous journals surgery Google Scholar indexed journals Albinism articles Albinism Research articles Albinism review articles Albinism PubMed articles Albinism PubMed Central articles Albinism 2023 articles Albinism 2024 articles Albinism Scopus articles Albinism impact factor journals Albinism Scopus journals Albinism PubMed journals Albinism medical journals Albinism free journals Albinism best journals Albinism top journals Albinism free medical journals Albinism famous journals Albinism Google Scholar indexed journals enterectomy articles enterectomy Research articles enterectomy review articles enterectomy PubMed articles enterectomy PubMed Central articles enterectomy 2023 articles enterectomy 2024 articles enterectomy Scopus articles enterectomy impact factor journals enterectomy Scopus journals enterectomy PubMed journals enterectomy medical journals enterectomy free journals enterectomy best journals enterectomy top journals enterectomy free medical journals enterectomy famous journals enterectomy Google Scholar indexed journals
Article Details
1. Introduction
Hermansky-Pudlak syndrome (HPS) was first described by F. Hermansky and P. Pudlak in 1959 [1]. The mode of inheritance is autosomal recessive. The characteristic main symptoms are bleeding tendency and oculocutaneous albinism with congenital nystagmus, iris transillumination, decreased visual acuity and reduced skin pigmentation [2]. Bleeding symptoms range from epistaxis, petechiae to easy bruising and may cause serious problems in the case of trauma or surgery. So far, in humans ten HPS types (HPS1-10) have been described, some of which are associated with pulmonary interstitial lung disease, granulomatous colitis and in case of HPS2 and 10 with immune deficiency [3]. Cause for the development of HPS is a malfunction of lysosome-related organelles such as delta granules and melanosomes which are vital for membrane and protein trafficking [4]. Platelet delta granules contain serotonin, calcium, ADP and polyphosphate. In healthy individuals the content of delta granules is secreted to enhance platelet adhesion and activation [5]. The malfunction of delta granules in platelets causes the bleeding tendency and the malfunction of melanosomes explains the albinism.
HPS is a rare disease with a prevalence of 1-9 per 1,000,000 individuals [3]. Prevalence is much higher in some populations and goes up to 1 in 1800 in Puerto Rico due to founder effects [6]. Pathogenic variants can be found in 10 HPS associated genes (HPS1, AP3B1, HPS3-6, DTNBP1, BLOC1S3 BLOC1S6, AP3D1). The HPS genes encode for multi-protein complexes called biogenesis of lysosome related organelles complex (BLOC). BLOC 3 contains the gene products of HPS1 and HPS4 [7]. HPS1 is located on chromosome 10 (10q24.2). The gene comprises 20 exons and codes for a 700 amino acid polypeptide with a predicted molecular weight of 79,3 kDa. There are 76 known variants associated with the HPS1 phenotype [3]. HPS1 is the most described genotype in patients. According to the literature HPS1 and HPS4 seem to display the most severe HPS phenotype regarding pulmonary interstitial lung disease and granulomatous colitis [3, 8]. Here, using panel sequencing we identified a novel essential splice site mutation and already reported pathogenic HPS1 variants in three patients with classic HPS phenotype.
2. Patients, Materials and Methods
2.1 Family A
Two brothers, 10 and 6 years old (A-II.1 and A-II.2) presented with oculocutaneous albinism and nystagmus at our outpatient clinic. In addition, both brothers showed easy bruising after minor trauma and prolonged bleeding after injuries. At the age of seven years patient A-II.1 underwent circumcision, surprisingly, without bleeding problems. So far, the younger brother (A-II.2) has not undergone any surgery. The non-consanguine parents (A-I.1 and A-I.2) were not affected clinically.
2.2 Family B
The 12 year old index patient (B-II.2) presented at our outpatient clinic with cutaneous albinism and had experienced prolonged bleeding after dental surgery. However, he did not show any bleeding symptoms during and after appendectomy. At the age of 11 years a rectal abscess was surgically removed. Afterwards he developed wound healing problems which lasted for about 12 months. His non-consanguine parents (B-I.1; B-I.2) and older brother (B-II.1) were not affected clinically.
2.3 Platelet count and platelet aggregometry analyses
Platelet count was measured using an automated cell counter (Sysmex KX-21 N, Norderstedt, Germany). Using citrate-anticoagulated blood samples platelet-rich plasma (PRP) and platelet-poor plasma (PPP) was prepared by centrifugation. Platelet counts in PRP were adjusted to a concentration of 250 G/L. Platelet aggregometry analyses were performed after stimulation with the platelet agonists: collagen (2 and 10 μg/mL; Takeda, Linz, Austria), adenosine diphosphate (ADP; 4 and 10 μmoL/L; Sigma-Aldrich, St. Louis, MO, USA), epinephrine (8μmoL/L; Sanofi-Aventis, Frankfurt, Germany), and ristocetin (1.2 mg/mL; American Biochemical and Pharmaceutical LTD, Frankfurt, Germany) using APACT 4004 (Labitec, Ahrensburg, Germany).
2.4 Flow cytometry analyses
Flow cytometry analyses were performed according to Lahav et al. [9] using FACSCalibur (Becton Dickinson, Heidelberg, Germany). Aliquots of diluted PRP (5 × 107 platelets/mL) were fixed and stained with FITC-labeled monoclonal surface antibody against CD41 (fibrinogen receptor GPIIb/IIIa-complex), CD42a (von Willebrand receptor GPIb/IX) and CD42b (von Willebrand receptor GPIb) (Coulter, Immunotech, Marseille, France), respectively. For VWF-binding and fibrinogen binding analyses, diluted PRP (5 × 107 platelets/mL) was stimulated with different concentrations of ristocetin (0–1 mg/mL) and ADP (0–2 μmoL/L) for 3 min at RT, respectively. Platelets were stained with FITC-labeled anti-VWF (Bio-Rad AbD Serotec, Puchheim, Germany) and Alexa Fluor 488-labeled anti-fibrinogen (Invitrogen, Waltham, MA, USA). For CD62 and CD63 expression analyses diluted PRP (5 × 107 platelets/mL) was stimulated with different concentrations of thrombin (0, 0.05, 0.1, 0.2, 0.5, and 1 U/mL; Siemens Healthineers, Marburg, Germany) in the presence of 1.25 mM Gly-Pro-Arg-Pro (Bachem, Bubendorf, Switzerland). Platelets were stained with monoclonal FITC-labeled anti-CD62 (P-selectin) and anti-CD63 antibodies (lysosomal membrane associated glycoprotein 3, LAMP-3; Immunotech, Marseille, France). Data of patients and controls (day control and 20 independent measurements from 6 controls as mean ± standard error of the mean, SEM) were analyzed using GraphPad Prism software (version 8, San Diego, CA, USA).
2.5 Molecular genetic analysis
Informed consent for molecular genetic analysis was given for all family members. Genomic DNA was extracted from EDTA-blood using standard procedures and the Blood and Cell DNA Kit from Qiagen (Qiagen GmbH, Hilden, Germany). For the affected patients we performed targeting enrichment (panel including all 10 HPS-genes) using Nextera Rapid Custom Enrichment Kit (Illumina) followed by sequencing on a MiSeq (Illumina). For data analyses SeqPilot (JSI medical systems) was used. Variants were exported and filtered according to allele frequency and serious consequences. For classification we used supporting software ALAMUT® (v.2.15), pathogenicity prediction (SIFT, MutTaster, PolyPhen2 and CADD), occurrence in population and disease databases (HGMD public version, Huizing HPS Mutation update [3]). Variants were classified in accordance to the ACMG (American College of Medical Genetics) guidelines [10]. To validate our findings and for family genotyping Sanger sequencing was performed.
3. Results
Both affected brothers from family A (A-II.1 and A-II.2) presented with normal platelet counts (394 G/l and 347 G/l, respectively). The patient from family B (B-II.2) also showed normal platelet count (404 G/l). In vivo bleeding time (Ivy) was performed for two patients A-II.1 and B-II.2, respectively and was severely prolonged (10 min and 10,5 min, respectively; normal <6 minutes). The brothers of family A showed impaired aggregation after stimulation with low dose collagen (2,0µg/mL), ADP and epinephrine. The patient from family B (B-II.2) reached almost normal values (Table 1).
In flow cytometric analyses the two brothers (A-II.1 and A-II.2) and the unrelated patient (B-II.2) showed severely reduced CD63 expression hinting to a delta granule secretion defect (figure 1D-F). All patients showed slightly reduced CD62 expression (figure 1A-C). Additional fibrinogen binding after ADP activation was slightly impaired for patient A II.1. Expression of CD42a, 42b, 41 and VWF-binding after stimulation with Ristocetin were normal compared to healthy controls for all patients (data not shown).
|
Stimulation |
A II.1 [%] |
A II.2 [%] |
B II.2 [%] |
Norm [%] |
|
Collagen (2,0µg/mL) |
36 |
29 |
64 |
>70 |
|
Collagen (10,0µg/mL) |
88 |
86 |
- |
>70 |
|
Ristocetin (1,2mg/mL) |
88 |
64 |
89 |
>85 |
|
ADP (4µmol/L) |
50 |
52 |
75 |
>70 |
|
ADP (10µmol/L) |
75 |
72 |
- |
>70 |
|
Epinephrine (8µmol/L) |
34 |
26 |
78 |
>70 |
Table 1: Platelet aggregation/agglutination analyses measured by light transmission.
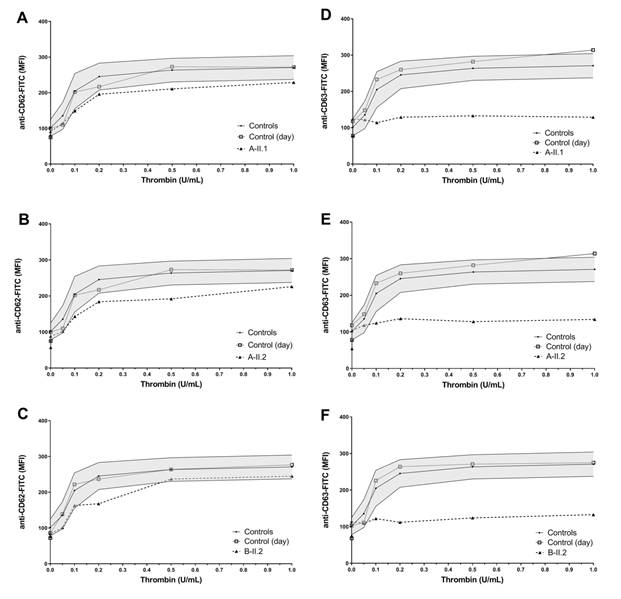
Figure 1: Platelet granule secretion stimulated with thrombin (concentrations: 0, 0.05, 0.1, 0.2, 0.5, and 1.0 U/mL) for all three patients measured by flow cytometry. A-C Slightly impaired α-granule secretion indicated by reduced platelet CD62 expression compared to the healthy controls/day control. D-F Severely impaired δ-granule secretion indicated by reduced platelet CD63 expression compared to the healthy controls/day control.
3.1 Molecular genetic analysis
We identified compound heterozygous pathogenic variants in the HPS1 gene (NM_000195.3) in all patients. Both affected brothers of family A presented with a novel essential splice site mutation (c.987+1[G>A]). The consequence of this mutation at the donor splice site of intron 11 is not predictable but leads very likely to skipping of exon 11. To our knowledge this mutation has not been described so far. The second variant was a one base pair deletion (c.1189[delC]) in exon 13. This leads to a frameshift with premature stop codon (p.Gln397Serfs*2). Sanger sequencing identified the mother (A-I.2.) as heterozygous carrier of the c.1189[delC] variant, no material was available from the father (A-I.1) (Pedigree figure 2). The same 1-bp-deletion (c.1189[delC]) was also identified in patient (B-II.2), compound heterozygous with another 1-bp-deletion (c.355[delC]; p.His119HisThrfs*5) in exon 5 leading also to a frameshift with premature stop codon. Sanger sequencing identified the father (B-1.1) as heterozygous carrier of the c.355[delC] variant whereas the mother (B-1.2) was a carrier of the c.1189[delC] variant. The older brother (B-II.1) showed wild type sequence at these positions (Pedigree figure 3). Results are summarized in Table 2.
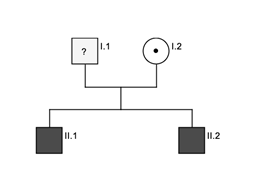
Figure 2: Pedigree family A. Black indicates affected patients, dot indicates heterozygous carrier.
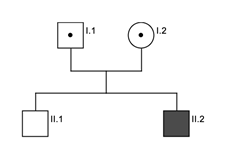
Figure 3: Pedigree family B. Black indicate affected patient, dot indicates heterozygous carrier and white indicates unaffected brother.
|
Patient |
Nucleotide |
Protein |
CADD-Score |
|
Family A |
|||
|
Patient 1 (A-II.1) |
c.987+1[G>A] c.1189[delC] |
Splice defect p.Gln397Serfs*2 |
28.2 25,5 |
|
Patient 2 (A-II.2) |
c.987+1[G>A] c.1189[delC] |
Splice defect p.Gln397Serfs*2 |
28.2 25,5 |
|
Mother (A-I.2) |
c.987+1[G=] c.1189[delC] |
p.Gln397Serfs*2 |
25,5 |
|
Family B |
|||
|
Patient (B-II.2) |
c.355[delC] c.1189[delC] |
p.His119Thrfs*5 p.Gln397Serfs*2 |
29.2 25,5 |
|
Father (B-I.1) |
c.355[delC] c.1189[C=] |
p.His119Thrfs*5 |
29.2 |
|
Mother (B-I.2) |
c.355[C=] c.1189[delC] |
p.Gln397Serfs*2 |
25,5 |
|
Brother (B-II.1) |
c.355[C=] c.1189[C=] |
||
Novel variant in bolt; CADD, Combined Annotation Dependent Depletion: a scaled C-score of greater of equal 10 indicates that these are predicted to be the 10% most deleterious substitutions, a score of greater or equal 20 indicates the 1% most deleterious.
Table 2: Pathogenic variants in HPS1 identified in the index patients and family segregation analysis.
4. Discussion
We report three patients with typical symptoms of HPS (oculocutaneous albinism, nystagmus and bleeding tendency). All patients showed pathological platelet delta granule secretion which leads to increased bleeding tendency. Using NGS the molecular genetic defect in these patients could be identified leading to the diagnosis HPS1 in these patients. To our knowledge the splice mutation (c.987+1[G>A]) has not been described yet. According to ACMD guidelines we classified the mutation as pathogenic due to serious consequences. The 1-bp-deletion at position c.1189 was identified in both families who are not related. This variant (rs281865084) has already been described several times [11-17] and has a relatively high minor allele frequency (MAF) in Europeans (ClinVar: MAF 0.000067) [3]. The other 1-bp-deletion (c.355del) found in family B has also been reported before [13, 16]. HPS1 is the most common type of HPS and is found in all ethnic backgrounds. Many patients with HPS1 develop pulmonary fibrosis at middle age [3]. Doubkova et al. and Thompson et al. described two compound heterozygous patients of older age who were compound heterozygous with the c.1189[delC] variant in combination with other variants leading to serious consequences (nonsense, frameshift). The reported patients developed lung fibrosis at the age of 57 and 40 years [11, 18]. Therefore, monitoring for pulmonal fibrosis yearly, beginning in adolescence, is recommended. Lung transplantation may become necessary for patients with HPS1 related pulmonary fibrosis. The impaired platelet function can lead to increased bleeding symptoms during and after surgery especially in case of surgeries in mucocutaneous regions or brain surgery. Therefore, the transfusion of platelet concentrates may become necessary depending on the kind of the surgery. Periodic screening for granulomatous colitis is also recommended since some HPS1 patients can develop this disorder [19, 20]. According to the literature reports it seems that the HPS1 and HPS4 (BLOC-3) defects lead to the most severe prognosis of the HPS types because of the increased risk for pulmonary fibrosis, granulomatous colitis and the increased bleeding complications. Furthermore, these patients can develop wound healing problems as observed in one patient (B-II.2) in this study.
5. Conclusion
Careful history regarding bleeding symptoms or wound healing problems from patients with albinism should be done before any surgical procedure is initiated. In addition, an early molecular genetic analysis should be performed to provide an adequate follow-up and therapy in case of complications during the further course of the disease. The identification of the molecular genetic defects in HPS will allow further insights in the pathophysiology of this disorder.
Acknowledgements
We thank Eileen Lerner for excellent technical assistance. The article processing charge was funded by the Baden-Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg in the funding programme Open Access Publishing.
Author Contributions
D.B. and F.S. contributed equally to the study design, performed molecular genetic analyses and data interpretation and wrote the manuscript. F.A. and A.S. took care of the patients and collected blood samples. S.F. performed DNA extraction. B.Z. contributed to the study design, supervision of the molecular genetic analyses and manuscript
editing. All authors critically reviewed the manuscript, with approval of the final draft.
References
- Hermansky F, Pudlak P. Albinism associated with hemorrhagic diathesis and unusual pigmented reticular cells in the bone marrow: report of two cases with histochemical studies. Blood 14 (1959): 162-169.
- Gahl WA, Brantly M, Kaiser-Kupfer MI, et al. Genetic defects and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky-Pudlak syndrome). The New England journal of medicine 338 (1998): 1258-1264.
- Huizing M, Malicdan MCV, Wang JA, et al. Hermansky-Pudlak syndrome: Mutation update. Human mutation 41 (2020): 543-580.
- Vincent LM, Adams D, Hess RA, et al. Hermansky-Pudlak syndrome type 1 in patients of Indian descent. Molecular genetics and metabolism 97 (2009): 227-233.
- Bowman SL, Bi-Karchin J, Le L, et al. The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic 20 (2019): 404-435.
- Witkop CJ, Nunez Babcock M, Rao GH, et al. Albinism and Hermansky-Pudlak syndrome in Puerto Rico. Boletin de la Asociacion Medica de Puerto Rico 82 (1990): 333-339.
- Wei ML. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment cell research 19 (2006): 19-42.
- Huizing M, Helip-Wooley A, Westbroek W, et al. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annual review of genomics and human genetics 9 (2008): 359-386.
- Lahav J, Jurk K, Hess O, et al. Sustained integrin ligation involves extracellular free sulfhydryls and enzymatically catalyzed disulfide exchange. Blood 100 (2002): 2472-2478.
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics 17 (2015): 405-424.
- Doubkova M, Trizuljak J, Vrzalova Z, et al. Novel genetic variant of HPS1 gene in Hermansky-Pudlak syndrome with fulminant progression of pulmonary fibrosis: a case report. BMC pulmonary medicine 19 (2019): 178.
- Griffin AE, Cobb BR, Anderson PD, et al. Detection of hemizygosity in Hermansky-Pudlak syndrome by quantitative real-time PCR. Clinical genetics 68 (2005): 23-30.
- Hermos CR, Huizing M, Kaiser-Kupfer MI, et al. Hermansky-Pudlak syndrome type 1: gene organization, novel mutations, and clinical-molecular review of non-Puerto Rican cases. Human mutation 20 (2002): 482.
- Lasseaux E, Plaisant C, Michaud V, et al. Molecular characterization of a series of 990 index patients with albinism. Pigment cell & melanoma research 31 (2018): 466-474.
- Oh J, Ho L, Ala-Mello S, et al. Mutation analysis of patients with Hermansky-Pudlak syndrome: a frameshift hot spot in the HPS gene and apparent locus heterogeneity. American journal of human genetics 62 (1998): 593-598.
- Sandrock K, Bartsch I, Rombach N, et al. Compound heterozygous mutations in 2 siblings with Hermansky-Pudlak syndrome type 1 (HPS1). KlinPadiatr 222 (2010): 168-174.
- Shotelersuk V, Hazelwood S, Larson D, et al. Three new mutations in a gene causing Hermansky-Pudlak syndrome: clinical correlations. Molecular genetics and metabolism 64 (1998): 99-107.
- Thompson G, Sekiguchi H, Chen D, et al. A 40-Year-Old Man With Albinism and Progressive Dyspnea. Chest 154 (2018): e143-e146.
- Girot P, Le Berre C, De Maissin A, et al. Crohn's-like acute severe colitis associated with Hermansky-Pudlak syndrome: A case report. World journal of gastroenterology 25 (2019): 1031-1036.
- Schinella RA, Greco MA, Cobert BL, et al. Hermansky-Pudlak syndrome with granulomatous colitis. Annals of internal medicine 92 (1980): 20-23.
