Other Driver Genes as Resistance Mechanisms of ALK Inhibitors in ALK-Rearranged Lung Adenocarcinoma
Article Information
Xiao-Cheng Lin1,#, Qian-Yu Wang1,#, Xu-Hui Guan2, Yu-Fa Li3, Hai-Yan Tu2, Hua-Jun Chen2, Jin-Ji Yang2,*
1Guangdong Cardiovascular Institute, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences. Guangzhou, China
2Guangdong Lung Cancer Institute, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Southern Medical University, China
3Department of Pathology, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Southern Medical University, China
#Contributed equally to this work.
*Corresponding Author: Jin-Ji Yang, Guangdong Lung Cancer Institute, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Southern Medical University, 510080, China.
Received: 31 August 2023; Accepted: 21 September 2023; Published: 12 January 2024
Citation: Xiao-Cheng Lin, Qian-Yu Wang, Xu-Hui Guan, Yu-Fa Li, Hai-Yan Tu, Hua-Jun Chen, Jin-Ji Yang. Other Driver Genes as Resistance Mechanisms of ALK Inhibitors in ALK-Rearranged Lung Adenocarcinoma. Journal of Cancer Science and Clinical Therapeutics 8 (2024): 70-79.
View / Download Pdf Share at FacebookAbstract
Introduction: Other dependent driver genes, including MET, EGFR and BRAF, can cause resistance to the first- and second-generation ALK inhibitors. In vitro studies have shown that individualized intervention against these resistance pathways may be an optional treatment. We aimed to explore the clinical characteristics of patients with other dependent driver genes as resistance mechanisms to ALK inhibitors.
Methods: We performed next-generation sequencing on tissue and liquid samples obtained at every treatment milestone from three ALK-rearranged patients with advanced lung adenocarcinoma receiving targeted therapy. FISH and IHC were underwent in some tissue samples to verify the existence of ALK rearrangement and MET amplification
Results: Three patients revealed other driver gene alteration after resistance to the first- and second-generation ALK inhibitors (one BRAF V600E mutation, one EGFR L858R mutation and one MET amplification). Combination targeted therapy could overcome the resistance of acquired other driver gene alteration. When BRAF V600E, EGFR mutations and MET amplification occurred as resistance mechanisms of ALK inhibitors, they performed gene heterogeneity. Pleural effusion only revealed BRAF V600E mutation in Patient 1 and EGFR L858R mutation only showed up in cerebrospinal fluid and peripheral blood by NGS in Patient 2. MET amplification individually occurred in adrenal gland in Patient 3.
Conclusions: BRAF V600E, EGFR mutations and MET amplification could be served as resistance mechanisms of targeted therapy in ALK-rearranged lung adenocarcinoma. Dabrafenib and trametinib might overcome such resistance in acquired BRAF V600E-mutant patients. Other driver genes as resistance mechanisms of ALK inhibitors may have heterogeneity.
Keywords
<p>Lung Cancer, ALK Rearrangement, ALK TKI, ALK-TKI Resistance.</p>
lung Cancer articles; ALK Rearrangement articles; ALK TKI articles; ALK-TKI Resistance articles
Lung Cancer articles Lung Cancer Research articles Lung Cancer review articles Lung Cancer PubMed articles Lung Cancer PubMed Central articles Lung Cancer 2023 articles Lung Cancer 2024 articles Lung Cancer Scopus articles Lung Cancer impact factor journals Lung Cancer Scopus journals Lung Cancer PubMed journals Lung Cancer medical journals Lung Cancer free journals Lung Cancer best journals Lung Cancer top journals Lung Cancer free medical journals Lung Cancer famous journals Lung Cancer Google Scholar indexed journals ALK Rearrangement articles ALK Rearrangement Research articles ALK Rearrangement review articles ALK Rearrangement PubMed articles ALK Rearrangement PubMed Central articles ALK Rearrangement 2023 articles ALK Rearrangement 2024 articles ALK Rearrangement Scopus articles ALK Rearrangement impact factor journals ALK Rearrangement Scopus journals ALK Rearrangement PubMed journals ALK Rearrangement medical journals ALK Rearrangement free journals ALK Rearrangement best journals ALK Rearrangement top journals ALK Rearrangement free medical journals ALK Rearrangement famous journals ALK Rearrangement Google Scholar indexed journals ALK TKI articles ALK TKI Research articles ALK TKI review articles ALK TKI PubMed articles ALK TKI PubMed Central articles ALK TKI 2023 articles ALK TKI 2024 articles ALK TKI Scopus articles ALK TKI impact factor journals ALK TKI Scopus journals ALK TKI PubMed journals ALK TKI medical journals ALK TKI free journals ALK TKI best journals ALK TKI top journals ALK TKI free medical journals ALK TKI famous journals ALK TKI Google Scholar indexed journals ALK-TKI Resistance articles ALK-TKI Resistance Research articles ALK-TKI Resistance review articles ALK-TKI Resistance PubMed articles ALK-TKI Resistance PubMed Central articles ALK-TKI Resistance 2023 articles ALK-TKI Resistance 2024 articles ALK-TKI Resistance Scopus articles ALK-TKI Resistance impact factor journals ALK-TKI Resistance Scopus journals ALK-TKI Resistance PubMed journals ALK-TKI Resistance medical journals ALK-TKI Resistance free journals ALK-TKI Resistance best journals ALK-TKI Resistance top journals ALK-TKI Resistance free medical journals ALK-TKI Resistance famous journals ALK-TKI Resistance Google Scholar indexed journals MET articles MET Research articles MET review articles MET PubMed articles MET PubMed Central articles MET 2023 articles MET 2024 articles MET Scopus articles MET impact factor journals MET Scopus journals MET PubMed journals MET medical journals MET free journals MET best journals MET top journals MET free medical journals MET famous journals MET Google Scholar indexed journals EGFR articles EGFR Research articles EGFR review articles EGFR PubMed articles EGFR PubMed Central articles EGFR 2023 articles EGFR 2024 articles EGFR Scopus articles EGFR impact factor journals EGFR Scopus journals EGFR PubMed journals EGFR medical journals EGFR free journals EGFR best journals EGFR top journals EGFR free medical journals EGFR famous journals EGFR Google Scholar indexed journals BRAF articles BRAF Research articles BRAF review articles BRAF PubMed articles BRAF PubMed Central articles BRAF 2023 articles BRAF 2024 articles BRAF Scopus articles BRAF impact factor journals BRAF Scopus journals BRAF PubMed journals BRAF medical journals BRAF free journals BRAF best journals BRAF top journals BRAF free medical journals BRAF famous journals BRAF Google Scholar indexed journals adenocarcinoma articles adenocarcinoma Research articles adenocarcinoma review articles adenocarcinoma PubMed articles adenocarcinoma PubMed Central articles adenocarcinoma 2023 articles adenocarcinoma 2024 articles adenocarcinoma Scopus articles adenocarcinoma impact factor journals adenocarcinoma Scopus journals adenocarcinoma PubMed journals adenocarcinoma medical journals adenocarcinoma free journals adenocarcinoma best journals adenocarcinoma top journals adenocarcinoma free medical journals adenocarcinoma famous journals adenocarcinoma Google Scholar indexed journals targeted therapy articles targeted therapy Research articles targeted therapy review articles targeted therapy PubMed articles targeted therapy PubMed Central articles targeted therapy 2023 articles targeted therapy 2024 articles targeted therapy Scopus articles targeted therapy impact factor journals targeted therapy Scopus journals targeted therapy PubMed journals targeted therapy medical journals targeted therapy free journals targeted therapy best journals targeted therapy top journals targeted therapy free medical journals targeted therapy famous journals targeted therapy Google Scholar indexed journals heterogeneity articles heterogeneity Research articles heterogeneity review articles heterogeneity PubMed articles heterogeneity PubMed Central articles heterogeneity 2023 articles heterogeneity 2024 articles heterogeneity Scopus articles heterogeneity impact factor journals heterogeneity Scopus journals heterogeneity PubMed journals heterogeneity medical journals heterogeneity free journals heterogeneity best journals heterogeneity top journals heterogeneity free medical journals heterogeneity famous journals heterogeneity Google Scholar indexed journals
Article Details
1. Introduction
Lung cancer is one of the most commonly diagnosed cancer and the leading cause of cancer-related deaths [1]. Rearrangement of the anaplastic lymphoma kinase (ALK) occur in 2-7% of lung adenocarcinoma and response to ALK inhibitors [2]. The first- and second-generation ALK inhibitor crizotinib and alectinib are recommended as fist-line treatment for ALK-positive advanced non-small-cell lung cancer (NSCLC), as it significantly prolonged the median progression-free survival (PFS) to 10.9 and 34.8 months, respectively [3-5]. And lorlatinib, considered as a third-generation TKI targeting ALK rearrangement, demonstrated a better efficacy. With a median follow-up time of 36.7 months, the 3-year PFS rate was 63.5% for and the median PFS of lorlatinib still has not been reached [6].
However, the patients who derive benefit initially will eventually develop resistance. Secondary mutations and ALK gene amplification are known mechanisms of ALK inhibitors resistance [7-9]. Otherwise, a few reports found that adopting another different driver gene, including MET, EGFR and BRAF, can also cause ALK-TKIs resistance [10-14]. In vitro studies have shown that individualized intervention against these resistance pathways may be optional treatment [13-15]. The treatment strategy and the clinical characteristics for patients after resistance to ALK-TKI was unclear, especially the population with other driver gene alteration as the resistance mechanisms of ALK-TKI.
Therefore, we retrospectively analyzed the clinical characteristics of ALK rearrangement patients with another driver gene alteration after resistance to ALK-TKI. We reported the clinical evidence of efficacy generated by a combination of alectinib, dabrafenib and trametinib targeting ALK and BRAF V600E. And we also reported the first clinical case with acquired EGFR L858R mutations after the treatment of alectinib and another special case with MET amplification.
2. Methods and Materials
2.1 Patients and data collection
We retrospectively reviewed patients with lung cancer who were diagnosed with ALK rearrangement of histological specimens at Guangdong Lung Cancer Institute from January 2018 to December 2020 (Figure 1). Three patients were treated and evaluated at our institute; meanwhile informed consent was used to collect tumor tissue, peripheral blood and cerebrospinal fluid. This study was approved by the institutional review board at the Guangdong Provincial People's Hospital. The follow-up time was from January 1st, 2018 to December 31th, 2022.
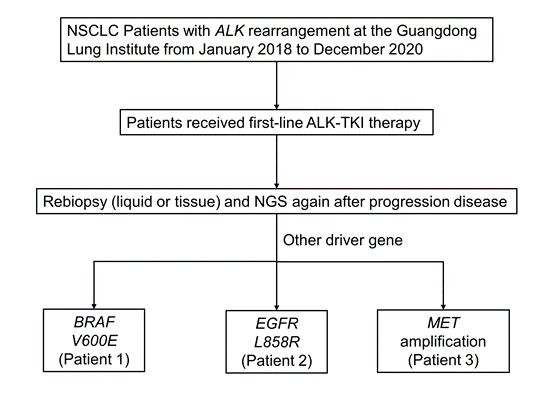
Figure 1: Flow chart of the study.
Abb: NSCLC, non-small cell lung cancer; NGS, next-generation sequencing.
2.2 Immunohistochemistry (IHC), Fluorescence in situ hybridization (FISH) and next-generation sequencing (NGS)
ALK rearrangement was performed by IHC using D5F3 (Ventana, Tucson, AZ) and by FISH using Vysis ALK Break-Apart FISH Probe (Abbott Molecular, Abbott Park, Illinois, USA) according the manufacturer’s instructions, respectively. NGS was performed by a Clinical Laboratory Improvement Amendments (CLIA)-certified testing center (Burning Rock Biotech, Guangzhou, China).
2.3 Response evaluation of treatment
Assessments of partial response (PR), stable disease (SD) or progression disease (PD) were according to the efficacy evaluation criteria of solid tumor (RECIST) version 1.1. PFS was defined from the beginning of targeted therapy to the date of tumor progression or the last follow-up.
3. Results
3.1 Characteristics and pathological features of patients
We selected patients with ALK rearrangement treated with ALK-TKI in Guangdong Provincial People’s Hospital January 2018 to December 2020 and found 3 patients who appear another driver gene alteration after resistance to ALK-TKI (Figure 1). The clinical information is shown in Table 1. All patients (3/3, 100%) are female and never smoked, with a great performance status and low tumor mutation burden. Distant multiple metastasis was found when they were diagnosed or relapsed. The pathology of them was all adenocarcinoma.
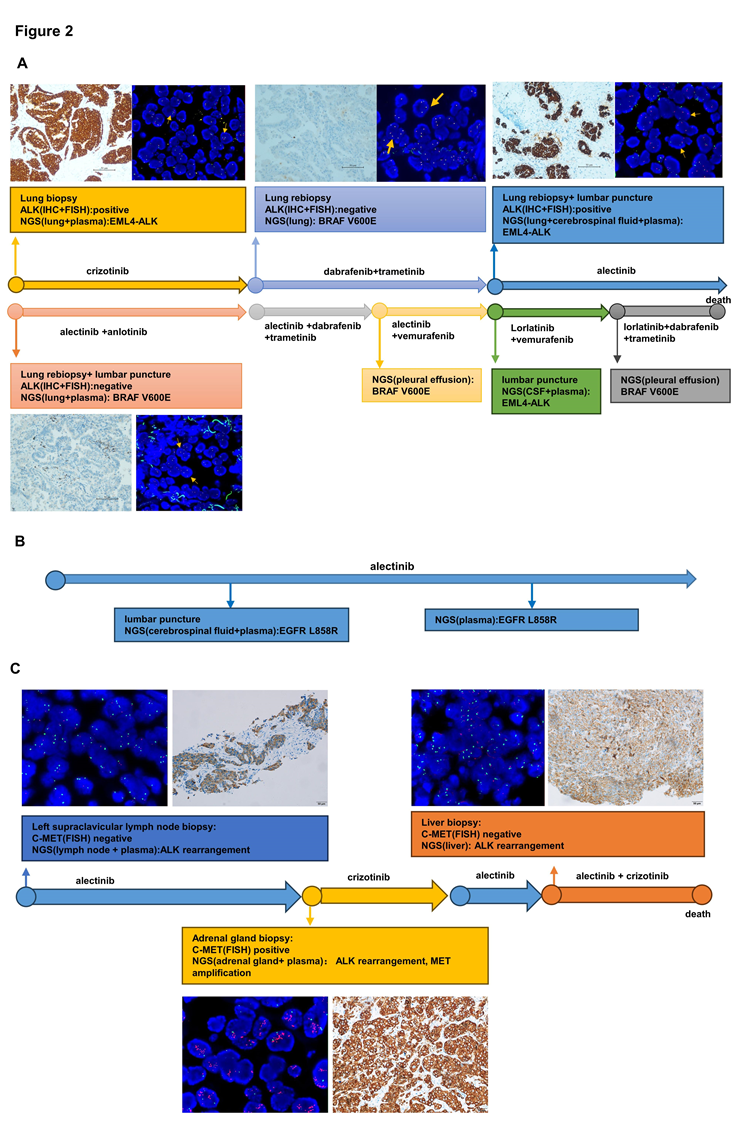
Figure 2: Validation of other driver gene and ALK rearrangement. (A) Treatment line of patient 1 and validation of BRAF V600E and ALK rearrangement by IHC, FISH and NGS. (B) Treatment line of patient 2 and validation of EGFR L858R by NGS of Patient 2. (C) Treatment line of patient 3 and validation of MET amplification by IHC, FISH and NGS of Patient 3. Abb: IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; NGS, next-generation sequencing.
3.2 Validation of ALK rearrangement and other driver gene
In our study, we performed NGS on tissue and liquid samples obtained at every treatment milestone from three ALK-rearranged patients receiving targeted therapy. FISH and IHC were undergone in some tissue samples to verify the existence of ALK rearrangement and MET amplification (Figure 2).
For patient 1, the tissue of the first lung biopsy performed ALK rearrangement positive by IHC, FISH and NGS. The second time lung biopsy showed an acquired BRAF V600E mutation was observed by NGS, but ALK rearrangement was negative by NGS, FISH or IHC. And the third time lung biopsy identified ALK rearrangement again by IHC and FISH, while the NGS analysis of tissue biopsy, peripheral blood, and cerebrospinal fluid re-identified EML4-ALK without BRAF V600E mutation. About the fourth time biopsy, ALK rearrangement disappeared and BRAF V600E mutation showed again. Then we found BRAF V600E mutation from pleural effusion and EML4-ALK from peripheral blood and cerebrospinal fluid by NGS.
For patient 2, after 9 months therapy of alectinib, EGFR L858R mutation was found in peripheral blood and cerebrospinal fluid by NGS. Then we found it again in peripheral blood by NGS 4 months later. And it disappeared in next time NGS of peripheral blood. In baseline, Patient 3 showed MET amplification negative by IHC and FISH using left supraclavicular lymph node. After resistance to alectinib, IHC, FISH and NGS all validated MET amplification positive by adrenal gland tissue. But MET amplification negative again in liver tissue in subsequent resistance.
3.3 Efficacy of combined targeted therapy after resistance to ALK-TKI
Patient 1 was treated with first-line crizotinib and received PR but brain metastases were found after 8.8 months (Figure 3A). An acquired BRAF V600E mutation was observed by NGS, but ALK rearrangement was negative by NGS, FISH or IHC. The patient was then switched to the combination therapy of dabrafenib and trametinib for 4.0 months, with the efficacy of SD. NGS analysis of tissue biopsy, peripheral blood, and cerebrospinal fluid re-identified EML4-ALK without BRAF V600E mutation. She started treatment with alectinib as third-line setting and achieved PR. After a PFS of 13.7 months, CT scan revealed an increase in the size of pulmonary lesions. The lung biopsy was performed again and NGS revealed BRAF V600E without EML4-ALK. Due to poor performance status, the patient was then received the combination therapy of alectinib and anlotinib before the molecular analysis came out, but progressed in 2.0 months. According to the latest NGS result, the patient was switched to the combination therapy of alectinib, dabrafenib and trametinib, and received PR after one month. However, she suffered from vomiting, nausea, and decreased appetite, therefore she discontinued alectinib from the second month. The patient benefitted from dabrafenib and trametinib about 3 months until the new lesion was found in brain finally. However, the patient refused to receive more biopsy, we recommended intercalated combination of alectinib and dabrafinib plus trametinib according to her previous NGS results. After intercalated therapy, lung lesion enlarged slowly and brain lesion shrank gradually, with the comprehensive evaluation of stable disease. After using combination therapy of alectinib, dabrafenib and trametinib for 19.1 months, she was evaluated as PD again because of the enlarged lung lesion, new pleural effusion and new brain metastases. NGS of pleural effusion and peripheral blood identified BRAF V600E mutation without EML4-ALK. According to the failure and side effect of dabrafenib and trametinib, she received alectinib and vemurafenib together and the CT scan showed lung lesion shrank obviously after one month of sixth-line treatment. But disease quickly progressed after 2.6 months and she started to take lorlatinib plus vemurafenib on the basis of the NGS result of peripheral blood and cerebrospinal fluid (EML4-ALK). Though PR again, pleural effusion became more and NGS showed BRAF V600E mutation again and no EML4-ALK. After taking lorlatinib, dabrafenib and trametinib about only 0.67 months, patient 1 dead because of the tumor progression (Figure 3B).
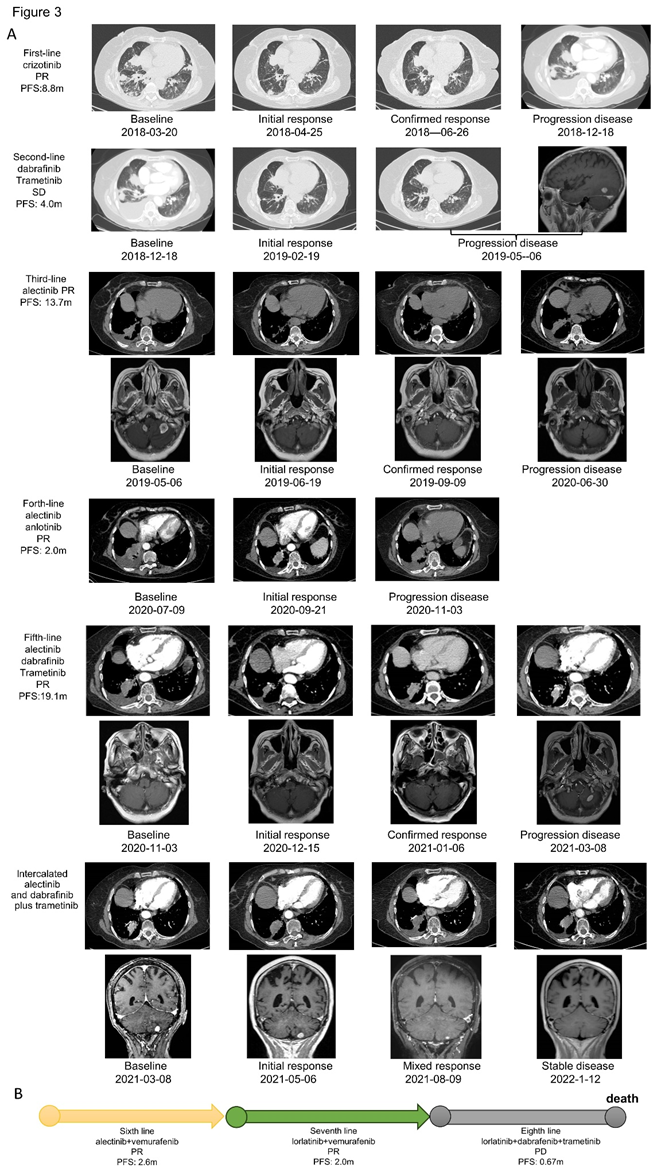
Figure 3: Treatment and efficacy of patient 1. (A) Response evaluation of treatment and radiographic imaging of patient 1. (B) Sixth to eighth line treatment and efficacy of patient 1.
Abb: PR, partial response; SD, stable disease; PD, progression disease; PFS, progression-free survival.
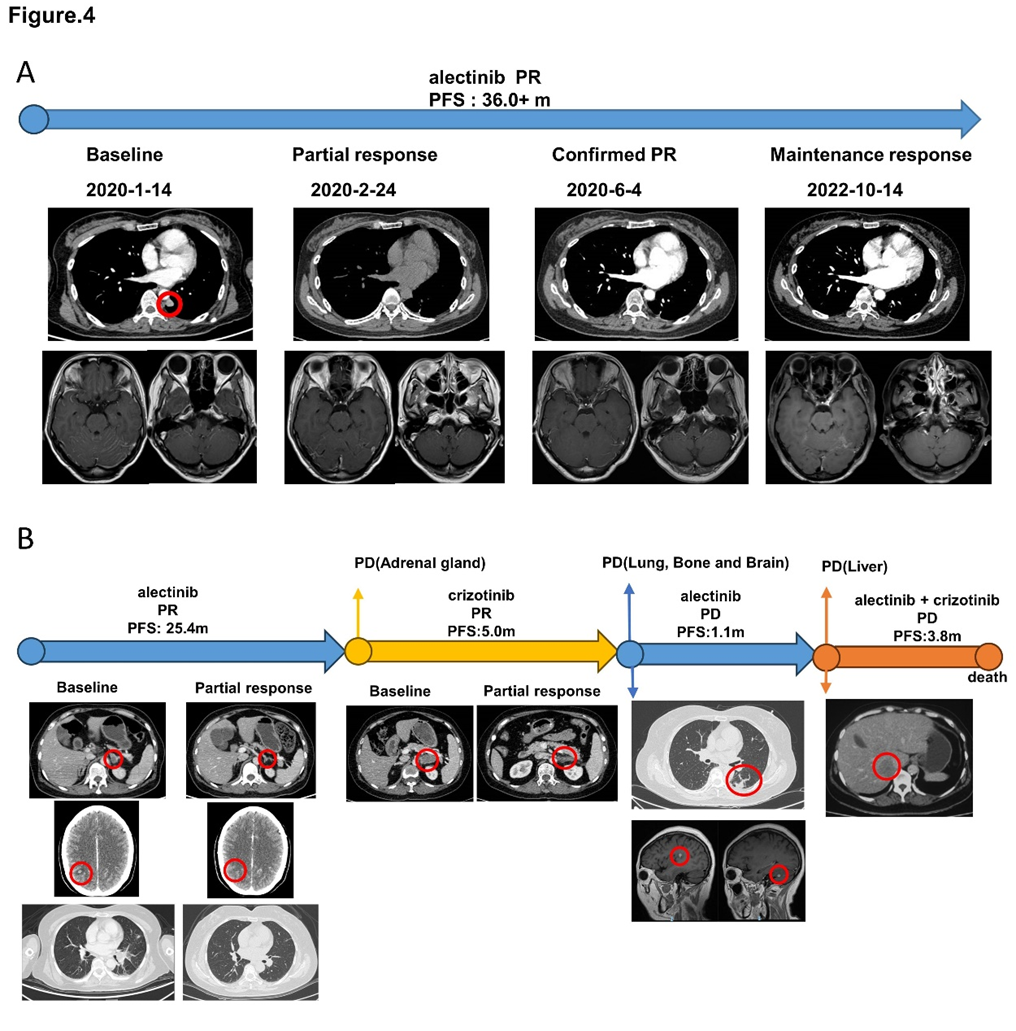
Figure 4: Treatment and efficacy of patient 2 and patient 3. (A) Response evaluation of treatment and radiographic imaging of patient 2. (B) Response evaluation of treatment and radiographic imaging of patient 3. Abb: PR, partial response; PD, progression disease; PFS, progression-free survival.
Patient 2 underwent radical surgical resection at other hospital and diagnosed with pT1N2M0 stage IIIA adenocarcinoma. Without any adjuvant treatment, after a disease-free survival of 28.0 months, new lesions were found in the left upper lobe, left lower pleura and leptomeningeal metastasis. Thus, she was diagnosed with a recurrence. Then, she underwent the wedge section of left upper lobe and pleural node biopsy and NGS revealed EML4-ALK. The patient was treated with alectinib and experienced significant improvement in headache and dizzy. After 8 months of treatment, magnetic resonance (MR) showed enhanced signal at cerebellar vermis. Molecular analysis of cerebrospinal fluid and peripheral blood revealed EGFR L858R mutation. However, the patient continued alectinib because she still had clinical benefit. We did lumber puncture again 4 months later, EGFR L858R was confirmed in peripheral blood but missed in cerebrospinal fluid. Then she continued alectinib, and new pulmonary nodule was showed in CT after 6 months. Considering the NGS of peripheral blood was negative and the patient was asymptomatic, she continued the treatment of alectinb. The disease was stable until follow-up time (Figure 4A).
Patient 3 was diagnosed with advanced lung cancer, with metastases of adrenal glands, liver, multiple bone, pleural effusion, peritoneal lymph nodes. IHC and FISH of supraclavicular lymph node and NGS of lymph node and peripheral blood all identified ALK rearrangement. She received alectinb as first-line treatment with the best therapeutic evaluation of partial response. CT indicated lesion of left adrenal gland enlarged after alectinib therapy for 25.4 months. Then she underwent biopsy of left adrenal gland. NGS of left adrenal gland showed not only ALK rearrangement but also MET amplification, which was identified again by IHC and FISH. Targeting to ALK rearrangement and MET amplification, crizotinib was selected as second-line therapy to her. Fortunately, the lesion of left adrenal gland shrank obviously and she obtain PR. About 5 months later, new lesions were found in lung, bones and brain. She underwent lumber puncture and the NGS of cerebrospinal fluid revealed ALK rearrangement and MET amplification while the peripheral blood just showed ALK rearrangement, without MET amplification. While waiting for the result of NGS, she refused to take chemotherapy and tried alectinib again. But PD quickly because of the liver lesion. The gene testing of the liver revealed ALK rearrangement and MET amplification was disappeared. She received crizotinib plus alectinb as fourth-line therapy but dead 3.8 months later because of the tumor progression. (Figure 4B)
3.4 Gene heterogeneity and dynamic changes of gene alteration
In our study, we found some gene heterogeneity and special dynamic changes of gene alteration. The dynamic gene alteration of patient 1 was that EML4-ALK and BRAF V600E appeared alternately, both in tissue or blood by NGS. (Figure 5A and B) When EML4-ALK rearrangement occurred, BRAF V600E mutation was disappeared. The same phenomenon showed up when they switched. And for patient 1, the dynamics changes of gene are consistent between tissue and blood. Otherwise, pleural effusion only showed BRAF V600E mutation while cerebrospinal fluid was only found EML4-ALK. EGFR L858R mutation was only occurred in liquid biopsy (peripheral blood and cerebrospinal fluid) but not in tissue for patient 2, which showed a situation of gene heterogeneity. Patient 3 identified MET amplification only in adrenal gland, not in lung and liver lesion (Table 2).
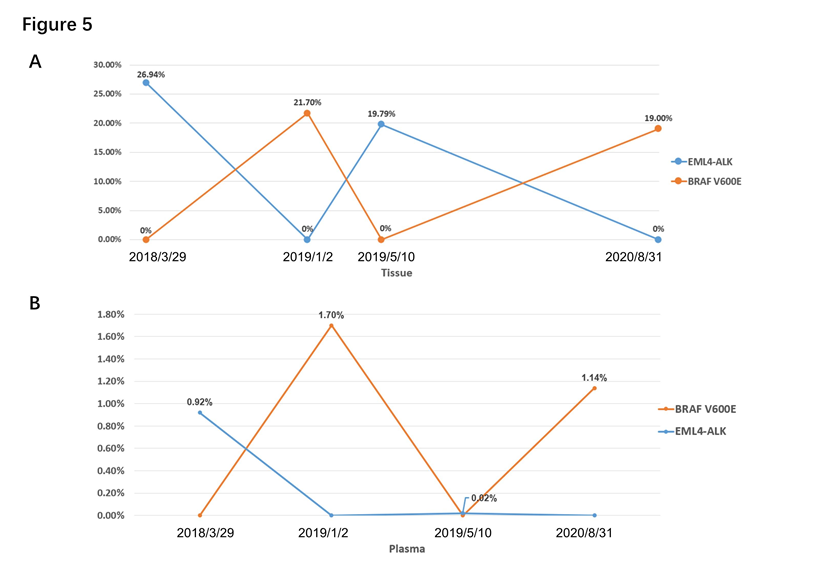
Figure 5: Dynamic changes of genes of patient 1. (A) Dynamic changes of genes in tissue by NGS. (B) Dynamic changes of genes in peripheral blood by NGS.
4. Discussion
In this study, we confirm that EML4-ALK lung cancer acquired resistance to crizotinib and alectinib due to other driver gene activation. We performed molecular analysis on tissue, plasma, pleura effusion or cerebrospinal fluid sample obtained at various treatment milestones of these three ALK positive NSCLC. At disease progression after treating with crizotinib or alectinib, our sequencing data revealed BRAF V600E, EGFR L858R and MET amplification in samples of 3 patients respectively.
Mechanisms of resistance to ALK TKI include ALK dependent secondary mutations and bypass signaling pathway activation [7]. Previous studies have demonstrated that BRAF mutation, including V600E, G15V and D587A mutations, could be a resistance mechanism in ALK positive NSCLC [7, 13]. It has been reported that acquired BRAF V600E in vitro responded to the triple combination of alectinib/dabrafenib/ trametinib. A few cases have been reported in previous studies that BRAF V600E mutation was showed after resistance to ALK-TKI [16, 17]. But these cases do not try the combined targeted therapy clinically. In our study, although no ALK mutation was detected after resistance to alectinib, the patient 1 continued alectinib in combination of dabrafenib and trametinib due to the efficacy of alectinib in central nervous system. Our case suggests triple combination might be an optional treatment of post-alectinib ALK positive patient with acquired BRAF V600E mutation, but the side effect should be paid more attention. Further investigations are needed to study the efficacy and safety of combination therapy.
Additionally, EGFR pathway activation also have been reported as a mechanism of resistance to ALK-TKI [12]. Sachiko et al. suggested that osimertinib combined with alectinib may be suitable treatment for post-alectinib patients with acquired EGFR mutation in cell experiment [12]. Otherwise, previous study indicated that EGFR 19del mutation was one of the resistance mechanisms to ALK-TKI [18, 19], and it seems not play the leading role. In our study, we first reported a case that EGFR L858R mutation showed up after resistance to ALK-TKI. Patient 2 had detectable EGFR L858R mutation by NGS after enhanced signal showed at cerebellar vermis.
We also found that other driver genes as resistance mechanisms of ALK inhibitors may have heterogeneity. Pleural effusion only showed BRAF V600E mutation while cerebrospinal fluid was only found EML4-ALK in Patient 1. EGFR L858R mutation was only occurred in liquid biopsy (peripheral blood and cerebrospinal fluid) but not in tissue for patient 2. For Patient 3, MET amplification only occurred in adrenal gland, not in lung and liver lesion. This appearance may indicate that when other driver genes mutate after resistance to ALK inhibitors, they may play a secondary role or less and these mutations may only occur in local tissues, showing heterogeneity.
Nevertheless, our study has some limitations. As a single-center retrospective study, there was a recall bias in our study. Limited by the low frequency and sample size, only 3 patients in our study had enough tissue samples to conduct NGS/IHC/FISH, hence, it was difficult to clarify the mechanism of how BRAF V600E, EGFR L858R and MET amplification affects the resistance to ALK inhibitors. Therefore, further explorations with a larger population to verify our conclusion and identify the main mechanisms from multidimension are warranted.
In a word, resistance to ALK-TKI is inevitable. With further research on the resistance mechanisms, especially other driver genes, the study for follow-up treatment are necessary. We present the first clinical evidence of sequential use of dabrafenib and trametinib in combination with alectinib and report the first clinical case that EGFR L858R mutation showed up after resistance to ALK-TKI. Furthermore, we also reveal other driver genes as resistance mechanisms of ALK inhibitors may have heterogeneity.
Acknowledgement
We thanks to all patients and family. We thank the staff at Pulmonary Oncology Division, Guangdong Lung Cancer Institute, Guangdong Provincial People’s Hospital and Guangdong Academy of Medical Sciences.
Declaration of Competing Interest
The authors have declared no conflicts of interest.
Funding
This work was supported by National Natural Science Foundation of China [Grant No. 81972164, JJ Yang], Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (Grant No. 2017B03031412)and High-level Hospital Construction Project [Grant No. DFJH201809, JJ Yang].
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71 (2021): 209-249.
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131 (2007): 1190-1203.
- Zhou C, Kim SW, Reungwetwattana T, et al. Alectinib versus crizotinib in untreated Asian patients with anaplastic lymphoma kinase-positive non-small-cell lung cancer (ALESIA): a randomised phase 3 study. Lancet Respir Med 7 (2019): 437-446.
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 377 (2017): 829-838.
- Mok T, Camidge DR, Gadgeel SM, et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann Oncol 31 (2020): 1056-1064.
- Solomon BJ, Bauer TM, Mok TSK, et al. Efficacy and safety of first-line lorlatinib versus crizotinib in patients with advanced, ALK-positive non-small-cell lung cancer: updated analysis of data from the phase 3, randomised, open-label CROWN study. Lancet Respir Med 11 (2023): 354-366.
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 6 (2016): 1118-1133.
- Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer Discov 8 (2018): 714-729.
- Fukuda K, Takeuchi S, Arai S, et al. Epithelial-to-Mesenchymal Transition Is a Mechanism of ALK Inhibitor Resistance in Lung Cancer Independent of ALK Mutation Status. Cancer Res 79 (2019): 1658-1670.
- Kim S, Kim TM, Kim DW, et al. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. J Thorac Oncol 8 (2013): 415-422.
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 18 (2012): 1472-1482.
- Arai S, Takeuchi S, Fukuda K, et al. Osimertinib Overcomes Alectinib Resistance Caused by Amphiregulin in a Leptomeningeal Carcinomatosis Model of ALK-Rearranged Lung Cancer. J Thorac Oncol 15 (2020): 752-765.
- Shi R, Filho SNM, Li M, et al. BRAF V600E mutation and MET amplification as resistance pathways of the second-generation anaplastic lymphoma kinase (ALK) inhibitor alectinib in lung cancer. Lung Cancer 146 (2020): 78-85.
- Dagogo-Jack I, Yoda S, Lennerz JK, et al. MET Alterations Are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer. Clin Cancer Res 26 (2020): 2535-2545.
- Pailler E, Faugeroux V, Oulhen M, et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer. Clin Cancer Res 25 (2019): 6671-6682.
- Sui A, Song H, Li Y, et al. BRAF V600E mutation as a novel mechanism of acquired resistance to ALK inhibition in ALK-rearranged lung adenocarcinoma: A case report. Medicine (Baltimore) 100 (2021): e24917.
- Urbanska EM, Sørensen JB, Melchior LC, et al. Changing ALK-TKI-Resistance Mechanisms in Rebiopsies of ALK-Rearranged NSCLC: ALK- and BRAF-Mutations Followed by Epithelial-Mesenchymal Transition. Int J Mol Sci 21 (2020): 2847.
- Hua G, Zhang X, Zhang M, et al. Real-world circulating tumor DNA analysis depicts resistance mechanism and clonal evolution in ALK inhibitor-treated lung adenocarcinoma patients. ESMO Open 7 (2022): 100337.
- Sánchez-Herrero E, Serna-Blasco R, Ivanchuk V, et al. NGS-based liquid biopsy profiling identifies mechanisms of resistance to ALK inhibitors: a step toward personalized NSCLC treatment. Mol Oncol 15 (2021): 2363-2376.
