NMF Clustering: Accessible NMF-based Clustering Utilizing GPU Acceleration
Article Information
Ted Liefeld1*, Edwin Huang1, Alexander T Wenzel1, Kenneth Yoshimoto2, Ashwyn K Sharma3, Jason K Sicklick4, Jill P Mesirov1,3, Michael Reich1
1University of California San Diego, Department of Medicine, School of Medicine, La Jolla, CA, 92093, USA
2University of California San Diego, San Diego Supercomputer Center, La Jolla, CA, 92093, USA
3University of California San Diego, Moores Cancer Center, La Jolla, CA, 92093, USA
4University of California San Diego, Departments of Surgery and Pharmacology, Moores Cancer Center,La Jolla, CA, 92093, USA
*Corresponding author: Ted Liefeld, University of California San Diego, Department of Medicine, School of Medicine, La Jolla, CA, 92093, USA.
Received: 28 November 2023; Accepted: 06 December 2023; Published: 21 December 2023
Citation: Ted Liefeld, Edwin Huang, Alexander T Wenzel, Kenneth Yoshimoto, Ashwyn K Sharma, Jason K Sicklick, Jill P Mesirov, Michael Reich. NMF Clustering: Accessible NMF-based Clustering Utilizing GPU Acceleration. Journal of Bioinformatics and Systems Biology. 6 (2023): 379-383.
View / Download Pdf Share at FacebookAbstract
Non-negative Matrix Factorization (NMF) is an algorithm that can reduce high dimensional datasets of tens of thousands of genes to a handful of metagenes which are biologically easier to interpret . Application of NMF on gene expression data has been limited by its computationally intensive nature, which hinders its use on large datasets such as singlecell RNA sequencing (scRNA-seq) count matrices. We have implemented NMF based clustering to run on high performance GPU compute nodes using CuPy, a GPU backed python library, and the Message Passing Interface (MPI). This reduces the computation time by up to three orders of magnitude and makes the NMF Clustering analysis of large RNA-Seq and scRNA-seq datasets practical. We have made the method freely available through the GenePattern gateway, which provides free public access to hundreds of tools for the analysis and visualization of multiple ‘omic data types. Its web-based interface gives easy access to these tools and allows the creation of multi-step analysis pipelines on high performance computing (HPC) clusters that enable reproducible in silico research for non-programmers.
Keywords
Gene Expression, ScRNA-seq, Matrix Factorization, Bioinformatics
Gene articles; scRNA-seq articles; NMF articles
Gene Expression articles Gene Expression Research articles Gene Expression review articles Gene Expression PubMed articles Gene Expression PubMed Central articles Gene Expression 2023 articles Gene Expression 2024 articles Gene Expression Scopus articles Gene Expression impact factor journals Gene Expression Scopus journals Gene Expression PubMed journals Gene Expression medical journals Gene Expression free journals Gene Expression best journals Gene Expression top journals Gene Expression free medical journals Gene Expression famous journals Gene Expression Google Scholar indexed journals ScRNA-seq articles ScRNA-seq Research articles ScRNA-seq review articles ScRNA-seq PubMed articles ScRNA-seq PubMed Central articles ScRNA-seq 2023 articles ScRNA-seq 2024 articles ScRNA-seq Scopus articles ScRNA-seq impact factor journals ScRNA-seq Scopus journals ScRNA-seq PubMed journals ScRNA-seq medical journals ScRNA-seq free journals ScRNA-seq best journals ScRNA-seq top journals ScRNA-seq free medical journals ScRNA-seq famous journals ScRNA-seq Google Scholar indexed journals Matrix Factorization articles Matrix Factorization Research articles Matrix Factorization review articles Matrix Factorization PubMed articles Matrix Factorization PubMed Central articles Matrix Factorization 2023 articles Matrix Factorization 2024 articles Matrix Factorization Scopus articles Matrix Factorization impact factor journals Matrix Factorization Scopus journals Matrix Factorization PubMed journals Matrix Factorization medical journals Matrix Factorization free journals Matrix Factorization best journals Matrix Factorization top journals Matrix Factorization free medical journals Matrix Factorization famous journals Matrix Factorization Google Scholar indexed journals Bioinformatics Analysis articles Bioinformatics Analysis Research articles Bioinformatics Analysis review articles Bioinformatics Analysis PubMed articles Bioinformatics Analysis PubMed Central articles Bioinformatics Analysis 2023 articles Bioinformatics Analysis 2024 articles Bioinformatics Analysis Scopus articles Bioinformatics Analysis impact factor journals Bioinformatics Analysis Scopus journals Bioinformatics Analysis PubMed journals Bioinformatics Analysis medical journals Bioinformatics Analysis free journals Bioinformatics Analysis best journals Bioinformatics Analysis top journals Bioinformatics Analysis free medical journals Bioinformatics Analysis famous journals Bioinformatics Analysis Google Scholar indexed journals NMF articles NMF Research articles NMF review articles NMF PubMed articles NMF PubMed Central articles NMF 2023 articles NMF 2024 articles NMF Scopus articles NMF impact factor journals NMF Scopus journals NMF PubMed journals NMF medical journals NMF free journals NMF best journals NMF top journals NMF free medical journals NMF famous journals NMF Google Scholar indexed journals GPU articles GPU Research articles GPU review articles GPU PubMed articles GPU PubMed Central articles GPU 2023 articles GPU 2024 articles GPU Scopus articles GPU impact factor journals GPU Scopus journals GPU PubMed journals GPU medical journals GPU free journals GPU best journals GPU top journals GPU free medical journals GPU famous journals GPU Google Scholar indexed journals GenePattern articles GenePattern Research articles GenePattern review articles GenePattern PubMed articles GenePattern PubMed Central articles GenePattern 2023 articles GenePattern 2024 articles GenePattern Scopus articles GenePattern impact factor journals GenePattern Scopus journals GenePattern PubMed journals GenePattern medical journals GenePattern free journals GenePattern best journals GenePattern top journals GenePattern free medical journals GenePattern famous journals GenePattern Google Scholar indexed journals RNA-Seq articles RNA-Seq Research articles RNA-Seq review articles RNA-Seq PubMed articles RNA-Seq PubMed Central articles RNA-Seq 2023 articles RNA-Seq 2024 articles RNA-Seq Scopus articles RNA-Seq impact factor journals RNA-Seq Scopus journals RNA-Seq PubMed journals RNA-Seq medical journals RNA-Seq free journals RNA-Seq best journals RNA-Seq top journals RNA-Seq free medical journals RNA-Seq famous journals RNA-Seq Google Scholar indexed journals Python articles Python Research articles Python review articles Python PubMed articles Python PubMed Central articles Python 2023 articles Python 2024 articles Python Scopus articles Python impact factor journals Python Scopus journals Python PubMed journals Python medical journals Python free journals Python best journals Python top journals Python free medical journals Python famous journals Python Google Scholar indexed journals
Article Details
1. Background
Non-negative matrix factorization (NMF) [1] has proved to be a very effective technique for decomposing array-based biological data into meaningful components. In gene expression analysis, NMF is used to describe the activity of tens of thousands of genes in a typical gene expression dataset in terms of a small number, k, of metagenes, i.e., positive linear combinations of genes [2]. Biological samples data can then be summarized as expression patterns of their metagenes. Compared with other dimension reduction techniques such as Principal Components Analysis (PCA), the components identified by NMF are more easily related to actual biological processes and mechanisms due to the non-negativity constraint.
However, the use of NMF has been limited by its computational requirements. NMF is a non-convex optimization problem that requires many iterations to converge to a solution, and the solution is not guaranteed to be a global minimum. Compounding this problem is that the optimal number of clusters, k, is seldom known in advance. Therefore, when NMF is used to cluster data, it is run many times with varying initial conditions over a range of values of k.
Cluster stability metrics such as the cophenetic correlation coefficient [3] or the silhouette score [4] are then used to identify the value of k that corresponds to the most stable clustering.
The result is that on top of the computational complexity of NMF, to perform clustering it must be run many thousands of times for a given analysis. With the current NMF-based methods it is not unusual to encounter analyses that require days to run on a dataset as small as 1000 cells by 5000 transcripts. For larger gene expression or single-cell RNA-Seq (scRNASeq) datasets, use of NMF becomes impractical, requiring potentially weeks of computing time on large-memory supercomputers.
Graphical Processing Units (GPUs) have become a popular and cost-effective way to speed up computation. GPUs are limited in memory compared to CPUs but are designed for processing large blocks of data in parallel and are particularly well-suited to matrix operations, which are the basis of the NMF algorithm. Via GPU application programming interfaces (APIs) such as the nVidia Compute Unified Device Architecture (CUDA) [5], general purpose algorithms can be executed in parallel using GPUs to speed computation. To further parallelize computation across multiple GPUs, parallel programs can be synchronized using the Message Passing Interface (MPI).
Mejía-Roa [6] demonstrated NMF-mGPU, a C-language implementation of NMF that used CUDA and MPI to perform multi-GPU computation of NMF. This implementation allowed processing up to 120 times faster than conventional single-CPU implementations and demonstrated the capability of processing matrices up to 54000 x 2000 in size. This implementation included the ability to split the NMF computation across multiple GPUs to permit the calculation to be performed on datasets larger than could be held in GPU memory. However, this program was not easily usable by many biomedical sciences researchers as it required specialized programming skills and access to an HPC cluster. .
GenePattern [7], www.genepattern.org, is a gateway providing free access to cloud-based and HPC systems. GenePattern includes access to hundreds of analysis tools (modules) and visualization tools for multiple genomic data types without requiring any programming skills. It has a web-based interface to provide easy access to these tools and allows the creation of multi-step reproducible in silico research workflows. These can be implemented as GenePattern analysis pipelines, or alternatively as Jupyter [8] Notebooks via the GenePattern Notebook Environment [9]. The publicly available GenePattern servers currently support thousands of users and log up to tens of thousands of analyses each month which are executed on clusters in the AWS Cloud or on the Expanse [10] supercomputer cluster at the UC San Diego Supercomputing Center (SDSC). GenePattern has long included an R-language implementation of the NMF
Clustering algorithm as described in Brunet et al. as the NMFConsensus GenePattern module. This implementation served as a baseline for performance comparison.
To bring the power of the Meija-Roja NMF-GPU implementation to non-programming investigators, we have reimplemented their algorithm using CuPy [11] an open-source library for GPU-accelerated computing with Python. It is freely available on GenePattern as the NMFClustering module at https://genepattern.ucsd.edu and is also distributed as both open source code and as a Docker [12] container. For HPC clusters, the Docker container can be cross-compiled into Singularity [13] if required. By wrapping this implementation in GenePattern, we have made it available to all investigators without the need for any programming skills and have also provided access to GPU cycles for the processing of their data.
2. Results
We illustrate the improved performance of NMFClustering, our CuPy based NMF-GPU implementation, relative to the previous generation of NMFConsensus implemented in the R programming language by demonstrating its ability to handle much larger datasets, and by demonstrating its improved performance on smaller datasets. It is not possible to do a true performance comparison on larger datasets as the R language NMFConsensus implementation requires in excess of 2 days, the time limit for GPU jobs on the Expanse HPC cluster, to analyze datasets larger than 1,000 cells/samples. We note that most single cell data sets are much larger than this.
For our test dataset, we use a single-cell RNA-Seq dataset derived from patient samples of gastrointestinal stromal tumors (GIST). The complete dataset is a count matrix of 77020 cells and 28561 transcripts. To create datasets small enough to run on the R-language NMFConsensus, we subsetted this dataset, creating smaller versions with 5,000 transcripts and cell counts between 20 and 1,000. Additional larger datasets with 8,000 to 32,000 cells were run through NMFClustering in a CUDA configuration using a single node and both single and 2 GPUs allocated. Finally two larger datasets of 64,000, and 77,020 (the complete dataset) cells were created and run using a CUDA configuration of 4 nodes and 4 GPUs.
Using these datasets we ran the NMFConsensus module on GenePattern’s public cloud based server, which launched AWS C5 compute nodes of various types through AWS Batch using the SPOT_CAPACITY_OPTIMIZED strategy. Datasets larger than 1,000 cells were not run using the CPU based algorithm as they were unable to complete the analysis below the 48 hour time limit for most jobs on the SDSC Expanse system. The Expanse GPU nodes have dual 20 core Intel Xeon processors with 384 GB of memory and four NVIDIA V100 processors (32 GB SMX2). Each dataset size was run 4 times and the average elapsed time used for the following analysis.
For the NMFClustering code we ran the same datasets as were used for NMFConsensus using the GPU nodes of the SDSC Expanse supercomputer. Using a single GPU configuration we ran up to 8,000 cells successfully. The largest datasets consistently failed with exhausted GPU memory for the single GPU configuration. Since the NMFClustering implementation could split the dataset to compute only a portion on each GPU at one time, using MPI to coordinate, we could reduce the impact of limited GPU memory by adding GPUs to the runtime configuration. Therefore for the largest 4 datasets (16,000 through 77,020 cells) we ran using a quad-GPU (4 GPU) configuration which successfully completed a NMF-GPU run in under 7 hours on the largest dataset. The dataset size and runtimes, averaged over 4 runs, are displayed below in figure 1.
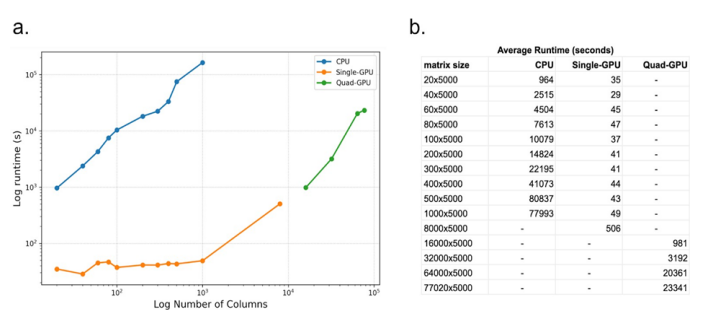
Figure 1: Plot (a) and table (b) of compute time vs number of cells (columns) for CPU-based NMFConsensus and multiple GPU configurations of NMFClustering. Each dataset contains 5000 rows and was run 4 times to ensure consistency of runtimes.
With 1000 cells (columns) and 5000 transcripts (rows), the largest dataset that could be computed on the CPU, the average execution time was 77993 seconds. The same dataset completed using NMFClustering on a single GPU in 49 seconds, 1588 times faster or in 0.062% of the time of the CPU version. The smallest improvement was for the smallest 20 sample dataset, where the GPU speedup was 27 times faster than the CPU version.
The NMFClustering (GPU-based) and NMFConsensus (R-based) implementations are available as modules on the public GenePattern server at https//genepattern.ucsd.edu. The R code for the NMFConsensus GenePattern module is available in github at https//github.com/genepattern/NMFConsensus. The Python code for the NMFClustering (GPU based) implementation is also available on github at https//github.com/genepattern/nmf-gpu.
Features
The NMFClustering module, and underlying Python code, allow the user to define the minimum and maximum values of k, the number of clusters. It also allows the user to select the number of NMF runs with varied initial conditions per value of k, called the “num clusterings” parameter as well as the maximum number of total iterations of NMF to run per clustering to prevent the process for running forever if it does not converge to a stable result. To control the convergence it provides parameters for the “stop convergence”, to specify how many "no change" checks are needed to stop NMF iterations before max iterations is reached along with the “stop frequency” parameter, which defines the frequency (number of NMF iterations) between ‘no-change’ checks. Finally the module also allows the user to define the maximum error difference for KL divergence.
For advanced users, the module allows the selection from three parallelization strategies. They are: (1) ‘serial’ in which no parallelization is used; (2) ‘kfactor’ where NMF runs on values of k are balanced sending the entirety of runs for one value of k to a single GPU; and (3) ‘Input matrix’ parallelization in which case the input matrix is broken up with each GPU computing on only a portion of the input as defined in Meija-Roja et al, 2015. The ‘input matrix’ serialization is required for datasets that are too large to fit in the relatively smaller memory space of the GPUs and is what we used for the larger datasets in the performance testing.
Outputs
For each value of k, NMFClustering outputs a consensus matrix, which contains the number of times each sample appears in the same cluster as each other sample. In a stable clustering, the same samples will cluster with each other a majority of the time. Consensus matrices are output in the GenePattern gct[14] tab delimited format. If the number of input columns is under 1,000 the module also provides sorted consensus matrices in gct format, and if less than 100 columns, the consensus matrices will be plotted and returned as a pdf file.
Documentation
Documentation for NMFClustering is available on github pages at https//genepattern.github.io/NMFClustering/v3/
Availability and Implementation:
NMFClustering is freely available on the public GenePattern server at https//genepattern.ucsd.edu. Code for the NMFClustering is available under a BSD style license on github at https:// github.com/genepattern/nmf-gpu.
Supplementary Information:
Supplementary data are available at Bioinformatics online and at
https://datasets.genepattern.org/?prefix=data/test_data/NMFClustering/.
Acknowledgements
The work was supported by the National Institutes of Health U24CA194107 to JPM, R50CA243876 to TL, F31CA257344 to ATW, R01CA226803 to JKS, and T32CA121938
fellowship to AKS. This work used the Expanse supercomputer at the San Diego Supercomputer Center through allocation MED220012 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by National Science Foundation (grant numbers #2138259, #2138286, #2138307, #2137603, and #2138296).
References
- Lee DD and Seung HS. Algorithms for non-negative matrix Adv Neur In 13 (2001): 556-562.
- Brunet Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A 101 (2004): 4164-4169.
- Farris On the Cophenetic Correlation Coefficient. Systematic Zoology. GenePattern 18 (1969): 279-285.
- Rousseeuw PJ. Silhouettes - a Graphical Aid to the Interpretation and Validation of Cluster-Analysis. J Comput Appl Math 20 (1987): 53-65.
- NVIDIA. CUDA Technology (2007). http://www.nvidia.com/CUDA.
- Mejia-Roa E et NMF-mGPU: non-negative matrix factorization on multi-GPU systems. Bmc Bioinformatics 16 (2015): 43.
- Reich M, et GenePattern 2.0. Nat Genet 38 (2006): 500-501.
- Ragan-Kelley M, et al. The Jupyter/IPython architecture: a unified view of computational research, from interactive exploration to communication and publication In (2014).
- Reich M, et al. The GenePattern Notebook Environment. Cell Syst 5 (2017): 149-151.
- Strande S, et al. Expanse: Computing without Boundaries. In, Practice and Experience in Advanced Research Boston, MA, USA: Association for Computing Machinery (2021): 1-4.
- Okuta R, et Cupy: A numpy-compatible library for nvidia gpu calculations. In, Proceedings of workshop on machine learning systems (LearningSys) in the thirty-first annual conference on neural information processing systems (NIPS) (2017).
- Boettiger An introduction to Docker for reproducible research. ACM SIGOPS Operating Sys Rev 49 (2015): 71-79.
- Kurtzer GM, Sochat V, Bauer MW. Singularity: Scientific containers for mobility of compute. Plos One 12 (2017): e0177459.
Supplementary Data:
NMFClustering User Interface
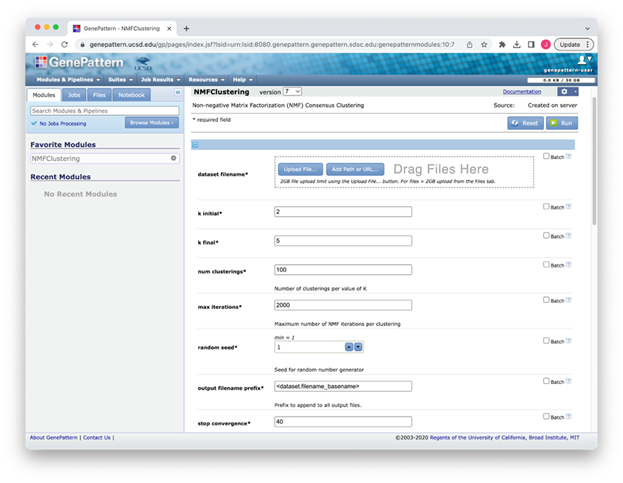
Figure S1: NMFClustering interface in GenePattern showing a subset of the available parameters
Example Consensus Matrix plot
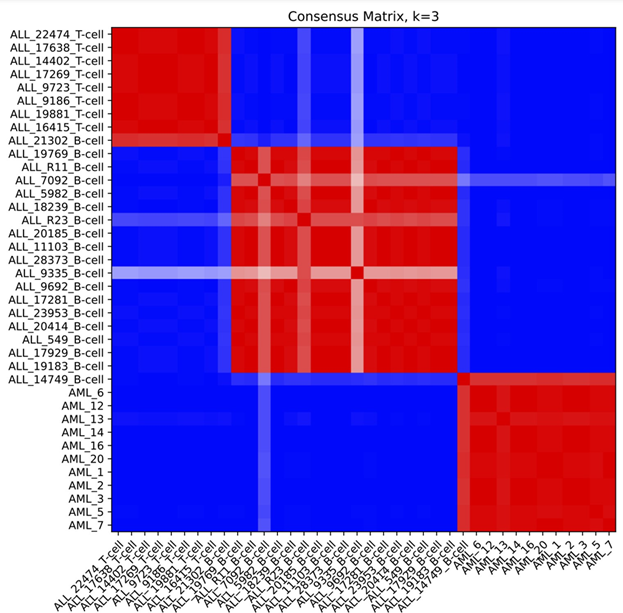
Figure S2: Example consensus matrix plot for k=3 using ALL_AML_data.gct dataset from the sample test data folder (below). Compare to figure 4, panel a in Brunet et al.
Sample Test Data
Test datasets are available at
https://datasets.genepattern.org/?prefix=data/test_data/NMFClustering/
