OmiCrisp: A CRISPR SARS-CoV-2 Test with Omicron Detection
Article Information
Suruchi Sharma1,2#, Manasa Bagur Prakash1,2#, Reety Arora1,2, Annes Siji1,2, Bidipta Roy3, Varsha Shridhar3, Nimisha Gupta4, Vijay Chandru1,5,6,7, Vaijayanti Gupta1,2*
1CrisprBits Private Limited, 3rd floor, Plot No.-3, F-301, Ashish Complex, LSC, New Rajdhani Enclave, East Delhi, Delhi-110092, India
2CrisprBits Laboratory, C-CAMP, NCBS-TIFR Campus, GKVK Post, Bellary Road, Bengaluru, Karnataka-560065, India
3Molecular Solutions Care Health, No.11, 1st floor, VCN Complex, 1st Main road, Seshadripuram, Bengaluru, Karnataka- 560020, India
4G-KnowMe Private Limited, Godrej Woodsman Estate, Bellary Road, Hebbal, Bengaluru, Karnataka- 560024, India
#Both authors contributed equally to the work
5National Center for Biological Sciences, Tata Institute of Fundamental Research, Bellary Road, Bengaluru- 560065
6Harvard T.H. Chan School of Public Health, Harvard University-Boston Campus, 677 Huntington Ave, Boston, MA 02115, United States
7ARTPark, Indian Institute of Science, Indian Institute of Science, Ground Floor, Entrepreneurship Building, Bengaluru- 560012, India
*Corresponding author: Vaijayanti Gupta, CrisprBits Private Limited, 3rd floor, Plot No.-3, F-301, Ashish Complex, LSC, New Rajdhani Enclave, East Delhi, Delhi-110092, India.
Received: 03 January 2024; Accepted: 11 January 2024; Published: 29 January 2024
Citation: Suruchi Sharma, Manasa Bagur Prakash, Reety Arora, Annes Siji, Bidipta Roy, Varsha Shridhar, Nimisha Gupta, Vijay Chandru, Vaijayanti Gupta. OmiCrisp: A CRISPR SARS-CoV-2 Test with Omicron Detection. Journal of Biotechnology and Biomedicine. 7 (2024): 67-81.
View / Download Pdf Share at FacebookAbstract
We have developed a CRISPR based assay that can detect the presence of SARS-CoV-2, in human as well as environmental samples, and predict if it is an Omicron or non-Omicron variant of the virus. This nucleic acid amplification-based test (NAAT) consists of two independent steps: a one-step multiplexed RT-PCR amplification using a standard kit, followed by CRISPR based detection that utilizes the trans-cleavage activity of the Cas12a enzyme. We have evaluated the performance of OmiCrisp in over 80 clinical samples and more than 160 sewage samples. We observed an agreement of >99% with the sequencing results in labeling SARS-CoV-2 positive samples as well as Omicron or non-Omicron call, in clinical cases. Similar accuracy was observed with qRT-PCR results in wastewater samples. Our OmiCrisp -like platforms can be developed quickly and can potentially complement sequencing for quick and rapid tracking of the transmission of emerging pathogenic variants.
Article Details
Introduction
Regular testing for timely isolation has been a crucial activity in the management of the SARS-CoV-2 pandemic. Nucleic acid-based detection assays, real-time PCR (rtPCR), ubiquitously proved to be the most effective way of testing for the containment of Covid19 pandemic. This encouraged the development of new ways of nucleic acid-based tests that are cost-effective, rapid, and instrument agnostic [1]. Furthermore, similar to all viruses, SARS-CoV-2 accumulates mutations in its genome over time that give rise to new variants of the virus. Some variants exhibit changes in transmission, infectivity, disease severity, response to therapy. Hence, it is important to track the transmission of the new SARS-CoV-2 variants for the effective management of the pandemic. So far, for SARS-CoV-2 variant tracking, most countries have relied on whole genome sequencing of the virus from patient samples. Indeed sequencing has been used to both understand the evolution of variants as well as track the variants by their signature mutations, once they have been sequenced [2]. Sequencing is expensive, time-consuming, and requires specialized instruments, and skilled personnel underscoring the need for a complementary rapid and cost-effective method for variant tracking. There are various cost-effective and rapid assays in published literature for genotyping of mutations [3, 4]. However, these are not being used in practice for tracking of SARS-CoV-2 variants because these methods require extensive design and validation efforts to develop, and hence not suitable for tracking of viruses which are evolving rapidly. Waste water based surveillance of the SARS-CoV2 concentrations, which enters the sewage through human discards, has been a complementary adjunct method to track the spread of the virus within a community [5], in addition to clinical testing. Testing samples of sewage can monitor community level transmissions and provide early warning signs for rapidly emerging variants especially since non-symptomatic people can also shed the virus [6]. Scientists have been looking for pathogens in sewage for environmental surveillance for the past decade. Indeed, in the case of polio virus, waste water surveillance is considered more effective than tracking symptomatic individuals [7]. While next generation sequencing is the only fool-proof method for identifying a novel viral strain as it evolves, it is expensive and unaffordable in public health settings to track the virus, especially in low income countries. In addition wastewater and other such mixed samples yield nucleic acids of inferior quality and yield that are not at par with the requirements of NGS tests.
CRISPR systems appear to be a promising technology for sequence specific detection of nucleic acids. There have been various reports demonstrating CRISPR based methods for SARS-CoV-2 detection based on Cas13, Cas12, and Cas9 that show the promise of being instrument minimalistic [8-19]. Some of them have also shown variant detection capabilities, but most of these variant tracking platforms have not shown the variant selectivity at higher viral loads making them of limited use [20, 14, 21]. Here we report the development OmiCrisp that can detect the presence of SARS-CoV2 in RNA extracted from a nasopharyngeal swabs as well as wastewater samples, and predict if it is an “Omicron” or “non-Omicron” lineage variant of the virus. This is a nucleic acid amplification-based test (NAAT). The amplification and detection are carried out in two independent steps in this assay. Amplification is done using a multiplexed RT-PCR method, followed by detection that relies on the trans-cleavage activity of the Cas12a enzyme [22, 23] (Figure 1a). Briefly, Cas12a enzyme binds to a guide RNA to make Cas12a:guide RNA complex. The guide RNA is customized to recognize the target DNA sequence -a sequence that one wants to detect. The Cas12a: guide RNA complex in the presence of target DNA makes a trimeric nucleoprotein complex, Cas12a: guide RNA: target DNA. This trimeric nucleoprotein complex has an endonuclease activity and it cleaves ssDNA irrespective of the sequence of ssDNA, this activity of the trimeric complex is known as trans-cleavage activity. The trans-cleavage activity of trimeric complex can be easily observed as an increase in fluorescence signal if the ssDNA that gets cleaved is labeled with a fluorophore and quencher pair at its termini, reporter ssDNA. In our assay, the detection reagent contain Cas12a, a guide RNA, and ssDNA reporter, when a sample to be tested contains the target DNA to be detected by the guide RNA; the Cas12a:guide RNA: target DNA forms resulting in trans-cleavage of the ssDNA reporter and an increase in fluorescence signal. Total turnaround time of the assay is 3 hours. The assay has been designed to detect ORF1ab gene, N gene, and S gene targets for SARS-CoV-2; while ribonuclease P_MRP subunit (POP7) transcript target has been used as human RNA control. OmiCrisp has been designed in such a way that the selectivity for variant prediction is retained at the highest possible viral load. Furthermore, we identified mutations in the virus, which can discriminate Omicron from non-Omicron variants and show 100% agreement with sequenced clinical samples. The same test shows 99% concordance with RT-PCR data, in identifying SARS-COV2 in sewage samples and higher accuracy for calling Omicron variants in waste water samples compared to the traditional RT-PCR. Finally, the test is still applicable in identifying all variants of concern of Omicron lineage that have appeared in India including the recently reported JN.1 (BA.2.86.1.1).
Materials and Methods
Reagents and Equipment
LbaCas12a, Alt-R™ L.b. Cas12a (Cpf1) Ultra, custom guide RNAs Alt-R® L.b. Cas12a crRNA, custom ssDNA_FQ reporter (/56-FAM/TT ATT /3IABkFQ/) were procured from IDT(Integrated DNA Technologies, USA). DNA oligonucleotides used as primers and synthetic templates were custom synthesized from Sigma (USA), and VNIR Biotechnologies (India). NEBuffer™ 2, and BSA (B9000S) were purchased from NEB(New England BioLabs, USA). The fluorescence measurements were done in VarioSkan LUX micro plate reader (Thermo Scientific, USA) in Corning® 96 Well Black Polystyrene Microplates (CLS3603-48EA, Sigma, USA) were used. The fluorescence measurements for OmiCrisp_v2 were acquired in Bio-Rad CFX96 Real-Time PCR machine (Bio-rad, USA). The amplification reactions were performed in Mastercycler Nexus Thermal Cycler™ (Eppendorf, Germany) and Applied Biosystems 2720 thermal cycler (Thermo Scientific, USA). SARS-CoV-2 synthetic RNA controls for reference strain (Wuhan-Hu-1, cat. 102024) and Omicron strain BA.1 (Omicron EPI_ISL_6841980, cat.105204) were procured from TwistBioscieces, USA. PCR master mix 2X, K01721, was procured from Thermo Scientific. For purification of PCR products QIAquick Gel Extraction Kit (cat.28704, Qiagen, Germany) was used.
Synthetic DNA template preparation
Synthetic DNA templates corresponding to S gene Omicron BA.1, and Sgene Omicron BA.2 were prepared by the overlap extension of the synthetic oligonucleotides. Briefly, the overlap extension mix was prepared by mixing overlapping oligonucleotides, see SI table 4 for sequences, at a concentration of 1 µM in 1X PCR master mix and overlap extension was performed in thermal cyclers with following conditions: Initial heat denaturation 95 °C for 5 minutes; 15 cycles of 90 °C 30 seconds heat denaturation, annealing at 60 °C for 30 seconds, extension 72 °C for 30 seconds; and final extension for 5 minutes at 72 °C. Rest of the synthetic templates were prepared by One step RT-PCR amplification from the corresponding synthetic RNA controls with the suitable primers at 250 nM concentration and 1*10^5 copies of synthetic RNA as template. The products obtained after overlap extension and one step RT-PCR were purified using a purification kit and quantified based on absorbance at 260 nm in NanoDrop One™ (Thermo Scientific, USA).
One step multiplexed RT-PCR
One step RT PCR was performed using commercially available one step RT PCR mix. The concentration of each primer in the reaction mix was about 250 nM. Following thermal cycling conditions were used for the one step RT-PCR: Step 1. primer incubation at about 25 °C for about 2 minutes, Step 2. reverse transcription at about 53 °C for about 10 minutes, Step 3. enzyme denaturation at about 95 °C for about 2 minutes, and Step 4. 35-40 cycles of amplification that includes incubation at about 95 °C for about 15 seconds for denaturation and combined extension/annealing at about 60 °C for about 1 minute. For OmiCrisp_v1 and OmiCrisp_v2 multiplexed One step RT PCR was done. For v1 two primer pairs (4 primers) corresponding to the ORF1ab and S gene were used as N gene primers were not designed for the assay. OmiCrisp_v2 used 4 primer pairs to amplify fragments of interest corresponding to ORF1ab, S gene, N gene, and human RNaseP; hence a total of 8 primers were added in a single tube.
Trans-cleavage assay
All the trans-cleavage assays for a given target template or sample were carried out using 25 nM of LbaCas12a, 25 nM of the indicated guide RNA, 50 nM ssDNAFQ reporter in a solution containing 50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, and 100 µg/ml BSA pH 7.9 (at 25 °C) at 37 °C. For the end-point assays the reactions were stopped after 1 hour of incubation by adding 10 µL of stop buffer, 250 mM EDTA and 37.5 mM Tris.Cl pH 7 at 25 °C to the 50 µL reaction mix. For real time assays, all the reactions were initiated by addition of the target samples, and it was ensured that the time difference between beginning of addition of sample and the start of the data acquisition is not more than 2 minutes. At the end of the reaction, after 1 hour of incubation, it was stopped by adding 10 µL of stop buffer containing 250 mM EDTA and 37.5 mM Tris.Cl pH 7 at 25 °C to the 50 µL reaction mix and fluorescence measurements were performed on a microplate reader. Fluorescence data (emission at 525 nM) was captured using either a plate reader or real time PCR machine. Data is shown in an arbitrary fluorescence unit (AU).
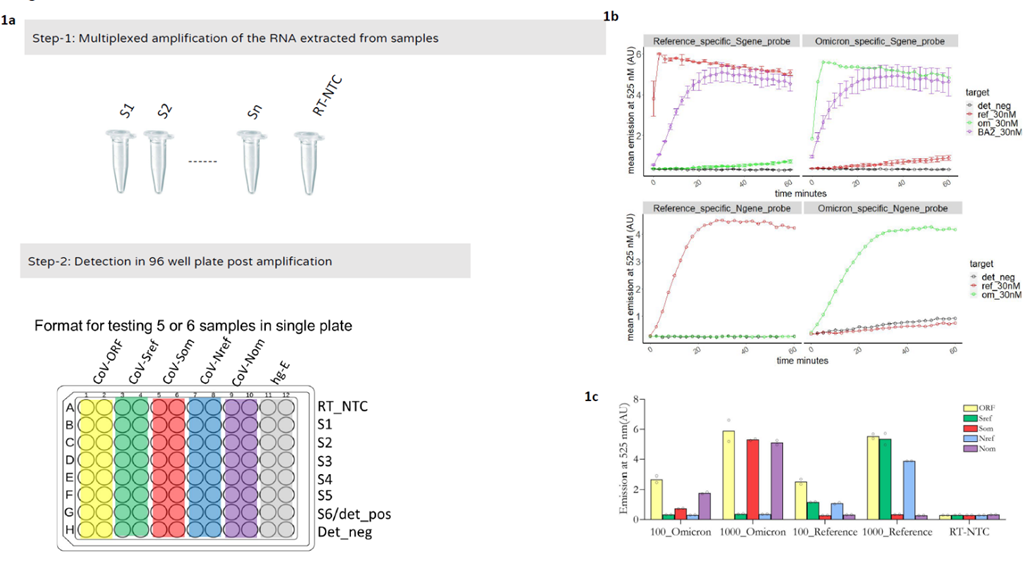
Detection step was carried out in a 96-well plate. The amplified product from the One step RT-PCR was used as an input for end-point trans-cleavage assays in the presence of 6 different detection reagents containing the following guide RNAs: 1. ORF1ab (ORF1ab_ref + ORF1ab_Om) 2. Sgene_omm 3. Sgene_ref 4. Ngene_ref 5. Ngene_om and 6. RNaseP. For each trans-cleavage reaction about 5 µL of the amplified sample was used and the reaction in each detection reagent was done in duplicate, resulting in a total of 12 independent trans-cleavage reactions for each sample to be tested (Figure 1a).
Clinical samples for validation
Clinical nasopharyngeal swab samples were obtained from two sources. (1) Strand Life Sciences which was mandated by the Technical Advisory Committee (TAC) of the State of Karnataka for Pandemic to initiate the genomic Surveillance of SARS-CoV-2 in various parts of Bengaluru city (Ref: CHO (PH)/PR/P-103/2021-22), helped us to cross validate OmiCrisp v1 of the assay. Ethics for sample collection and research was provided by the institutional ethics committee at HCG (HealthCare Global), and (2) The Institutional Ethics Committee, inStem, provided the approval for archival and access of clinical samples from the biorepository at inStem (inStem/IEC-19/01/27E, inStem/IEC-17/001). The Institutional Biosafety Committee provided permission (inStem/G-141(3)-17/CJ) for the utilization of the samples for clinical research. Permission to conduct and support research on SARS-CoV2 has been provided to inStem by the Review Committee on Genetic Manipulation (RCGM), Department of Biotechnology, Ministry of Science and Technology, Government of India (approval number: BT/IBKP/035/2019).
Environmental samples for validation
Extracted RNA from wastewater samples were obtained from Molecular Solutions Care Health LLP, Bangalore. These samples were collected as part of the Precision Pandemic Health Initiative, a community based wastewater surveillance project implemented in various parts of Bangalore city. Sample collection sites included open storm water drains. Collection, transportation and RNA extraction protocols are described in Basu et. al., 2022 with one modification: the RNA extraction kit used was NeoDx Total Nucelic Acid Extraction kit (NeoDx, Bangalore). All extracted RNA from retrospective samples were stored at -20C until further use. For the CRISPR based assay, data was acquired for all 3 markers ORF1ab, S gene and N gene using OmiCrisp_v2 for the experimental samples as well. However, the Q493R, G496S mutations in the S gene were no longer valid for a sample to be classified as Omicron positive, hence S gene was not analyzed in the study. For the wastewater samples, only ORF1ab and N gene markers were used to identify positive samples.
RT-PCR for cross validation
All environmental samples were initially tested using RT-PCR at Molecular Solutions Care Health LLP laboratory using the SARS CoV2 quantitative kit (GenePath Technologies, Pune).These kits are referred to as RT-PCR Kit 1 in the manuscript. Omicron variant calls were cross-validated with the CoViDelta kit, (GenePath Technologies, Bangalore) and is referred as RT-PCR Kit 2 in the manuscript.
Data analysis and interpretation
Threshold signal estimation
The threshold signal is important for labeling a given signal as unambiguously positive without requiring a subjective decision of the analyst. The threshold signal was calculated as described below. Every time a trans-cleavage assay was performed, negative controls were included for each guide RNA used for the detection in trans-cleavage assay. The negative controls had all the reagents except the sample, added to them. Signals from negative controls were used to estimate the threshold signal. The signal from the negative controls of all the guide RNAs are pooled into one set that was labeled as “pooled negative controls”. The median and interquartile range of the “pooled negative controls” was calculated using standard formulas. First, outliers, which could arise from random contamination, were removed from the “pooled negative controls”. Upper limit of outliers was calculated using equation Eq.1.
Upper limit of outlier = median negative control signal + 1.5*Interquartile range (IQR)---------Eq.1
All the negative control points with signals higher than the Upper limit of outlier were removed. In case of a situation where more than 20 % of the negative controls were higher than the upper limit of outlier, the entire assay was discarded and repeated with fresh reagents. After removal of outlier points standard deviation was calculated of the remaining “pooled negative data set” using standard formulas and the threshold signal (Thresh_signal) was calculated using equation Eq.2.
Thresh_signal = 3.6* Standard deviation of pooled negative data set after outlier removal---Eq.2
RT-NTC check
The RT-NTC control is where no sample was added at the step of amplification to the RT-PCR reaction. The presence of signal higher than threshold signal in the RT-NTC for any guide RNA indicated contamination. A criterion for the acceptable limit of contamination, RT-NTC cut off, was set. If for any guide RNA the signal in RT-NTC was more than RT-NTC cut off, the data for the guide RNA was not used for analysis. The RT-NTC cut off was calculated using equation Eq.3.
RT-NTC cut off = median of pooled negative controls + 1.5*Thresh_signal-----------------------Eq.3
Unreliable data removal
In order to interpret results of the assay for a given sample, first the reliability of the signals obtained after trans-cleavage reaction with all guide RNAs was evaluated. The reliability of the signals was estimated from the variations in technical duplicates of the trans-cleavage assay for each guide for each sample. And any data point, above the threshold signal (as per Eq.1), is to be considered a positive signal. If any such positive signal data point showed large variations in duplicates, they were considered unreliable and were excluded from the data analysis. In order to remove the positive signal data points with large variation in signal in technical duplicates, percentage relative standard deviation (%RSD) for technical duplicates was calculated using equation Eq.4.
%RSD for one guide for a given sample = (Standard deviation of duplicate*100)/(average of duplicate)------------------------------------------------------------------------------------------------------------Eq.4
All the data points with %RSD more than 20% were removed from the analysis. The data points with human gene guide RNA %RSD more than 20% were not removed from the analysis. In those cases where after removing the unreliable data, the data for less than three SARS-CoV-2 guides was left, the assay for that sample was repeated. Note: the exception was applied to two samples used for OmiCrisp_v1 validation. The S om guide of sample 12 and ORF1ab guide of S40 had %RSD of 28 and 20 %, respectively. We did not have sufficient sample for the repeat; however, the signal of both the duplicates were approximately 10 times higher than the background, so it was reasonable to believe that these signals were not unreliable.
Labeling the signal as positive or negative
In order to label a signal as positive or negative, the average signal of all the technical duplicates left after the unreliable data removal was estimated. Next, the background subtracted signal was estimated for each guide RNA used in the assay for each sample tested. The background subtracted signal (back_sub_signal) for all these average signals was calculated using equation Eq.5.
back_sub_signal = average signal of a guide for a sample - average signal of the corresponding guide for RT-NTC-------------------------------------------------------------------------------------------------Eq.5
If the background subtracted signal was greater than threshold signal it was considered positive signal else it was considered negative.
SARS-CoV-2 positive versus negative prediction for clinical samples
- For the clinical samples, those that did not report “positive” signal for any of the three SARS-CoV-marker genes (Orf1a, S gene or N-gene), were labeled as “SARS-CoV-2 negative”. For the sewage samples, those that did not report “positive” for either of the two SARS-CoV-marker genes (Orf1a or N gene), were labeled “SARS-CoV-2 negative”
- The clinical samples that showed positive signal for only one SARS-CoV-2 gene (out of ORF, S, and N) or sewage samples that showed only one marker (out of ORF or N gene) were labeled as “SARS-CoV-2 negative” with a remark saying, “one gene positive repeat to confirm”.
- The samples that showed positive signal for at least two out of three SARS-CoV-2 genes (out of ORF, S, and N) for clinical samples or both positive signal genes (ORF and N gene) in sewage samples, were labeled as “SARS-CoV-2 positive”.
Notably, if either one or both guides of a guide pair for a gene showed a positive signal it was counted as only 1 gene positive. For example, the S gene had two guides: Sref and Som. S gene was counted as one gene positive if either one or both the S gene guides showed a positive signal. Similarly, the N gene has two guides: Nref and Nom. The N gene was counted as one gene positive if either one or both the N gene guide RNA showed a positive signal.
SARS-CoV-2 positive vs negative prediction in wastewater samples
In case of sewage samples, since the signal was 4-5 fold lower than clinical samples, as expected in RNA extracted from a mixed matrix like waste water compared to nasopharyngeal swab, we considered the signal from the negative controls of all the guide RNAs from different plates and 3 SD above the mean was considered the cut off for positive signal for threshold. All negative controls were found within this threshold. Therefore, for individual markers, if a signal was above 3 standard deviations of the negative control signal for the corresponding marker, calculated across all runs, it was labeled as “positive”, else negative”. If only of the two markers, either Orf1ab or the N gene signal alone was found positive, then the sample was labeled ambiguous.
Omicron versus non-Omicron prediction
Variant prediction using S gene guide RNA pair
Based on the signals observed for the S gene guide pair, Som and Sref , following two situations were possible. The S gene guide pair signal was only followed for clinical samples S gene variant was not discriminatory for sewage samples.
- In the case of clinical samples, where the back_sub_signal for both Som and Sref signals were not available, variant prediction was not made with S gene saying “insufficient data for prediction of variant”.
- For clinical samples where both SOm and Sref back_sub_signal were available, Som/Sref was calculated using equation 6. If, Som/Sref >0.5, the sample was labeled as “Omicron”, else it is labeled as “Not-Omicron”.
Som/Sref = back_sub_signal of Som/ back_sub_signal of Sref---------------------------Eq.6
Variant prediction using N gene guide pair
Based on the signals observed for the N gene guide pair, Nom and Nref, following two situations were possible.
- For samples where the back_sub_signal for both Nom and Nref signals were not available, variant prediction was not made with N gene saying “insufficient data for prediction of variant”.
- For samples where the back_sub_signal for both Nom and Nref were available, Nom/Nref was calculated using equation Eq.7. If, Nom/Nref >1, the sample was labeled as “Omicron”, else it was labeled as “Not-Omicron”.
Nom/Nref = back_sub_signal of NOm/ back_sub_signal of NOm---------------------------------Eq.7
- For Sewage samples, only the N gene was used to label samples as Omicron positive or negative. if the signal was “positive” for NOm above the Threshold signal for the markers (no background subtraction was performed), it was labeled Omicron, else it was labeled “Not-Omicron”. There were no samples where both Nom and Nref were positive, which would be ambiguous call.
Final variant prediction was done as described below
- In clinical samples, if only one of the N or S gene could be used for variant prediction, the results corresponding to the variant prediction of that gene was reported.
- If both N and S genes could be used for prediction and the prediction was same for both, the corresponding result was reported.
- If N and S gene variant predictions do not match the result reported was, “N and S gene predictions do not match, possible contamination or mixed samples repeat with freshly collected samples.”
- If neither N or S gene could be used for variant prediction the result was, “assay needs to be repeated because the data was not sufficient for variant prediction.”
Human gene signal analysis
In clinical samples If the signal for human gene was positive the result reported was “human gene detected” else the result reported was “human gene not detected”. All the samples showed good fluorescence which was above the threshold in the RNase P reaction for sewage samples. If the sample was SARS-CoV-2 negative and human gene was not detected, the remark was added “assay needs to be repeated” with fresh sample or higher amount of sample. For wastewater samples, human gene signal as an indicator of sample integrity was deprioritized.
Results and Discussion
Guide design
To develop a nucleic acid-based assay for the detection of Omicron variant, we identified mutations that were specific to Omicron variant of SARS-CoV-2 and were not associated with any other SARS-CoV-2 variant identified by WHO; i.e. Alpha, Beta, Delta, Epsilon, Eta, Gamma, Iota, Kappa, Lambda, Mu, Theta, and Zeta. We enlisted mutations in the S gene of the Omicron variant that were present at a frequency higher than 90%, and not present in any of the aforementioned non-Omicron variants of SARS-CoV-2 at a frequency higher than 0.1 % (SI Table S1). Next, we selected the subset of these mutations that were present in all the three variants of Omicron, BA.1, BA.2, and BA.3, identified at that time of the work. This analysis was done manually from the data available at www.outbreak.info website on 6 January 2022. We continued to track the selected mutations in subsequent lineages that appeared in India, including BA.5, BA.2.75, XBB.1, XBB.1.5 and the recent JN.1 (lineage- BA.2.86.1.1) for the validity of our assays and found our Orf1ab and N gene markers conserved both for the identification of SARS-CoV2 and for the Omicron lineage variants (Supplementary Figure S1). To design a Cas12a trans-cleavage assay that can detect single nucleotide variations in a sequence, the variant should be present within the 6 bases of PAM site [23]. We examined the sequence of shortlisted mutation/s and identified the ones that matched the criteria for developing the Cas12a trans-cleavage-based assay for single nucleotide variation detection. We planned to develop an assay in which the variant prediction is based on the presence of a signal as opposed to absence of signal; hence, we designed a pair of guide RNAs to detect the sequences harboring each of these selected mutation/s. One of the guides of a pair is labeled as Reference-specific guide and another one is labeled as Omicron-specific guide. The Reference-specific guide RNA is designed to give a positive signal in trans-cleavage assay if the selected mutation/s is absent in the target, and the Omicron-specific guide RNA is designed to give a positive signal in a trans-cleavage if the selected mutation/s is present in the target. (Figure 1a). To test the selectivity of the designed guide RNA pairs to discriminate Omicron from non-Omicron variants experimentally, the target region that harbors the mutation/s of interest, was amplified using one step RT-PCR with a suitable primer from synthetic RNA controls of indicated SARS-CoV-2 variant. The amplified product was used as target or input for the trans-cleavage assay. A guide pair qualified for being able to differentiate the Omicron from not Omicron targets, if a trans-cleavage assay done with the Omicron-specific guide showed at least 5 fold higher signal in presence of the target amplified from Omicron synthetic RNA control than in the presence of the target amplified from reference synthetic RNA control after 1 hour of the assay. Also, the trans-cleavage assay done using the “Reference-specific guide” showed at least a 5 fold higher signal in the presence of the target amplified from reference synthetic RNA control than in the presence of the target amplified from Omicron synthetic RNA control after 1 hour of the assay. Some of these designed guide pairs were indeed found to be able to differentiate Omicron from non-Omicron synthetic controls. Our selected guides showed the ability to differentiate Omicron from non-Omicron in end-point assays where we used ~1*105 copies of synthetic RNA control as input (Figure 1b, c). However, at higher target concentrations the rates of trans-cleavage are expected to be faster and the reactions for both Reference-specific and Omicron-specific guides may reach saturation after 1 hour of the assay, irrespective of the input RNA sample being Omicron or non-Omicron. Hence, it was important to evaluate if the selectivity of the guide RNA will be retained at higher input RNA concentrations in these end-point assays. The input to trans-cleavage assay is a DNA fragment amplified from the input RNA sample after RT-PCR amplification step. The maximum amount of the target DNA that can be expected at the end of RT-PCR amplification is equal to primer concentration used in the RT-PCR. In OmiCrisp, 250 nM of each primer is used for amplification; hence the highest possible concentration of the amplified product expected is 250 nM. 5 µL of the amplified product is employed in about 50 µL of reaction for trans-cleavage assay, so the highest possible concentration of the target DNA expected is 25 nM in the trans-cleavage reaction. So, it is reasonable to believe if a guide pair showed selectivity in the trans-cleavage assay performed in the presence of 30 nM of the synthetic DNA targets as inputs, the selectivity would be retained for any amount of viral RNA present in the unknown sample to be tested by the two-step assay. Synthetic DNA templates corresponding to the amplicon region harboring the mutation/s of interest were synthesized as described in the materials and methods section. The trans-cleavage assays were done using these synthetic DNA targets (30 nM) as input for the selected guide pair. The guide RNA pair was defined as suitable for the end-point assay if a trans-cleavage assay done with Omicron-specific guide showed at least 5 fold higher signal in the presence of Omicron synthetic DNA target (30nM) than in the presence of the reference synthetic DNA target (30 nM) after one hour of the assay, and trans-cleavage assay done with Reference-specific guide showed at least 5 fold higher signal in the presence of Reference synthetic DNA target (30 nM) than in the presence of the Omicron synthetic DNA (30 nM) after one hour of the assay. Two guide pairs that retained selectivity at high target DNA concentration: one pair targets a region on S gene and another targets the region on N gene (Figure 1b,c). The S gene guide pair that we selected targets a region on S gene that harbors a set of three mutations; Q493R, G496S, and Q498R. The BA.2 variant of Omicron does not harbor the G496S mutation. Interestingly, we observed that both the reference-specific and Omicron-specific guide of this guide pair can induce trans-cleavage in the presence of the synthetic DNA template corresponding to BA.2. So, the samples that will show trans-cleavage in the presence of both Omicron- and Reference specific guides will still be Omicron. We will have to keep testing for all the evolving variants what the results will be if they do not harbor all three mutation/s. See, methods sections for the detailed calculations for the variant prediction of the assay. The guide pair for the N gene that we selected targets a region on the N gene that harbors mutation NΔ31-33 (Figure 1b). In the first version of OmiCrisp, OmiCrisp_v1, we did not include the N gene guide pair for detection. As described ahead, upon completion of the validation of OmiCrisp_v1 we observed that S gene gives lower or undetectable signals at low sample loads, see figure 2, hence compromising the sensitivity of assay of SARS-CoV-2 prediction. To overcome this limitation, we included a guide pair for the N gene in our version 2 of the OmiCrisp (Figure 1c).
Figure 1: a) Two step workflow involving amplification and detection of the OmiCrisp assay. Detection reagents are color coded based on the guide sequence present in the detection mix. The RT_NTC is a no sample RT-PCR amplification reaction that is added to all the wells of the indicated row. S1-S6 are RT-PCR amplified product of the clinical samples. Det_pos is synthetic template for testing the quality of detection reagents. Det_neg is nuclease free water added to all the wells of the indicated row b) Trans-cleavage assay done at saturated concentration of the indicated DNA template with indicated guide RNA. This shows the selectivity of the guide pair for N and S gene to discriminate Omicron variant from non-Omicron variant is retained at saturating target DNA concentrations. c) The analytical validation of the OmiCrisp with specified synthetic RNA as input at indicated concentrations. Note: OmiCrisp_v1 did not have Ngene_om and Ngene_ref guide RNAs containing detection reagents, hence only detection was done in presence of only four different detection reagents for it.
Primer design
We designed the primers that could amplify the regions of interest, harboring the target of the guide RNA to be used for detection of S, ORF1ab, and N genes. To ensure that these primers will be applicable for the majority of SARS-CoV-2 variants, we designed consensus primers for a group of 23 variants of SARS-CoV-2 that represent measure mutation constellations of SARS-CoV-2, using NCBI primer BLAST (SI Table S2 for the variants selected for primer design). To estimate the applicability of these primers for Indian isolates of SARS-CoV-2, we aligned these primers with all the complete genome sequences of SARS-CoV-2 Indian isolates deposited at NCBI. We estimated the frequency of isolates that have mis-matches with primer. We observed that 99 % of these isolates have no mis-matches with the designed primers. The sequence of primers and the frequency of isolated mis-matches for each prime is provided in SI Table S3. Also, the N gene primer was modified to insert a PAM site in the amplicon because the mutation/s of interest in the N gene did not have a PAM site closer than 6 bases from it.
Assay validation
Figure 1a shows the workflow of the two-step assay. Step-1 or the multiplexed amplification was carried out in PCR tubes. RT-NTC control is included at this step where nuclease free water is added to the indicated tube instead of a sample. Step-2 or detection step was carried out in a 96-well plate (Figure 1a). Briefly, a fragment of interest of ORF1ab, N, S, and human RNaseP gene was amplified from the sample to be tested using about 5 µL of the sample to be tested in 70 µL of the reaction volume using multiplexed one step RT PCR as described in materials and methods section. In the second step, 5 µL of the amplified product was used as an input for end-point trans-cleavage assay in the presence of 6 different detection reagents containing the following guide RNAs: 1. ORF1ab (ORF1abref + ORF1abom) 2. Som 3. Sref 4. Nref 5. Nom_om and 6. RNAseP (SI table 5 for guide RNA sequences). Trans-cleavage reaction in the presence of each guide RNA was done in duplicate, resulting in a total of 12 independent trans-cleavage reactions for each sample to be tested. OmiCrisp_v1 did not have Nom and Nref guide, hence detection was done in presence of only four different detection reagents leading to total 8 independent trans-cleavage for each sample.
Analytical validation
For analytical validation a known number of copies of a given synthetic RNA control was used as a sample to be assayed. As can be seen in Figure 1C when 100 copies of either Omicron or Reference synthetic RNA control samples were used as input to the assay a detectable signal higher than RT-NTC was observed at the end of the assay; hence the limit of detection of the assay was expected to be at least 100 copies of RNA. It was observed that the Reference synthetic controls samples showed more than 2 times higher signal in the trans-cleavage reactions that had Reference-specific guides (Sref or Nref) than in the trans-cleavage reactions that has Omicron-specific guide of corresponding gene (Som or Nom), and Omicron synthetic controls samples showed more than 2 times higher signal in the trans-cleavage reactions that had Omicron-specific guides (Som or Nom) than in the trans-cleavage reactions that had Reference-specific guide of corresponding gene (Sref or Nref). Hence, the N gene and S gene guide pair were able to discriminate the Omicron synthetic RNA from reference synthetic RNA at all the concentration of input RNA tested using the assay (Figure 1b, c).
Clinical validation
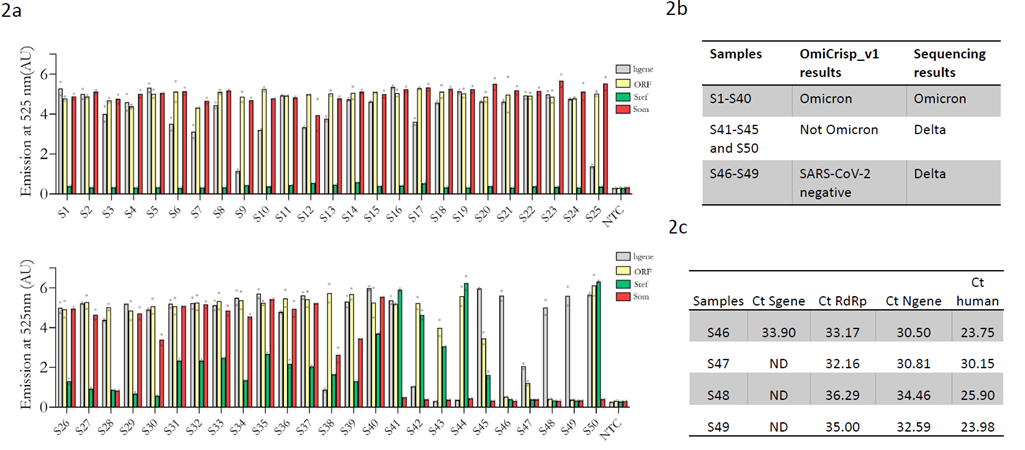
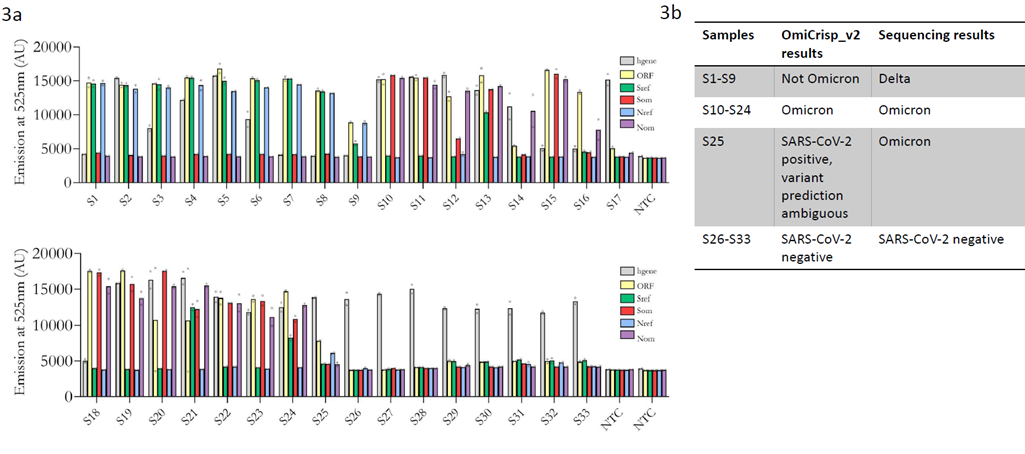
We validated the OmiCrips_v1 in RNA samples extracted from nasopharyngeal swabs that were previously sequenced and identified as SARS-CoV-2 positive. These blinded identity samples were provided by Strand Life Sciences, an accredited NGS genomic surveillance center in Bengaluru, India. As mentioned previously, the first version of OmiCrisp, OmiCrisp_v1, comprised four guide RNAs. Three of them are for SARS-CoV-2: ORF1ab, S gene Omicron-specific, S gene reference-specific, and one for human gene: RNase P gene. For clinical validation 5 µL of extracted RNA from a clinical sample was used as an input to the assay. Out of the 50 samples tested, only 46 were detected as SARS-CoV-2 positive by OmiCrisp_v1; therefore, the sensitivity of the assay for detecting SARS-CoV-2 was 92 %. Among the 46 samples detected as SARS-CoV-2 positive by OmiCrisp_v1, 40 were identified as Omicron and 6 were identified as non-Omicron. All the samples identified as Omicron by OmiCrisp_v1 were indeed Omicron, the sample identified as “non-Omicron” were Delta (non-Omicron) as per sequencing results. Therefore, the specificity of OmiCrisp_v1 for distinguishing Omicron from non-Omicron variants is 100 % (Figure 2a, 2b). OmiCrisp_v1 had a good selectivity to call out Omicron from non-Omicron variants; however, in order to improve the sensitivity of calling SARS-CoV-2 positive, we closely looked at the data of 4 samples that were identified as SARS-CoV-2 negative. We observed that two of these samples have unambiguously higher signals over no template controls for ORF1ab gene. But because the S gene signal was low, these samples were identified as negative (Figure 2a). Next, we estimated the Ct values of all the four samples using a commercially available rtPCR kit that targets S gene, N gene, and RdRP gene (Figure 2c). Interestingly, in three out of these four samples S-gene was not detectable, and the Ct values of the N-gene and RdRp were higher than 32 in these samples. This indicates lower viral load or degraded RNA in these samples. Based on the above data, to enable detection at viral lower loads, we introduced two changes to OmiCrisps_v1 to build the improved OmiCrisp_v2. First, we increased the number of PCR cycles from 35 to 40 at the amplification step, and we introduced a new SARS-CoV-2 at a q N-gene to the assay. We validated the Omcrisp_v2 on a total of 33 RNA samples extracted from nasopharyngeal swabs. This validation was done at inStem and conducted by the inStem technicians (Bengaluru, India) in a blinded fashion. The unprocessed data with blinded sample identities was handed over to CrispBits by InStem (Figure 3a). CrisprBits analyzed the data and made predictions based on OmiCrisp_v2 (Figure 3b). We developed an automated pipeline for the data analyses of OmiCrisp assay that is independent of technician’s subjective error. The details are in the method section. Out of 33 samples 25 samples were SARS-CoV-2 positive and 8 samples were SARS-negative. OmiCrisp_v2 correctly identified the positive sample as SARS-CoV-2 positives and negative samples as SARS-CoV-2 negative. Hence, the specificity and sensitivity of OmiCrisp_v2 assay in identifying SARS-CoV2 in this validation study were both at 100 %. Out of 25 positive samples, 9 samples were Delta variants of the SARS-CoV-2 virus and OmiCrisp_v2 correctly identified them as Non-omicron. Among the remaining 16 samples that were Omicron 15 were accurately identified as Omicron. One of the Omicron samples was labeled as ambiguous by OmiCrisp_v2 because it was identified as Omicron with S gene guide pair and as non-Omicron as N gene guide pair. The validation results indicate that the OmiCrisp platform is suitable for tracking the Omicron variant of SARS-CoV-2 variant in clinical samples.
Figure 2: Clinical validation of OmiCrisp_v1. a) Bar graph showing the fluorescence intensity of the indicated probe or guide RNA for each sample. The height of the bar represents the background subtracted mean intensity of the technical duplicates in arbitrary fluorescence units (AU), and the empty circle overlaid on bars indicate the background subtracted intensity of the individual technical duplicates. b) Tabular summary of the clinical validation, showing the comparison between sequencing and Omcrisp_v1. c) qRT-PCR analysis of the sample identified as SARS-CoV-2 negative by the OmiCrisp_v1.
Figure 3: Clinical validation of OmiCrisp_v2. a) Bar graph showing the average fluorescence intensity of the indicated probe or guide RNA for each sample. The height of the bar represents the mean intensity (arbitrary fluorescence unit (AU) of the technical duplicates, and the empty circle overlaid on bars indicate the intensity of the individual technical duplicate. b) Tabular summary of the clinical validation, showing the comparison between sequencing and OmiCrisp_v2.
*validation was performed in an external laboratory where the qPCR instrument was used to measure end point fluorescence of the CRISPR assay
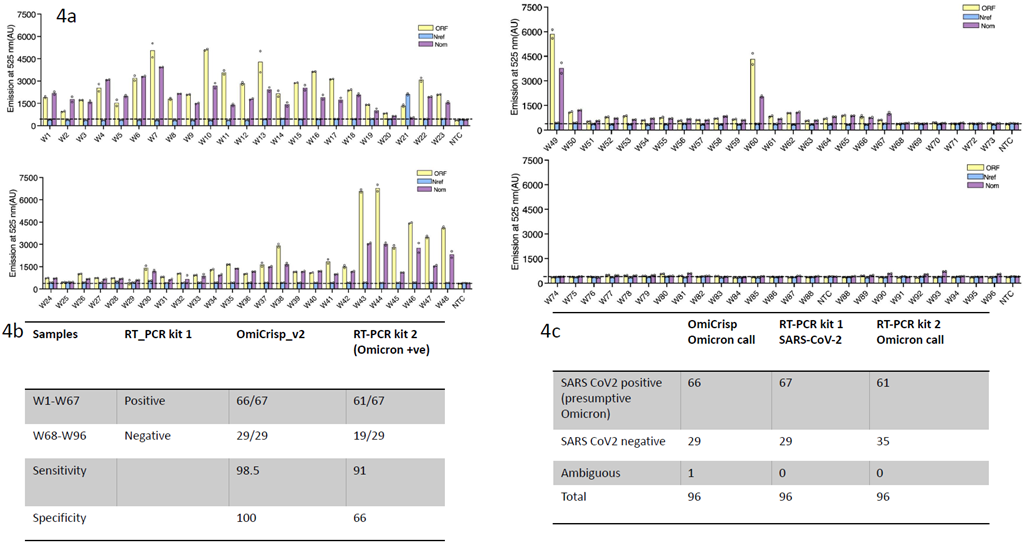
Figure 4: OmiCrisp_v2 testing on wastewater samples. a) Bar graph showing the fluorescence intensity of the indicated probe or guide RNA for each sample. The height of the bar represents the mean intensity of the technical duplicates, and the empty circle overlaid on bars indicate the intensity of the individual technical duplicate. b) Tabular summary showing the comparison between RT-PCR kit 1 and 2 and Omcrisp_v2. The table shows the concordance between the positive and negative samples called by various kits. The sensitivity and specificity of the OmiCrisp v2 test and the corresponding Omicron RT-PCR test are listed. c) Tabular summary showing total samples tested and their results in each test.
Waste-water sample validation for surveillance
To validate if our OmiCrisp could detect the virus, including the Omicron variant in pooled environmental samples such as waste water, we validated our test first in 166 waste-water samples, collected and tested (via qRT-PCR) simultaneously by a surveillance group in Bengaluru. Due to the constant evolution of the virus, the Omicron distinctive mutations in the S gene could not be used for wastewater surveillance study which was performed on samples collected between December 2021 and Jan 2023. Hence for the surveillance study, we restricted our analysis to the Orf1ab and the N gene mutations, with the N gene mutations discerning the Omicron variant from previous lineages of SARS-CoV-2. We first standardized our assay on 70 archived samples from 1 month to 6 months old. A total of 40 RT-PCR positive and 30 RT-PCR negatives for SARS-CoV-2 samples were selected (Figure S2). Samples were selected such that they reflect a range of quality. Some representative samples from good (Ct<25), medium (Ct 25-32)and poor (Ct 32-35) quality were selected and run using our OmiCrisp v2 assay, The assay detected 38/40 positive samples and 28/30 negative samples accurately. Since the signals for sewage samples are several folds (4-10) fold lower than clinical samples, we fine-tuned our algorithm to set our positive signal as 3 standard deviations (+3 SD) of pooled NTC signals for a marker, across all the batches of samples (See Methods). We then confirmed our OmiCrisp v2 assay in a fresh set of 96 samples- 67 positive samples and 29 negative samples (collected between October 2022 - Feb 2023). These samples were simultaneously cross validated for the presence or absence SARS-CoV-2 (TruNAT, CoviSure) as well as Omicron lineage confirmation (CoviDelta kit). We observed that 66 of 67 RT-PCR positive samples were also labeled positive by OmiCrisp v2 (Figure 4a,b,c). The single sample with an ambiguous result, 27DESWFM001, had one marker detected as positive (N) and one as negative (Orf1ab). All 29 RT-PCR negative samples were also confirmed as “negative” by the OmiCrisp v2 assay. Thus validation on sewage samples resulted in 98.5% sensitivity and 100% specificity for detecting SARS-CoV2 by our assay. Since sewage samples yield inferior quality of RNA, confirming their identity by sequencing was not feasible and 75% samples failed sequencing. Hence to confirm the identity of the Omicron variants in these 96 samples, they were tested using the CoviDelta kit which could distinguish the Omicron variant from the previously identified Delta lineage. At first glance, concordance was only 87.5% (84/96) for the Omicron variant detection between OmiCrisp v2 and the CoviDelta qRTPCR platforms. 10 samples were called as Omicron positive with RT-PCR, by virtue of any one of two markers (RdRp, and N gene) showing Ct< 35. However, all these 10 samples were negative not only by OmiCrisp v2, but also were called as negative using the previously described Covisure / TruNAT RT-PCR kits for SARS-CoV-2 detection. The two discrepant samples that were called negative by CoviDelta but positive by OmiCrisp v2 were also found positive by the previously described CoviSure/ TruNAT tests. Thus we observe high concordance between the RTPCR based CoviSure / TruNat qPCR platforms and CRISPR based OmiCrisp v2, than two RTPCR platforms themselves.
Conclusions
We developed a CRISPR based assay, OmiCrisp, that can predict the presence of Omicron or non-Omicron variant in extracted RNA samples. We validate the results of the assay on RNA samples extracted from human nasopharyngeal swabs as well as from environmental samples like sewage effluents. We observed that OmiCrisp is highly reliable in identifying the variants in clinical samples when compared to sequenced samples. The assay interpretation is streamlined, easily automated, and does not require any subjective decisions to be made by technicians performing the assay. The clinical validation of the assay was performed at an external laboratory indicating that the assay is amenable to technology transfer. The data from the study on detecting the virus and the Omicron variants in sewage samples indicate equally compelling application of the test in environmental surveillance. Kumar et al have also reported the development of a CRISPR systems-based platform for SARS-CoV-2 variant detection that is based on using FnCas9, named RAY [14]. This is an endpoint assay, and in any end-point assay, specificity is compromised as well as the selectivity of the variant can be lost if the concentration of input RNA is high. However, the validation for the study was performed on samples with Ct values above 22, indicating higher concentration of target DNA in these samples. The Ct values in samples from SARS-CoV-2 positive patients for ~20-50 % of samples is below 22 [24, 25]. We have designed OmiCrisp to ensure the variant selectivity will not be compromised at very high viral loads (Figure 1b). Another interesting CRISPR based platform for variant detection is miSHERLOCK which is based on Cas13a, where the input is saliva samples from prospective patients and nucleic acid extraction is done automatically in the device that is included in the platform. However, the platform performance has not been shown in clinical samples, and the variant calling specificity has not been tested at higher concentrations of RNA [26]. Fasching et al identified a novel Cas12a enzyme, CasDx1, for improved SNP detection and showed its application in SARS-CoV-2 variant detection [20]. Again, there is no explicit experiment evidence showing the selectivity of guide RNAs at higher concentrations on the input samples. The agreement of sequencing and the assay for the variant identification varies from 83% (Alpha) - 97 % (WT). OmiCrisp is a CRISPR based assay that can be used for the detection of SARS-CoV-2 Omicron variants. OmiCrisp depends on the presence of a positive signal to call out a sample either Omicron or non-Omicron that ensures that the assay is more reliable for variant prediction even in samples with lower viral load. An assay that labels a sample as Omicron or non-Omicron based on absence of signal from a given gene will have very low specificity for Omicron. For example, an RT-PCR that identifies non-Omicron based on absence of S gene signal would have an identified sample 46, 47, 48 used in OmiCrisp_v1 validation as Omicron whereas in reality they were non-Omicron (Figure 2c) [27, 28].
The design, development and validation of a nucleic acid-based assay, like OmiCrisp, for variant detection has rapid turnaround time and relatively lower investment costs in the R&D. Hence, these types of assays can complement ongoing sequencing efforts in a facility for quick screening of samples before selecting them for sequencing or to validate suspicious sequencing results. Furthermore, the OmiCrisp assay can be done with relatively lower quality and quantity of nucleic acid samples than that required for NGS. This is particularly exemplified in case of wastewater samples which are bound to be poor quality and as expected, majority daily sequencing. Our data on OmiCrisp shows that OmiCrisp and similar CRISPR based assays are a choice for screening of variants in relatively poor-quality samples that fail in NGS. From the onset, environmental samples, specifically wastewater samples, have shown great promise in getting the early warning of the upcoming peak [29]. Furthermore, waste water surveillance is not dependent on access to healthcare and testing capacity of clinical labs [30]. However, an environmental sample exposed to various conditions that can degrade nucleic acids are not expected to yield very high-quality RNA, and as well as represent a pooled matrix, where the signal will be quite diluted. This is evident in that the signal is at least 4-5-fold higher in clinical nasopharyngeal samples compared to sewage samples. Indeed we also observe that in sewage samples, the human gene RNA, shows degradation in a number of samples, even though the signal from viral gene RNA is significantly above background. Consequently we do not consider human RNA control as an indicator of sample integrity. As such, sewage samples cannot yield highly pure or concentrated samples as input. While our study successfully uses conventional RT-PCR for amplification of nucleic acid, followed by CRISPR detection, the latter can be coupled with isothermal amplifications like RT- LAMP (loop mediated isothermal amplification) or RT-RPA (recombinant polymerase amplification) which tolerate nucleic acids from crude extracts and relatively poor quality. At the same time the assay is sensitive enough to detect the virus in a pooled matrix. Thus, OmiCrisp is an example of an accurate, rapid and cost-effective platform that can be helpful in tracking fast evolving pathogens. We postulate that OmiCrisp and similar CRISPR assays can be utilized for monitoring of pathogens and their variants in both clinical and environment samples for variant tracking, and thus extending their use to pandemic surveillance.
Acknowledgements
We acknowledge the C-CAMP-InDx Program (Bengaluru, India) for providing us resources and technical support for analytical and clinical validation of the OmiCrisp assay. The validation of the OmiCrisp has been conducted with the significant contribution and expertise of the DBT-inStem biorepository and the COVID testing laboratory (Bengaluru, India). STRAND life sciences (Bengaluru, India) provided validation support for the work. The testing of validation of the OmiCrisp assay on environmental sewage samples was supported by a grant from ACT COVID Response Collective, IN COVID Support FZE and Blockchain for Impact (formerly CryptoRelief). We acknowledge Mr Lalith Kishore (CEO-InDx), Dr. Pooja Agrawal (Program Director- Biomedical Research and Innovation, Blockchain For Impact), and Dr. Chitra Pattabhiraman (ART-Park) for their scientific inputs and discussions.
References
- Vindeirinho JM, Pinho E, Azevedo NF, et al. SARS-CoV-2 Diagnostics Based on Nu-cleic Acids Amplification: From Fundamental Concepts to Applications and Beyond. Front Cell Infect Microbiol 12 (2022).
- Vandenberg O, Martiny D, Rochas O, et al. Considerations for diag-nostic COVID-19 tests. Nat Rev Microbiol 19 (2021): 171-183.
- Little S. Amplification-Refractory Mutation System (ARMS) Analysis of Point Mutations. Curr Pro-toc Hum Genet 7 (1995): 9.8.1-9.8.12.
- Morlan J, Baker J, Sinicropi D. Mutation Detection by Real-Time PCR: A Simple, Robust and Highly Selective Method. PLoS ONE 4 (2009): e4584.
- Environmental surveillance for SARS-COV-2 to complement public health surveillance-Interim Guidance (2022), World Health Organization. Environmental surveillance for SARS-COV-2 to complement public health surveil-lance – Interim Guidance (who.int)
- Li X, Kulandaivelu J, Guo Y, et al. SARS-CoV-2 shedding sources in wastewater and implications for wastewater-based epidemiology. J Hazard Mater 432 (2022): 128667.
- Klapsa D, Wilton T, Zealand A, et al. Sustained detection of type 2 poliovirus in London sewage between February and July, 2022, by enhanced environmental surveillance. Lancet 400 (2022): 10362 (1531-1538).
- Azhar Mohd, Phutela R, Kumar M, et al. Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens Bioelectron 183 (2021): 113207.
- Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol 38 (2020): 870-874.
- Ding X, Yin K, Li Z, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun 11 (2020): 4711.
- Fozouni P, Son S, Díaz de León Derby M, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184 (2021): 323-333.
- Guo L, Sun X, Wang X, et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov 6 (2020): 1-4.
- Joung J, Ladha A, Saito M, et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N Engl J Med 383 (2020): 1492-1494.
- Kumar M, Gulati S, Ansari AH, et al. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. ELife 10 (2021): e67130.
- Lucia C, Federico P-B, Alejandra GC. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas 12. (2020).
- Nouri R, Tang Z, Dong M, et al. CRISPR-based detection of SARS-CoV-2: A review from sample to result. Biosens Bioelectron 178 (2021): 113012.
- Patchsung M, Jantarug K, Pattama A, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng 4 (2020): 1140–1149.
- Safari F, Afarid M, Rastegari B, et al. CRISPR systems: Novel approaches for detection and combating COVID-19. Virus Res 294 (2021): 198282.
- Wang R, Qian C, Pang Y, et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 172 (2021): 112766.
- Fasching CL, Servellita V, McKay B, et al. COVID-19 Variant Detection with a High-Fidelity CRISPR-Cas12 Enzyme. J Clin Microbiol 60 (2022): e00261-22.
- Niu M, Han Y, Dong X, et al. Highly Sen-sitive Detection Method for HV69-70del in SARS-CoV-2 Alpha and Omicron Variants Based on CRISPR/Cas13a. Front Bioeng Biotechnol 10 (2022): 831332.
- Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360 (2018): 436-439.
- Li S-Y, Cheng Q-X, Wang J-M, et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 4 (2018): 1-4.
- Buchan BW, Hoff JS, Gmehlin CG, et al. Distribution of SARS-CoV-2 PCR Cycle Threshold Values Provide Practical Insight into Overall and Target-Specific Sensitivity Among Symptomatic Patients. Am J Clin Pathol Aqaa 133 (2020).
- Platten M, Hoffmann D, Grosser R, et al. SARS-CoV-2, CT-Values, and Infectivity-Conclusions to Be Drawn from Side Observations. Viruses 13 (2021): 1459.
- de Puig H, Lee RA, Najjar D, et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv 7 (2021): eabh2944.
- Thermo Fisher S. Thermo Fisher Scientific Confirms Detection of SARS-CoV-2 in Samples Containing the Omicron Variant with its TaqPath COVID-19 Tests. (2021).
- Vanaerschot M, Mann SA, Webber JT, et al. Identification of a Polymorphism in the N Gene of SARS-CoV-2 That Adversely Impacts Detection by Reverse Transcription-PCR. J Clin Microbiol 59 (2020): e02369-20.
- Zhu Y, Oishi W, Maruo C, et al. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci Total Environ 767 (2021): 145124.
- Kirby AE, Walters MS, Jennings WC, et al. Using Wastewater Surveillance Data to Support the COVID-19 Response - United States, 2020-2021. MMWR Morb Mortal Wkly Rep 70 (2021): 1242-1244.