P22 Phage Shows Promising Antibacterial Activity under Pathophysiological Conditions
Article Information
Logan Gildea1, Joseph A. Ayariga2,*, Boakai K. Robertson3, Robert Villafane3
1The Microbiology Program, College of Science, Technology, Engineering and Mathematics (C-STEM), Alabama State University, Montgomery, AL, 36104
2The Biomedical Engineering Program, College of Science, Technology, Engineering and Mathematics (C-STEM), Alabama State University, 1627 Hall Street, Montgomery, AL, 36104
3Department of Biological Sciences, Microbiology PhD. Program, College of Science, Technology, Engineering and Mathematics (C-STEM), Alabama State University, 1627 Hall Street Montgomery, Alabama 36104
*Corresponding author: Joseph A. Ayariga, The Biomedical Engineering Program, College of Science, Technology, Engineering and Mathematics (C-STEM), Alabama State University, 1627 Hall Street, Montgomery, AL, 36104. USA
Received: 02 February 2022; Accepted: 09 February 2022; Published: 10 February 2022
Citation:
Logan Gildea, Joseph A. Ayariga, Boakai K. Robertson, Robert Villafane. P22 Phage Shows Promising Antibacterial Activity under Pathophysiological Conditions. Archives of Microbiology and Immunology 6 (2022): 81-100.
View / Download Pdf Share at FacebookAbstract
The prevalence of multidrug resistant bacterial diseases is a major global health risk. Multidrug resistant bacterial diseases are prevalent, and the need for novel methods of treatment is essential to the preservation of public health. Annually foodborne pathogens cause 1.35 million infections and 26,500 hospitalizations in the United States alone. Foodborne pathogens such as Salmonella spp. are a major threat to public health. Bacteriophages offer a unique method for the treatment of these multidrug resistant bacteria. We studied the infection dynamics of a potential mono-phage therapy of Salmonella typhimurium under various pathophysiological conditions. Furthermore, we determined the resistance dynamics of Salmonella typhimurium against P22 phage treatment. We also determined synergy with antibiotics such as ampicillin and kanamycin. This research helps to further define and show the versatility of bacteriophages as potential novel treatment methods.
Keywords
Bacteriophage therapy, antimicrobial resistance, Salmonella, antibiotic synergy
Bacteriophage therapy articles, antimicrobial resistance articles, Salmonella articles, antibiotic synergy articles
Article Details
1. Introduction
The discovery of antibiotics revolutionized modern medicine; however, as we progress into the mid-21st century, the prevalence of antibiotic-resistant bacteria is a major threat to public health. The massive and sometimes inappropriate use of antibiotics has led to the increased prevalence of multidrug-resistant bacterial strains that present treatment challenges in the clinical setting [1-4]. Antibiotic-resistant bacterial infections account for roughly 2.8 million infections and 35,000 deaths annually in the United States alone [2]. These infections pose serious threats to public health and are also a major economic burden costing an estimated 7.7 billion dollars in medical costs annually [5]. A major hazard to public health is the increasing prevalence of antibiotic and multidrug-resistant foodborne pathogens. As these infections are becoming increasingly prevalent, the development of new antibiotic agents has declined, creating a major problem in the field of medicine [6, 7].
One of the most prevalent foodborne pathogens is Salmonella, which can be found in numerous food products such as poultry, seafood, and other fresh and processed meats [8-11]. Annually Salmonella spp. accounts for 1.35 million infections, 26,500 hospitalizations, and 420 deaths in the United States alone. An increased prevalence of antibiotic-resistant Salmonella strains could potentially result in an increase rate of mortality making it critical to develop new novel treatment strategies. Salmonella typhimurium has been of significant relevance due to the reported prevalence of antibiotic-resistance in this species [12, 13]. The Centers for Disease Control (CDC) reported in 2019 that Salmonella typhimurium accounted for 59% of ampicillin-resistant infections [2]. Continued mass usage of general antibiotics could drastically increase the prevalence of antibiotic resistant S. typhimurium as well as other Salmonella species [5, 14].
A potential alternative to antibiotic treatment is the utilization of phage therapy as a novel therapeutic treatment [15-18]. Bacteriophages, commonly referred to as phages, are prokaryotic viruses that infect bacterial species with high specificity. An important distinction of phages is their lifestyle which can either be lytic or lysogenic. The use of phages featuring the lytic phenotype is advised for the development of therapeutics [19, 20]. The host specificity of phages is a major benefit of phage therapy in comparison to traditional treatment methods such as antibiotics. The high host specificity of phages allows them to only interact with the target pathogen while leaving natural microbiota and eukaryotic cells unaffected [21, 11]. An additional benefit of phage therapy is the suggested synergy between phages and antibiotics that allows for co-therapy [15-18]. Co-therapy refers to the use of both antibiotics and phages in synonymy for the treatment of resistant or reoccurring bacterial infections. This method relies on the inability of the bacterial species to rapidly develop resistance to both treatment methods and utilizes the synergy that is sometimes present between antibiotics and phages [22-24]. Co-therapy has been suggested in literature as a potentially viable method for the treatment of multidrug-resistant bacterial strains [15-18].
Phage therapy is a strong potential candidate for the treatment of foodborne pathogens such as Salmonella typhimurium. The lytic Salmonella phage P22 has been widely studied but requires further study to be utilized as a standard treatment against Salmonella typhimurium [25, 26, 11]. Our study provides insight into the pathophysiological characteristics that the phage P22 must withstand to be considered a viable therapeutic against Salmonella typhimurium. Salmonella typhimurium typically colonizes within the small or large intestines after the pathogen is consumed [28]. The variation of pH throughout these organs is a challenge in the development of antibacterial agents [29, 30]. Determination of P22’s efficacy in pathophysiological conditions similar to the small and large intestines is critical to its progression as a potential therapeutic. Designing a phage therapy method for Salmonella typhimurium would also require further understanding of phage-host interactions in disease-mimicking conditions.
In this study, we have performed phage-bacteria kinetic studies under several pathophysiological conditions related to Salmonella typhimurium infections. These pathophysiological conditions include acidic pH and stationary phase. We utilized Salmonella typhimurium (referred to as BV4012) as the model organism for this study. This strain has a generation time of 30 minutes, allowing for kinetics under pathophysiological conditions to be examined rapidly. We show the ability of the phage P22 as an effective antibacterial agent against Salmonella typhimurium under low pH and stationary conditions. Additionally, we showed synergy with ampicillin and kanamycin. Most importantly, we showed that P22 could prevent the growth of Salmonella typhimurium under pathophysiological conditions, encouraging further study of P22 and other similar phages as potential antibacterial agents.
2. Methods
2.1 Soft Agar Overlay and Spot Assays for Phage Enumeration
To detect and quantify the phage P22, the soft agar overlay technique was utilized. Soft Agar was prepared by adding agar (0.8%) to Luria-Bertani (LB) media. (BD Difco, USA) This suspension was autoclaved and allowed to cool to 45 °C in a water bath. 1 mL of late log phase BV4012 bacterial culture (OD > 1) and 100 µL of P22 phage dilutions were added to 5 mL of the soft agar and was poured onto a Luria agar plate. The soft agar was allowed to cool to room temperature and solidify. These plates were then incubated at 37 °C overnight.
For verification of phage titers, spot assays were conducted using late log phase Salmonella typhimurium BV4012 (OD > 1). These cultures were plated on a LB agar plate to form a bacterial lawn. P22 phage samples were serially diluted and spotted onto the bacterial lawn. Plates were incubated at 37 °C for 24 h, and the number of plaques formed at various dilutions was quantified
2.2 Phage Amplification and Maintenance
P22 phage was amplified on S. typhimurium host. Liquid amplification was carried out by the addition of a P22 sample at 10 MOI to a log phase culture (OD between 1 and 2) and incubated for 12-24 h at 37 °C with rotary shaking at 180 rpm. Following incubation cultures were then centrifuged at 4,000 g for 10 minutes, and the supernatant was collected. For plate amplification, a soft agar overlay containing log phage S. typhimurium (OD 1-2) was prepared, and the P22 phage sample (10 MOI) was spotted on top of the soft agar. The plates were incubated for 12-24 h at 37 °C. The soft agar overlay was used to determine phage titers [31].
2.3 Bacterial Growth Kinetics
To study bacterial growth kinetics, LB media was prepared and inoculated with 50 μL of lag-phase secondary bacterial culture (OD<1) per 10 mL of the media. The cultures were then incubated at 37 °C with rotary shaking at 180 rpm. Measurements of bacterial density (OD) were taken periodically over the span of 48 h [31].
2.4 Bacterial Growth Kinetics in the Prescence of P22 Phage- pH and Stationary Phase
To study how pathophysiological conditions affect infection dynamics, phage kinetics assays were performed to observe phage kinetics at acidic pH and stationary phase. To study the effects of pH on phage infection dynamics, a phage kinetics assay was performed at varied media pH. The pH of the media was reduced from 6.8 utilizing Hydrochloric acid to either 6.0 or 5.5. Bacteria was inoculated into the media at pH 6.8, 6.0, and 5.5. Phage treated samples were inoculated with P22 phage at a MOI of 10 at the beginning of the experiment (lag phase).
To examine phage kinetics in the stationary phase, BV4012 (Salmonella typhimurium) was grown in LB media until the OD was 10. The stationary phase cultures were then either treated with the phage P22 at 10 MOI or left untreated. OD values were obtained periodically over the span of 48 h.
2.5 Antibiotic Minimum Inhibitory Concentration (MIC) Determination
The MICs of ampicillin and kanamycin were determined to establish the antibacterial activity of the respective antibiotics against Salmonella typhimurium strains (BV4012). Antibiotic stock solutions of 50 ug/mL in sterile ddH2O were made for these sets of experiments. Stocks were stored in a -20 °C refrigerator until usage. Several dilutions were prepared in serial fashion (50 μg/mL, 5 μg/mL, 0.5 μg/mL, 0.05 μg/mL). A 96 well plate with 100 μL of Salmonella typhimurium culture with an OD of 0.1 in each well was inoculated with either one of the antibiotic dilutions or left untreated. The plate was incubated for 48 h at 37 °C and growth was measured through absorbance at 600 nm. The lowest concentration of antibiotic without bacterial growth was termed as the minimum inhibitory concentration (MIC) [31].
2.6 Antibiotic Growth Kinetics
To study the effects of the antibiotic ampicillin or kanamycin on Salmonella typhimurium growth. LB media was prepared and inoculated with 50 μL of log phase secondary bacterial culture per 10 mL of the media. The cultures were then incubated at 37 °C with rotary shaking at 180 rpm. The OD of the bacteria was periodically measured against a media blank. The final concentrations of antibiotics in treated samples were either 50 μg/mL or 5 μg/mL were added at the beginning of the experiment (lag phase). To study the possible synergetic relationship between P22 phage and the antibiotics used, cultures were co-treated with an antibiotic and the P22 phage at MOIs of 10, 1, and 0.1 [31].
2.7 Phage Rechallenge Experiments
To determine the susceptibility of bacterial populations and developed resistance after exposure to the phage P22, BV4012 cultures were seeded into a 96 well plate (Fisherbrand™, FisherScientific, USA) and inoculated initially with P22, ampicillin, or kanamycin. The OD600 was recorded over a period of 48 h. Bacterial cultures that showed developed resistance following 48 h of exposure to phage or antibiotic treatment were treated again with P22. The OD600 was recorded following rechallenge for 24 h.
2.8 Phage-Antibiotic Synergy Assay
To study the potential synergy between ampicillin or kanamycin and the phage P22, the checkerboard assay was utilized. Salmonella typhimurium strain (BV4012) was seeded into a 96 well plate at an OD of 0.01 (50 μL/10 mL of media). Plate cultures were then inoculated with antibiotic and phage at respective concentrations and incubated at 37 °C while shaking at 180 rpm for 24 h. OD readings were taken at 600 nm using a plate reader (Biotek Cytation 3 Cell Imaging System). Fractional Inhibitory Concentrations (FIC) values were calculated using the formula ∑FIC = FICAB + FICP22 = (CAB/MICAB) + (CP22/MICP22) [31, 32]. In this calculation, MICAB and MICp22 are representative of the MIC values of the antibiotic used and the MIC value of P22. Values <0.5 were defined as synergistic, values between 0.5 and 4 as no interaction or additive, and values over four as antagonism.
2.9 Phage Stability Assay
To study the stability of the phage P22 at varying pH, a 108 solution of P22 phage at pH 6.8, 6.0, and 5.5 were incubated at 37 °C. Samples of the solutions were collected and titrated against S. typhimurium.
2.10 SEM Observation
BV4012 samples were treated with P22 phage, and the morphology and surface microstructure of the bacteria observed using scanning electron microscopy. Untreated BV4012 served as control. Prior to microscopy, the specimens were dried in a vacuum, and sprayed with gold using EMS Quorum (EMS 150R ES) ion-sputtering instrument and observed through Analytical Scanning Electron Microscope (SEM) (JEOL JSM-6010LA, Japan) installed with IntouchScope software.
Sample preparation for SEM was carried out as described by Li et al., 2021 [41] with few modifications. In short, the specimens for SEM were fixed with 10% formaldehyde solution at room temperature for 10 min, washed with PBS solution thrice, and dehydrated serially in 50%, 70%, and 95% absolute ethanol solutions for 10 min each. Finally, the specimens were dried in a vacuum, and sprayed with gold using EMS Quorum (EMS 150R ES) ion-sputtering instrument and observed through Analytical Scanning Electron Microscope (SEM) (JEOL JSM-6010LA, Japan) installed with IntouchScope software.
2.11 Statistical Analysis
All experiments were performed on independent biological replicates. Statistical significance was determined for control and experimental groups using paired sample t-test. Data points were excluded if contamination was identified.
3. Results
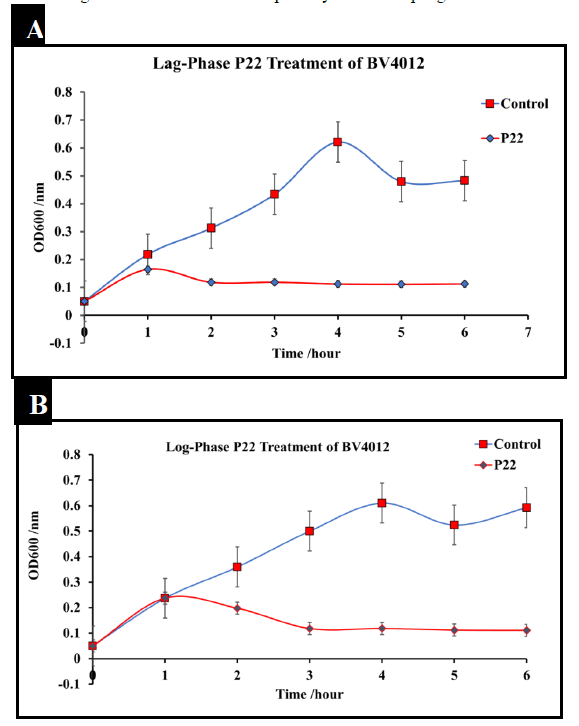
The phage P22 was amplified to 109 pfu/mL. To determine the effect of P22 phage on Salmonella typhimurium culture (BV4012), cultures were infected with P22 phage at the MOI of 10 either in the lag-phase or in the mid-log-phase. The optical density (OD) was recorded using a spectrometer (Molecular Devices SpectraMax® ABS Plus) at various time intervals. S. typhimurium cultures treated with P22 phage showed a significant reduction in OD in the mid-log phase as well as an inhibition of growth at the lag phase within the initial 8 h following infection (Fig.1). Bacteria commonly develop resistance after extended exposure to agents such as antibiotics and other various antimicrobials. Our kinetics studies followed S. typhimurium strains development of resistance against P22 phage treatment. Bacterial regrowth was observed 48 h after treatment with P22 phage suggesting the development of resistance against P22 by both strains of S. typhimurium. Bacterial strains were rechallenged with an identical P22 phage treatment at the MOI of 10 following 48 h to observed susceptibility. An observed decrease in optical density indicated that there was no significant reduction in susceptibility to the P22 phage treatment.
Figure 1: Treatment of S. typhimurium at lag and log phase with P22 phage. S. typhimurium cultures (BV4012) were treated with P22 phage at either the lag or log-phase. Three replicate OD600 were recorded hourly for six hours to examine the effect of P22 phage treatment on bacterial growth. (A) Lag-Phase P22 phage treatment of BV4012. (B) Log-Phase P22 phage treatment of BV4012.
3.2 SEM analysis of P22 Phage treatment of S. typhimurium
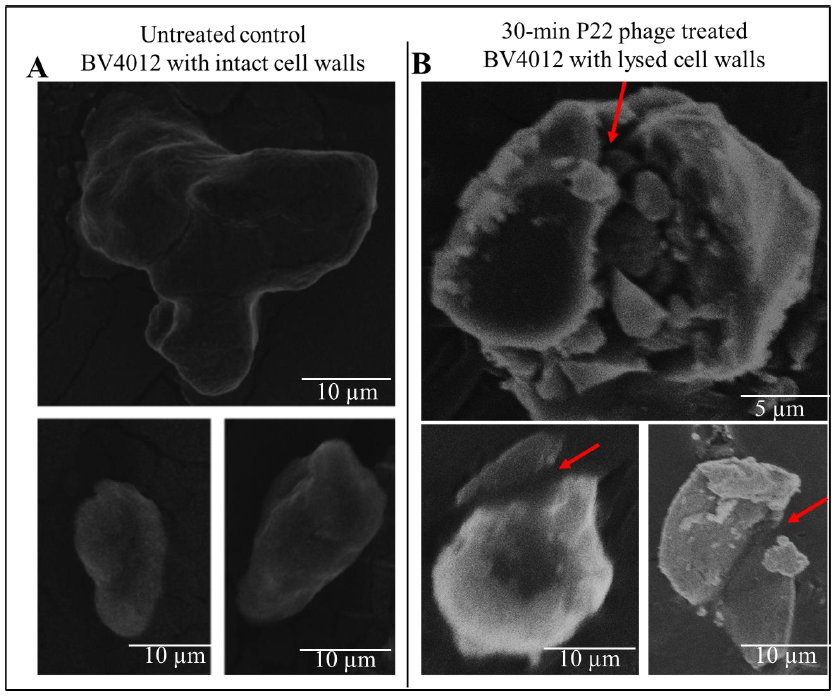
To verify the lytic effect of P22 phage on S. typhimurium, BV4012 samples were infected with P22 phage and allowed to incubate at 37 °C. Samples were then subjected to SEM analysis. As shown in the untreated control samples (Fig. 2A), the morphologies of the bacteria shown unbroken cell wall, hence the absence of lysis. The P22 phage treated samples however showed bacteria with broken cell walls (Fig. 2B) indicating the lysis of BV4012.
Figure 2: SEM images of S. typhimurium (BV4012) depicting the comparison between untreated and P22 phage treated samples. (A) Untreated BV4012 S. typhimurium visualized through Scanning Electron Microscopy (SEM), depicting unbroken cell wall of S. typhimurium. (D) P22 treated BV4012 S. typhimurium visualized through Scanning Electron Microscopy (SEM) shows lysed cell walls. Red arrows shows cell wall lysis.
3.3 P22 Phage Efficacy Under Varied pH
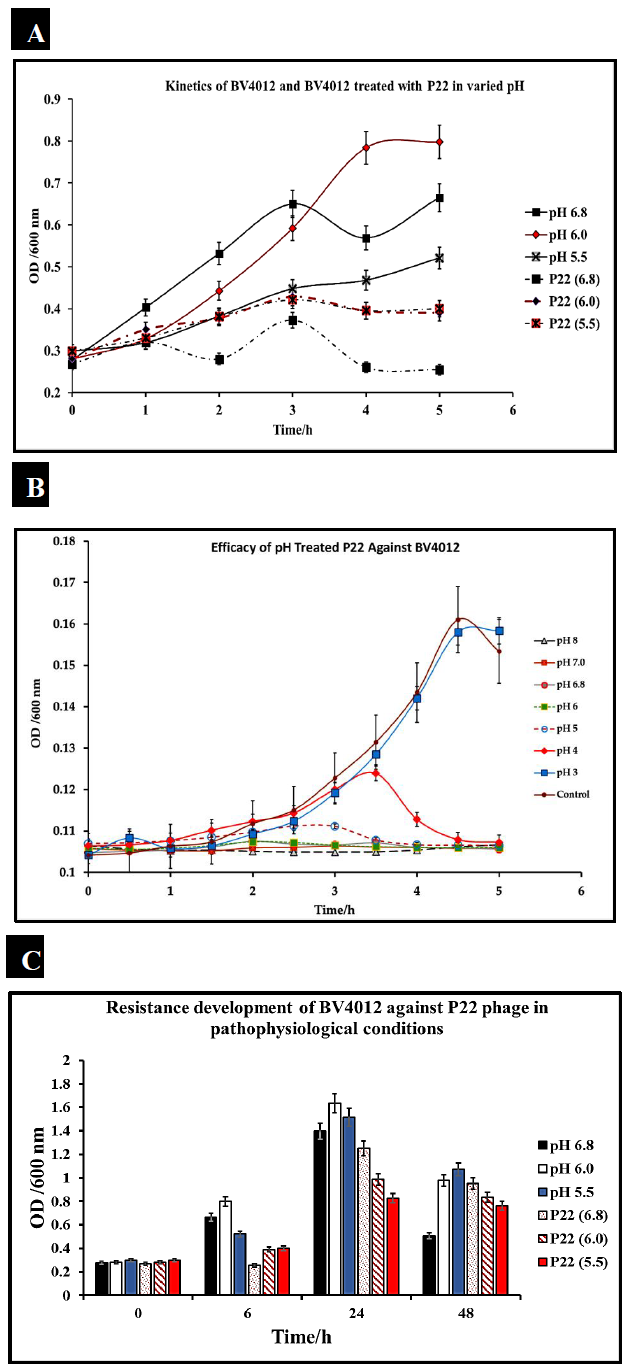
S. typhimurium infections are typically localized in the small and large intestine of the human body. This environment is characterized by acidic pH [33]. To consider P22 phage as a potential therapeutic, the phage must be able to function within these environmental constraints. We evaluated the efficiency of our phage P22 under pathophysiological conditions (pH ranging from 5-7) consistent with that present in human intestines. We cultured S. typhimurium at the pH of 5.5, 6.0, and 7 and measured optical density over 5 h to confirm bacterial growth at the pH of 5.5, 6.0, and 6.8. S. typhimurium (BV4012) cultures were then seeded into media at pH 5.5, 6.0, or 6.8 and then treated with P22 phage at 10 MOI. Optical densities recorded over 5 h determined that there was no functional loss of the phage in the acidic conditions and remained effective in reducing the growth of S. typhimurium (Fig.3A).
To further examine the effect of pH on P22 phage functionality we exposed P22 phage alone to pH conditions ranging from 3 to 8 and then examined their efficacy against BV4012 in standard conditions. This study would determine that P22 could retain functionality and lyse BV4012 after exposure to pH conditions ranging from 8 down to 4. This data suggests that P22 is stable at these conditions as well as not susceptible to pH associated with pathophysiological conditions (Fig.3B).
An important aspect of bacterial infection is the persistence of infections and the potential development of resistance associated with this persistence. We observed this development of resistance for 48 h to determine if there was any association between pH and rate of resistance development (Fig.3C). Our results suggest that 24 h after treatment, there is a slight resurgence of growth, suggesting resistance development. However, preceding results 48 h after treatment still suggest that P22 is effective in reducing S. typhimurium growth.
Figure 3: P22 phage efficacy against S. typhimurium under varied pH. S. typhimurium was cultured in varied pH conditions of 5.5, 6.0, and 6.8 and then treated with P22 phage. Three replicate OD600 were recorded hourly for 5 hours. (A) Efficacy of P22 against BV4012 in varied pH. BV4012 was cultured in varied pH condition of 5.5, 6.0, and 6.8 and then treated with P22 phage. Three replicate samples OD600 were recorded hourly for 5 hours (B). Efficacy of pH treated P22 against BV4012. P22 phage was exposed to pH varying from 3-8 and then introduced into BV4012 cultures. The OD600 was recorded hourly for 5 hours. (C). Resistance Development of BV4012 against P22 in pathophysiological conditions. BV4012 was cultured in pH of 5.5, 6.0, and 6.8 and then treated with P22 phage. Three replicate OD600 were recorded over a 48-hour period.
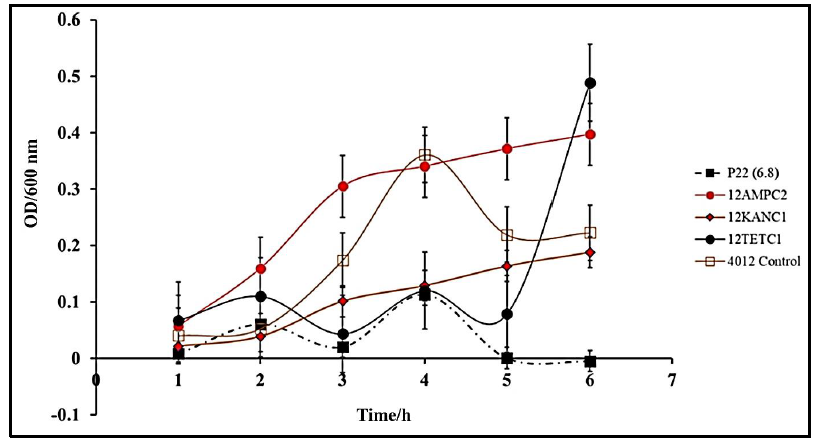
3.4 P22 Phage Effective Against S. typhimurium in Stationary Phase
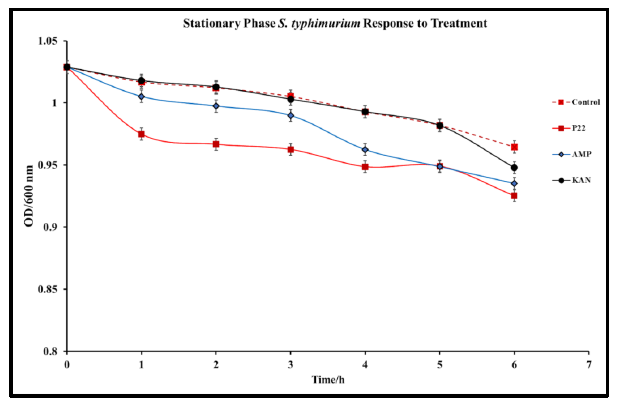
In cases of infection, following the log phase of S. typhimurium, a stationary phase represented by slowed growth rate and often a non-replicative state occurs. This state is typically conducive to the formation of biofilms within the intestinal system [34]. This state presents a major issue for antibiotic treatment methods that are only effective against bacteria in the replicative state [35]. Understanding P22 phage activity against S. typhimurium in the stationary phase is an important aspect of determining the viability of the phage against S. typhimurium infections. To determine the efficacy of P22 phage against S. typhimurium in the stationary phase, S. typhimurium cultures were grown to an OD>10. The stationary S. typhimurium cultures were then infected with P22 phage, and optical density was observed over 24 h. Significant reduction in OD was observed in phage treated samples 24 h after treatment (Fig. 4). This study confirms the antibacterial capabilities of P22 phage against S. typhimurium at the stationary phase. Following 48 h of incubation, no regrowth was detected, suggesting that S. typhimurium did not develop resistance.
3.5 Efficacy of Ampicillin and Kanamycin Against S. typhimurium
Salmonella infections are conventionally treated with a combination of common antibiotics including ampicillin or kanamycin but developed resistance to these treatments has become more prevalent [36]. To compare conventional antibiotic treatment to our novel phage treatment, comparative kinetic studies were conducted. Firstly, we determined minimal inhibitory concentrations for Ampicillin (0.5 μg/mL) and Kanamycin (0.5 μg/mL). Cultures were treated with the MIC of the respective antibiotic at the lag phase. Kinetics following treatment with either P22 phage at MOI of 1, ampicillin, or kanamycin revealed that the antibacterial function of P22 was significantly higher than that of either antibiotic account for a drastic decrease in the OD600 of S. typhimurium (Fig.5).
Figure 5: Comparative Efficacy of P22 phage and Antibiotic Treatments of S. typhimurium (BV4012) at normal physiological pH. BV4012 was cultured and treated with either P22, ampicillin, kanamycin, or tetracycline. Three replicate samples OD600 were recorded hourly for 6 hours to observe bacterial growth.
3.6 Synergistic Characteristics of Phage-Antibiotic Co-Therapy
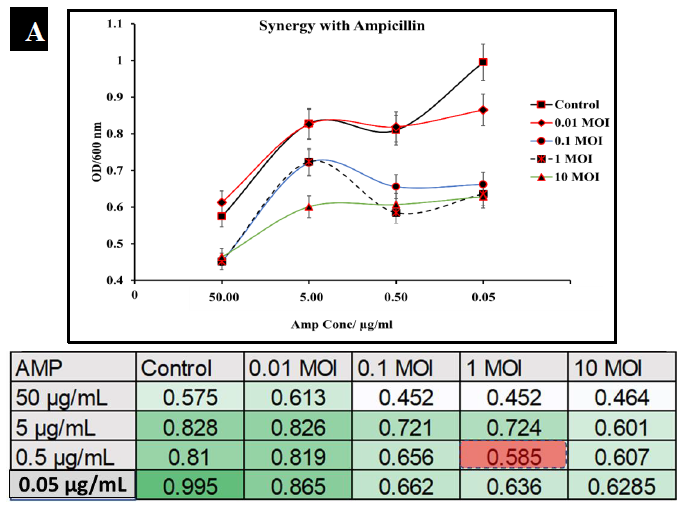
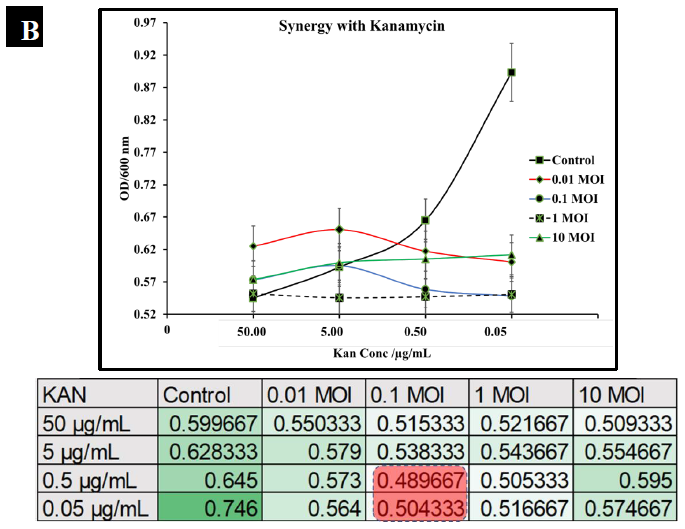
To observe the potential synergistic characteristics of phage-antibiotic co-therapy against S. typhimurium, checkerboard assays were conducted. S. typhimurium cultures (BV4012) were treated with antibiotics at concentrations of 50 μg/mL, 5 μg/mL, 0.5 μg/mL, or 0.05 μg/mL and P22 phage at MOIs of 10, 1, 0.1, or 0.01 during the lag phase. The optical density of the samples was recorded over 24 h. In comparison to ampicillin treated S. typhimurium cultures, cultures treated with ampicillin and P22 at MOIs of 0.1 and 1 resulted in greater inhibition of growth than ampicillin alone over the span of 24 h (Fig. 6A.). In comparison to kanamycin treated S. typhimurium cultures, cultures treated with kanamycin and 0.1 or 1 MOI of P22 resulted in greater inhibition of growth than kanamycin alone over the span of 24 h (Fig. 6B.). These results suggest a synergistic interaction between the ampicillin with the P22 phage at these concentrations. This is novel in the P22 phage system and similar work has been demonstrated in numerous other studies of bacterial infections such as P. aeruginosa and S. aureus [37, 38]. The correlation between increased growth reduction and MOIs of 0.1 and 1 of P22 suggest phage-antibiotic synergy. These antibiotics might affect cell lysis through targeting the cell membrane of Salmonella cells. This mechanism paired with the lytic nature of P22 provides synergistic killing activity against S. typhimurium. A major benefit of co-therapy that is demonstrated through these results is the reduced development of resistance to both antibacterial agents present when the bacteria are treated within the lag phase.
Figure 6A: Synergistic analysis of ampicillin and P22 phage co-treatment of S. typhimurium. Data presented as mean ±Standard deviation in the chart, whereas the table shows the checkerboard representation of the chart. The best synergistic effect was observed at MOI of 1 of P22 phage and 0.5 µg/mL of ampicillin co-treatment.
3.7 P22 Phage exhibits bacteriostatic behavior against Antibiotic-Resistant S. typhimurium
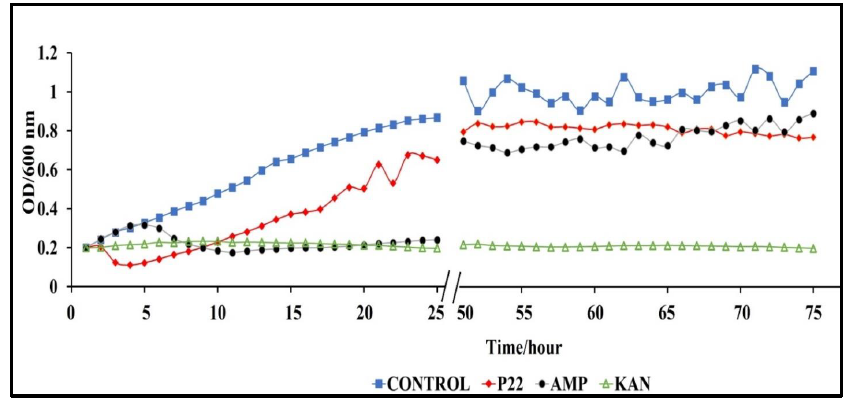
A major benefit of phages, over antibiotics, is their ability to complete lysis of antibiotic resistant bacterial strains [39-41]. In terms of clinical application, this significant role in the fight against antibiotic resistant bacterial infections. We conducted rechallenge experiments to observe the ability of our phage P22 to lyse S. typhimurium cultures with developed resistance to ampicillin or kanamycin. We inoculated BV4012 and BV4013 lag-phase cultures with either P22 phage, ampicillin or kanamycin and monitored the OD of the culture over 48 h until the OD began to steadily rise illustrating a developed resistance to the treatment. Once resistance was developed all cultures were inoculated with P22 phage at an MOI of 1 and the OD was measured over the preceding 24 h. Following 74 total h of incubation, we determined that the rechallenge of the cultures with P22 phage resulted in a significant reduction in the rate of S. typhimurium growth (Fig. 7). These results allowed us to infer that P22 phage can lyse the S. typhimurium cultures which developed resistance to ampicillin or kanamycin treatments.
Figure 7: Rechallenge of S. typhimurium (BV4012) with P22 phage over 74 h. BV4012 was first treated with P22 phage, ampicillin, or kanamycin. Upon development of resistance, signified by increase OD600 after 25 hours, all samples were rechallenged with P22 phage and the OD600 was recorded hourly for 24 hours following treatment.
4. Discussion
The development of novel treatment methods against pathogenic bacteria is essential to the preservation of public health. Bacteriophages have been shown to be promising as potential antibacterial agents against numerous pathogenic bacteria, including Salmonella spp., Pseudomonas spp., and Staphylococcus spp. [37, 38, 42]. While many phages have shown efficacy as potential therapeutics, extensive study of the effects of pathophysiological conditions on the efficacy of phages is limited. This study serves to determine the ability of P22 phage to maintain its functionality under several conditions associated with S. typhimurium infections in the small and large intestines. Understanding the limitations and boundaries of phages, such as P22 phage is essential to their application as therapeutic and preventative treatments.
For phage therapy to be successful, phages must retain their lytic function against their host in pathophysiological conditions and infect non-replicative stationary phase bacteria. This study focuses on S. typhimurium, a prevalent foodborne pathogen with an increased rate of antibiotic resistance over recent history [1-3]. There is an inherent need for the development of new therapy against S. typhimurium infections, since the efficacy of antibiotics is threatened and the potential for outbreak of antibiotic-resistant strains increases [1, 2, 10]. The phage P22, whose natural host is S. typhimurium, has been widely studied and utilized in some applications as a prophylactic [4, 42-46]. P22 exhibits a lytic lifestyle that is ideal for use as a potential therapeutic against S. typhimurium as well as been characterized as a having notable stability against several environmental conditions [4, 45]. The phage P22 is reliant on its incredibly stable tailspike protein (TSP) for the initiation of infection and lysis of S. typhimurium [44].
In this study, we first wanted to examine the kinetics associated with P22 phage infection of S. typhimurium at various growth stages, including lag-phase and log-phase. Our results illustrate the timeline of P22 phage infection and the associated reduction of bacterial growth and persistence in standard conditions. Treatment of S. typhimurium cultures at all three stages of growth resulted in a significant decrease in bacterial growth (Fig. 1A, 1B, 6A, 6B). These results serve to first prove the lytic ability of P22 phage against our target bacteria and secondly set a baseline for comparison of P22 phage performance in pathophysiological conditions. The lysis of S. typhimurium was demonstrated via scanning electron microscopy (Fig. 2B).
An important aspect in the study of any new novel agent is its comparison to standard therapeutic methods that are currently in place. In this study, we examined the comparative kinetics of P22 phage and two standard antibiotics: ampicillin and kanamycin, against S. typhimurium. An important distinction between bacteriophages and antibiotics is the versatility and abundance of phages in nature [47, 48]. Antibiotics, being man-made compounds require tedious design and development while also having low specificity and limited classes. There are estimated to be approximately 1031 bacterial and archaeal viruses present in the biosphere with over 500 of these phages’ genomes having been fully sequenced [49].
The ability to withstand acidic pH conditions is a major requirement of any antibacterial agent. The proposed use of P22 as a therapeutic against S. typhimurium requires it to withstand the pH conditions of the gastrointestinal tract where this bacterium typically colonizes [27, 28]. P22 phage was able to retain lytic function and reduce bacterial growth at pHs of 5.5, 6.0, and 6.8 (Fig. 3A). These pHs are consistent with the physiological conditions associated with the small and large intestines of humans. More importantly, these results confirm the ability of P22 phage to withstand physiological conditions found within these organs and retain its antibacterial function. Our study went further to also determine the pH limitations of P22 phage. It was determined that P22 phage even after extended exposure to media at pH 4-8 was able to retain antibacterial function against S. typhimurium (Fig. 3B). The pH threshold of P22 phage was determined in this study as well and suggested a loss of functionality at a pH of 3 (Fig. 3B). Resistance development was also examined in this study over a 48-h period revealing that after 48 h of P22 phage treatment, there was a slight resurgence of growth across all pH levels (Fig. 3C). Resistance development was significantly higher at the pH of 5.5 and 6.0 in both treated and untreated samples suggesting a correlation between resistance development and acidic pH. The development of resistance by bacteria when exposed to acidic pH has been recreated in studies of several other bacteria [50, 51]. Overall, our study concludes that P22 phage is effective in drastically reducing the growth of S. typhimurium within the first 24 h of administration even in acidic conditions which are similar to the physiological conditions of the gastrointestinal tract.
Another pathophysiological condition that was also examined in this study is the efficacy of P22 against S. typhimurium in the stationary phase. Infections of the intestinal system by S. typhimurium in the non-replicative stage is a major concern in the clinical setting [52]. Stationary phase bacterial infections typically are associated with the development of biofilms and antibiotic resistance [34]. Considering this, it is essential to develop methods for these difficult to treat infections and our results suggest that P22 phage shows potential as a co-therapy or alternative treatment (Fig 4). Kinetic study of P22 phage treated stationary-phase S. typhimurium shows that P22 phage inhibited bacterial growth at a rate higher than that of ampicillin or kanamycin over a 6-h period. Extended kinetics over a 48-h period confirmed that P22 treatment also did not result in regrowth of S. typhimurium.
Results show that P22 phage drastically stunted bacterial growth and nearly eliminated S. typhimurium completely after 6 h (Fig 5). In comparison to ampicillin and kanamycin, P22 phage performed significantly better in reduction of S. typhimurium. In this study we also examined the potential synergy between P22 phage and the antibiotics ampicillin and kanamycin. The synergistic characteristics of phage-antibiotic co-therapy has been suggested in numerous studies and is a promising incorporation of phage therapy to combat resistant infections [22-24, 53]. Our results suggest that P22 at MOIs of 1 and 0.1 paired with ampicillin or kanamycin reduces growth of S. typhimurium at a rate greater than either antibiotic individually (Fig.6).
While phages such as P22 show promise as potential therapeutics, it is important to consider the ability of bacterial species to develop resistance against phage treatments [54, 55]. Our studies did determine that there were cases of partial resistance against P22 by S. typhimurium. This resistance was observed in the form of increased bacterial growth after extended exposure of the bacteria to P22. The mechanism of resistance was not clear and requires further study. Phage resistance is mediated by bacteria in several ways including but not limited to receptor mutation, blockage of phage DNA injection, hypermutable loci, cleavage of phage DNA, abortive infection, R-M system, DISARM system, or CRISPR-Cas system [56-59].
A limitation of our study is the use of fast-growing laboratory use S. typhimurium (BV4012). While this strain is ideal for our study, it may not translate to naturally occurring S. typhimurium infections with variations in growth rate. It was demonstrated that P22 is effective against S. typhimurium, however after extended exposure, the development of resistance raises concerns about the use of P22 phage and the threat of infection resurgence. Further study of the resistance mechanisms possessed by S. typhimurium against P22 phage is essential to gaining a full understanding of this interaction. While we showed that P22 is effective in vitro against S. typhimurium cells, we did not explore phage delivery methods in vivo. The delivery of P22 phage to the location of infection is essential to the success of therapy, and an area that requires further study. Another major challenge of phage therapy is the human immunological response. Studies show that there is an immunological response to phage treatments and that this response can be in ways synergistic in the elimination of acute and chronic bacterial infections [60-63].
5. Conclusion
In summary, we demonstrated the efficacy of the phage P22 under pathophysiological conditions associated with S. typhimurium. These conditions included lag-phase, log-phase, and stationary phase of S. typhimurium as well under various pH ranges (4-8). Additionally, we showed that P22 phage is effective killing S. typhimurium, and the phage seemed to perform comparably to standard antibiotic’s such as ampicillin and kanamycin in killing S. typhimurium. We examined the synergistic potential of P22 phage at MOIs of 10, 1, 0.1 and 0.01 with ampicillin or kanamycin at concentrations of 50 μg/mL, 5 μg/mL, 0.5 μg/mL, 0.05 μg/mL against S. typhimurium cultures. The best synergistic effect was observed at phage MOI of 0.1 and kanamycin concentrations of 0.5 and 0.05 µg/mL. Whereas the best synergistic effect was observed at MOI of 1 of P22 phage and 0.5 µg/mL of ampicillin co-treatment.
References
- (2020). Antimicrobial Resistance.
- Antibiotic Resistance threats in the United States (2019).
- DM L. Has the era of untreatable infections arrived? Journal of Antimicrobial Chemotherapy 64 (2009): 29-36.
- Williams J, Venkatesan K, Ayariga JA, Jackson D, Wu H & Villafane R. A genetic analysis of an important hydrophobic interaction at the P22 tailspike protein N-terminal domain.Arch Virol,163 (2018): 1623-1633.
- Slayton RB, Toth D, Lee BY, Tanner W, et al. Vital Signs: Estimated Effects of a Coordinated Approach for Action to Reduce Antibiotic-Resistant Infections in Health Care Facilities - United States.MMWR. Morbidity and mortality weekly report64 (2015): 826-831.
- Ardal CBM, Laxminarayan R, McAdams D, Outterson K et al. Antibiotic Development- Economic, regulatory, and societal challenges. National Review Microbiology (2019).
- Sharland MGS, HB, Moja L, Pulcini C, Zeng M et al. Eml expert committee and antibiotic Working Group. Essential Medicines list becomes a global antibiotic stewardship tool. Lancet Infectious Disease 19 (2019): 1278-1280.
- Bryan FL. Foodborne Diseases in the United States Associated with Meat and Poultry.J Food Prot1 February 43 (1980): 140-150.
- Park SY, Woodward CL, Kubena LF, Nisbet DJ, Birkhold SG & Ricke SC.Environmental Dissemination of FoodborneSalmonellain Preharvest Poultry Production: Reservoirs, Critical Factors, and Research Strategies,Critical Reviews in Environmental Science and Technology38 (2008):73-111.
- Heinitz ML, Ruble RD, Wagner DE, Tatini SR; Incidence ofSalmonellain Fish and Seafood.J Food Prot 63 (2000): 579-592.
- Ayariga JA, Gildea L, Wu H. and Villafane R. The ℇ34 Phage Tailspike Protein: An in vitro characterization, Structure Prediction, Potential Interaction with S. newington LPS and Cytotoxicity Assessment to Animal Cell Line (2021).
- Anderson ES and Lewis MJ Drug resistance and its transfer in Salmonella typhimurium.Nature206 (1965): 579-83.
- Medalla F, Gu W, Mahon BE, Judd M, Folster J, Griffin PM & Hoekstra RM. Estimated Incidence of Antimicrobial Drug-Resistant Nontyphoidal Salmonella Infections, United States, 2004-2012.Emerg Infect Dis23 (2016): 29-37.
- White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, et al. The Isolation of Antibiotic-Resistant Salmonella from Retail Ground Meats. N Engl J Med 345 (2001): 1147-1154.
- Lin DM, Koskella B & Lin HC. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8 (2017): 162-173.
- D'Accolti M, Soffritti I, Mazzacane S & Caselli E. Bacteriophages as a Potential 360-Degree Pathogen Control Strategy. Microorganisms 9 (2021).
- Lukacik P, Barnard TJ & Buchanan SK. Using a bacteriocin structure to engineer a phage lysin that targets Yersinia pestis. Biochem Soc Trans 40 (2012): 1503-1506.
- Ghosh C, Sarkar P, Issa R & Haldar J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol 27 (2019): 323-338.
- Nicholas H. Mann,The potential of phages to prevent MRSA infections, Research in Microbiology, 159 (2008): 400-405.
- Hobbs Z, Stephen T. Abedon, Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’,FEMS Microbiology Letters 363 (2016): 047.
- van de Putte P, Cramer S & Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage Mu.Nature286 (1980):218-222.
- Comeau AM, Tetart F, Trojet SN, Prere MF & Krisch HM. Phage-Antibiotic Synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth 2 (2007): e799.
- Kamal F & Dennis JJ. Burkholderia cepacia complex Phage-Antibiotic Synergy (PAS): antibiotics stimulate lytic phage activity.Appl Environ Microbiol 81 (2015): 1132-1138.
- Segall AM, Roach DR & Strathdee SA. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy.Curr Opin Microbiol51 (2019): 46-50.
- Marietto Gonçalves GA, Lima ETL, Donato TC, Rocha TS, Cisneros Álvarez LE, Sequeira JL & Andreatti Filho RL. Eradication of Salmonella Typhimurium in broiler chicks by combined use of P22 bacteriophage and probiotic.Microbiology Research2 (2011): e2.
- Ahm J, Kim S, Jung LS, Biswas D. In Vitro Assessment of the Susceptibility of Planktonic and Attached Cells of Foodborne Pathogens to Bacteriophage P22-MediatedSalmonellaLysates.J Food Prot1 December 76 (2013): 2057-2062.
- McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils.J Cell Biol 123 (1993): 895-907.
- Mayer EA, Naliboff BD & Chang L. Evolving pathophysiological model of functional gastrointestinal disorders: implications for treatment.Eur J Surg Suppl 587 (2002): 3-9.
- Cho HB, Ahn JH, Yang HG, Lee J, Park WJ & Kim YW. Effects of pH and NaCl on hydrolysis and transpeptidation activities of a salt-tolerant gamma-glutamyltranspeptidase from Bacillus amyloliquefaciens S0904.Food Sci Biotechnol30 (2021): 853-860.
- Porter EM, van Dam E, Valore EV & Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5.Infect Immun65 (1997): 2396-2401.
- Kalapala YC, Sharma PR & Agarwal R. Antimycobacterial Potential of Mycobacteriophage Under Disease-Mimicking Conditions.Frontiers in microbiology11 (2020): 583661.
- Odds FC.Synergy, antagonism, and what the chequerboard puts between them.J. Antimicrob. Chemother52 (2003): 1.
- Garai P, Gnanadhas DP & Chakravortty D. Salmonella enterica serovars Typhimurium and Typhi as model organisms: revealing paradigm of host-pathogen interactions.Virulence3 (2012): 377-388.
- Harrell Jaikin E, Hahn Mark M, D’Souza Shaina J, Vasicek Erin M, Sandala Jenna L, et al. Salmonella Biofilm Formation, Chronic Infection, and Immunity Within the Intestine and Hepatobiliary Tract. Frontiers in Cellular and Infection Microbiology 10 (2021): 910.
- Nathan C. Fresh approaches to anti-infective therapies.Sci Transl Med4 (2012): 140sr2.
- Voogd CE, van Leeuwen WJ, Guinée PAM,et al.Incidence of resistance to ampicillin, chloramphenicol, kanamycin, and tetracycline among Salmonellaspecies isolated in The Netherlands in 1972, 1973 and 1974.Antonie van Leeuwenhoek43 (1977):269-281.
- Oechslin F, Piccardi P, Mancini S, Gabard J, Moreillon P, Entenza JM, et al. Synergistic Interaction Between Phage Therapy and Antibiotics Clears Pseudomonas Aeruginosa Infection in Endocarditis and Reduces Virulence. J. Infect. Dis 215 (2017): 703-712.
- Chhibber S, Kaur T and Kaur S. Co-Therapy Using Lytic Bacteriophage and Linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS One 8 (2013): e56022.
- Burrowes B, Harper DR, Anderson J, McConville M, Enright MC.Bacteriophage therapy: potential uses in the control of antibiotic-resistant pathogens,Expert Review of Anti-infective Therapy9 (2011):775-785.
- Lehman SM. Mearns G, Rankin D, Cole RA, Smrekar F, Branston SD, et al. Design and Preclinical Development of a Phage Product for the Treatment of Antibiotic-ResistantStaphylococcus aureusInfections.Viruses11 (2019): 88.
- Graham F Hatfull,Rebekah M Dedrick,Robert T Schooley. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annual Review of Medicine73 (2022): 1.
- WasehS, Hanifi-MoghaddamP, ColemanR, MasottiM, RyanS, et al.Orally Administered P22 Phage Tailspike Protein ReducesSalmonellaColonization in Chickens: Prospects of a Novel Therapy against Bacterial Infections. PLOS ONE 5 (2010): e13904.
- Villafane R & King J. Nature and distribution of sites of temperature-sensitive folding mutations in the gene for the P22 tailspike polypeptide chain.J Mol Biol204 (1988): 607-619.
- Venza Colon CJ, Vasquez Leon AY & Villafane RJ. Initial interaction of the P22 phage with the Salmonella typhimurium surface.P R Health Sci J23 (2004): 95-101.
- Salgado CJ, Zayas M & Villafane R. Homology between two different Salmonella phages: Salmonella enterica serovar Typhimurium phage P22 and Salmonella enterica serovar Anatum var. 15 + phageepsilon34.Virus Genes29 (2004): 87-98.
- Fane B, Villafane R, Mitraki A & King J. Identification of global suppressors for temperature-sensitive folding mutations of the P22 tailspike protein.J Biol Chem266 (1991): 11640-11648.
- Edwards RA, Rohwer F.Viral metagenomics.Nat. Rev. Microbiol3 (2005):504-510.
- Martha RJ Clokie, Andrew D Millard, Andrey V Letarov & Shaun Heaphy. Phages in nature, Bacteriophage 1 (2011): 31-45.
- Hatfull GF. Bacteriophage genomics.Curr Opin Microbiol11 (2008): 447-453.
- Xiong M, Bao Y, Xu X, Wang H, Han Z, Wang Z, et al. Helix-coil conformation transitionable polypeptide. Proceedings of the National Academy of Sciences 114 (2017): 12675-12680.
- de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, et al. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant.J Biol Chem270 (1995): 23937-23940.
- Monack DM, Mueller A and Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nature Reviews Microbiology 2 (2004): pp.747-765.
- Ryan EM, Alkawareek MY, Donnelly RF, Gilmore BF. Synergistic phage-antibiotic combinations for the control ofEscherichia colibiofilmsin vitro,FEMS Immunology & Medical Microbiology, Volume 65 (2012): 395-398.
- Hyman P & Abedon ST. Bacteriophage host range and bacterial resistance.Adv Appl Microbiol,70 (2010): 217-248.
- Labrie SJ, Samson JE, Moineau S.Bacteriophage resistance mechanisms.Nat. Rev. Microbiol8 (2010):317-327.
- Azam AH, Tanji Y.Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy.Appl. Microbiol. Biotechnol103 (2019):2121-2131.
- Turkington CJR, Morozov A, Clokie MRJ, Bayliss CD.Phage-Resistant Phase-Variant Sub-populations Mediate Herd Immunity Against Bacteriophage Invasion of Bacterial Meta-Populations.Front. Microbiol10 (2019): 1473.
- McCallin S, Oechslin F. Bacterial Resistance to Phage and Its Impact on Clinical Therapy. In: Górski A, Miedzybrodzki R, Borysowski J. (eds) Phage Therapy: A Practical Approach. Springer, Cham. (2019).
- Wright RCT, Friman V-P, Smith MCM, Brockhurst MA.Resistance evolution against phage combinations depends on the timing and order of exposuremBio10 (2019):e01652-19.
- Ooi ML, Drilling AJ, Morales S, Fong S, Moraitis S, MacIas-Valle L, et al.Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to staphylococcus aureus.JAMA Otolaryngol. Head Neck Surg145 (2019):723-729.
- Leung CY, Weitz JS.Modeling the synergistic elimination of bacteria by phage and the innate immune system.J. Theor. Biol429 (2017):241-252.
- Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, et al.Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen.Cell Host Microbe22 (2017): 38.e-47.
- Waters EM, Neill DR, Kaman B, Sahota JS, Clokie MRJ, Winstanley C, et al.Phage therapy is highly effective against chronic lung infections withPseudomonas aeruginosa.Thorax72 (2017):666-667.
- Ayariga JA, Gildea L. and Villafane R ℇ34 Phage Tailspike Protein is Resistant to Trypsin and Inhibits and Salmonella Biofilm Formation (2021).
- Ayariga JA, Venkatesan K, Ward R, Wu H, Jackson D and Villafane R. Initiation of P22 infection at the phage centennial. Frontiers in Science, Technology, Engineering and Mathematics 2 (2021): 64-81.
- DP Pires, LDR Melo, D Vilas Boas, S Sillankorva, J Azeredo. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Current Opinion in Microbiology 39 (2017): Pages 48-56.
- Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, Fairman JW,et al. Structural engineering of a phage lysin that targets gram-negative pathogens. Proc Natl Acad Sci U S A 109 (2012): 9857-9862.