Network Meta-Analysis on the Mechanisms underlying Type 2 Diabetes Augmentation of COVID-19 Pathologies
Article Information
Ryan J. Kim1,2, Mohammed AS Khan3, Maryam Khan4, Sulie L. Chang*, 1, 2
1Institute of NeuroImmune Pharmacology (I-NIP), Seton Hall University, 400 S Orange Ave, NJ 07079, USA
2Department of Biological Sciences, Seton Hall University, 400 S Orange Ave, NJ 07079, USA
3Department of Anesthesia, Critical Care and Pain Medicine, Massachusetts General Hospital, Harvard Medical School, Shriners Hospital for Children and Department of Neurosurgery, Brigham and Women's Hospital, Boston, MA, USA
4Brandeis University, Waltham, MA 02453
*Corresponding author: Sulie L. Chang, Institute of NeuroImmune Pharmacology, Seton Hall University, 400 S Orange, Ave NJ 07079, USA
Received: 31 August 2023 Accepted: 06 September 2023 Published: 07 December 2023
Citation: Ryan J. Kim, Mohammed AS Khan, Maryam Khan, Sulie L. Chang. Network Meta- Analysis on the Mechanisms underlying Type 2 Diabetes Augmentation of COVID-19 Pathologies. Archives of Microbiology and Immunology. 7 (2023): 372-383.
View / Download Pdf Share at FacebookAbstract
Coronavirus disease-2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. SARS-CoV-2 virus is internalized by surface receptors, e.g., angiotensin-converting enzyme-2 (ACE2). Clinical reports suggest that non-insulin dependent diabetes mellitus (DM-II) may enhance COVID-19. This bioinformatics study investigated how DM-II augments COVID-19 complications through molecular interactions with cytokines/chemokines, using QIAGEN Ingenuity Pathway Analysis (IPA) and CLC Genomics Workbench 22 (CLCG-22). “(Iβ-CG) RNA-sequencing of (Iβ-CG) through CLCG-22 (SRA SRP287500) were analyzed to identify differential expression of (Iβ-CG). IPA’s QIAGEN Knowledge Base (QKB) was also used to retrieve 88 total molecules shared between DM-II and SARS-CoV-2 infection to characterize and identify Iβ-CG, due to close association with DM-II. Molecules directly associated with ACE2 and cytokines/chemokines were also identified for their association with SARS-CoV-2 infection. Using IPA, it was found that 3 Ib-CG (SCL2A2, PPARγ, and CPLX8) are common in both diseases that were downregulated by DM-II. Their downregulation occurred due to increased activity of cytokines/chemokines and ACE2. Collectively, this network meta-analysis demonstrated that interaction of SARS-CoV-2 with ACE2 could primarily induce endothelial cell dysfunction. Identification of common molecules and signaling pathways between DM-II and SARS-CoV-2 infection in this study may lead to further discovery of therapeutic measures to simultaneously combat both diseases.
Keywords
SARS-CoV-2 Infection; DM-II; insulin; pancreas; Isit -cell genes; cytokine storm; ACE2 receptors
Article Details
1. Introduction
Diabetes mellitus (DM) is a common metabolic disorder that results from abnormalities in insulin secretion, prompting an imbalance of glucose levels in the bloodstream. A common affected cell type, known as the islet b-cells, are endocrine cells that are located mainly in the periphery of the pancreas. These cells are responsible for the production of insulin [1,2]. Regarding recent trends in the rise of DM cases, DM in older populations has increased from approximately 30 million in the 1990’s to over 170 million worldwide in 2010, and eventually surpassing the estimated mark of 365 million at nearly 425 million as of the year of 2020 [3,4,5]. DM is clinically defined as type 1 (DM-I) and type 2 (DM-II) diabetes. DM-I is a genetic metabolic disorder that occurs at a young age due to chronic inflammation of the islets b-cells in the pancreas. Autoantibodies attack the islet b-cells of the pancreas, resulting in an irretrievable loss of islet b-cell mass, and subsequent inability to produce insulin [6]. In contrast, DM-II, shortened for non-insulin dependent Diabetes mellitus, is caused by low-grade inflammation, sedentary lifestyle, lack of regular exercise and is related to unresponsiveness to insulin by target organs [6,7].
Despite the clinical differences, increased levels of proinflammatory cytokines in the pancreas are the crucial mediators of islet b-cell destruction in both DM-I and DM-II [7]. These diabetic pathologies lead to insulin resistance, which arises from low-grade inflammation, dysregulation of metabolic processes, oxidative stress, and mitochondrial dysfunction. Acute or chronic metabolic and physiological changes occur due to biochemical changes such as hyperglycemia, dyslipidaemia, and accumulation of adipose tissue mass which are closely associated with the pancreatic decompensation due to the loss of islet b-cell function. Clinical reports have also shown that patients with a history of pre-diabetes are at high risk, and display susceptibility to SARS-CoV-2 due to the pathology of Coronavirus disease-2019 (COVID-19) [8,9,10,11]. COVID-19 is an inflammatory disease that results from the entry of SARS-CoV-2 into host cells through its specific receptors [12,13].
SARS-CoV-2 is an enveloped positive single-stranded RNA virus, which is transmitted through respiratory droplets that are exhaled from the cough or sneeze of infected persons [5]. Initial entrance of SARS-CoV-2 into the cells of the respiratory system commonly results in symptoms such as fever, cough, and sore throat; in severe cases, SARS-CoV-2 can also generate harmful immune responses by generating increased levels of proinflammatory cytokines [5,14] mostly by immune cells [15,16,17,18]. The virus itself is enveloped by a nucleocapsid containing essential spike proteins (S1 and S2) that aid in its recognition and binding to host-membrane bound protein receptors, chiefly the angiotensin-converting enzyme 2 (ACE2) receptor [19,20,21,22]. Initial studies have discovered that these ACE2 receptors are expressed in type I and type II pneumocytes, which are epithelial cells of the lung [5]. ACE2 expression may therefore be correlated with SARS-CoV-2 infectibility, as the lung is a major target organ of infection [23,24]. However, RNA-sequence analyses show that ACE2 are also expressed in human endocrine cells, primarily dealing with islet b-cells and other pancreatic cells [25]. In addition to the ACE2 receptor, the entry of SARS-CoV-2 into the cells may also occur through neuropilin (NRP-1), and other carbohydrate and protein receptors (e.g., CD13/APN - aminopeptidase N and CD26/DPP4 - dipeptidyl peptidase 4). These receptors have binding affinity to either N-terminal domains (NTD) or C-terminal domains (CTD) of spike proteins of SARS-CoV-2 [26,27,28,29,30]. Although DPP4 plays an integral role in both DM-II and SARS-CoV-2 infection, the role of CD13/APN in DM-II is emerging gradually [26,27,30].
Meta-analyses, using various bioinformatics tools such as QIAGEN Ingenuity Pathway Analysis (IPA) and the CLC Genomics Workbench 22 have increasingly been used in conducting a comprehensive systemic review of evidence-based literature [31,32,33]. These bioinformatics tools allow the linking of molecular interactions for novel studies. In this study, COVID-19 was referred to as SARS-CoV-2 infection in using QIAGEN Knowledge Base (QKB), and non-insulin dependent diabetes mellitus was referred to as DM-II. This study performed a network meta-analysis to examine the hypothesis that SARS-CoV-2 infection activates inflammatory molecules through ACE2 receptors, as well as that DM-II augments SARS-CoV-2 infection pathology to further increase the expression of both pancreatic molecules and islet b-cell genes (Ib-CG). In this in-silico study, the molecular mechanisms underlying DM-II augmentation of SARS-CoV-2 infection pathology were investigated through the identification and interactions of overlapping molecules in inflammatory and diabetes signaling pathways. Additionally, the mediatory role of the ACE2 receptors in the pancreas was also studied, specifically in Ib-CG under the influence of DM-II and SARS-CoV-2 infection.
2. Materials and Methods
2.1 Ingenuity Pathway Analysis Software
The Ingenuity Pathway Analysis (IPA) software, licensed from QIAGEN, was used for this study. IPA consists of the QIAGEN Knowledge Base (QKB) and various IPA bioinformatics tools/features including My Pathway, Grow, Compare, Molecule Activity Predictor (MAP), and Upstream Regulator. The QKB curates and stores publications for reference. Data on the molecules associated with DM-II and SARS-CoV-2 infection, including their gene IDs and abbreviated names, were retrieved from the QKB. Data had been retrieved from QKB from 21 June 2021 to 23 February 2022.
2.2 My Pathway: Molecules Relationship Mapping and Connectivity
My Pathway was used to customize networks between biological molecules or diseases of interest. This tool was implemented to identify the biological relationships between the molecules of DM-II and SARS-CoV-2 infection. This feature contains a Grow tool, which was used to determine the full list of molecules that have a direct relation with DM-II and SARS-CoV-2 infection.
2.3 Canonical Pathway Analysis
For Canonical Pathway Analysis, a gene expression analysis tool was incorporated in the study to analyze the biological relations between the 88 molecules, which were associated with both DM-II and SARS-CoV-2 infection. Canonical Pathway Analysis was utilized to determine the rankings of canonical pathways based on their association with the 88 molecules that were added to Core Analysis. Diseases most associated with the 88 molecules were also retrieved from this analysis. An identical analysis was also conducted for the 127 significantly expressed Ib-CG.
2.4 Molecule Activity Predictor (MAP)
The MAP tool was used to determine the upstream or downstream effects of the simulated increase or decrease of a molecule of interest on other molecules or diseases. Red-colored molecules denote a simulated increased measurement of a molecule or disease of interest. Orange and blue-colored molecules denote subsequent activation and inhibition of neighboring molecules or diseases, respectively.
2.5 Pathway Explorer tool
Pathway Explorer was used to find shared molecules between DM-II and SARS-CoV-2 infection. For instance, among the 980 molecules associated with DM-II and 855 molecules associated with SARS-CoV-2 infection, a total of 105 were identified to be shared between these two diseases.
2.6 QIAGEN CLC Genomics Workbench 22
Under the license for CLC Workbench 22 (QIAGEN, Redwood City, CA), RNA-sequence analysis, as well as analyses on differential expressions of Ib-CG between SARS-CoV-2 affected and mock treated patients, were conducted. Publicly available datasets from Gene Expression Omnibus (GEO) by the National Center for Biotechnology Information (NCBI) were retrieved and analyzed. This study analyzed pancreas samples, using accession code SRP287500.
2.7 RNA-Sequence-Analysis
CLC Genomics Workbench 22 was used to analyze RNA samples for expression levels of individual Ib-CG from pancreas samples of SARS-CoV-2 infected and mock treated patients. The tools used on CLC Genomics Workbench 22 were “PCA for RNA-seq” and “Differential Expression in Two Groups.
2.8 Differential Expression Between Two Groups
CLC Genomics Workbench 22 was also used to compare different expression levels of the same genes between two different sample sets. Statistical data was collected based on the significant differences in the expression of 127 b-cell genes between SARS-CoV-2 infected and mock treated pancreas samples, by virtue of p-values.
3. Results
IPA was used to identify the molecules associated with DM-II and SARS-CoV-2 infection by retrieving the molecules from IPA’s QIAGEN Knowledge Base (QKB). DM-II was found to be associated with 980 molecules, and SARS-CoV-2 infection was found to be associated with 855 molecules. Upon identification of the full list of molecules associated with DM-II and SARS-CoV-2 infection, My Pathway was used to identify a total of 105 molecules that were shared between DM-II and SARS-CoV-2 infection. Gene expression profile with accession SRP287500 (Tang et al., 2021) was obtained from the Gene Expression Omnibus (GEO) database of the National Center of Biotechnology Information (NCBI). The GEO accession code SRP287500 was employed to download the sequence read archive (SRA) from CLC Genomics Workbench 22 (QIAGEN, Redwood City, CA). RNA sequencing was then performed with respect to the Ib-CG samples that had either been infected with SARS-CoV-2 or mock treated [25].
Quality check of the sequencing was performed using the Principal Component Analysis, which confirmed variations in expression of molecules or genes in the data set. Differential Expression Between Two Groups analysis was employed to identify the differentially expressed genes (DEG) in SARS-CoV-2 infected and mock treated samples. A total of 26,057 genes were found to undergo expression changes, 88 of which had matched with the total 105 molecules that had prior been retrieved in IPA’s QKB. The remaining 17 molecules which had not matched with the RNA-sequenced data from SRP287500 were removed from subsequent analyses, as they were identified to not be affected by SARS-CoV-2 infection in the collected Ib-CG (Figure 1). The remaining 88 molecules, which were in common with those retrieved from RNA-sequencing of Ib-CG, revealed that there was susceptibility of Ib-CG expression to SARS-CoV-2 infection; this study therefore took the RNA-sequence analysis findings to further investigate the potential Ib-CG expression changes that may also result from the onset of DM-II, prior to SARS-CoV-2 infection. Table 1 lists the symbols, full names, and molecule family of each of the 88 molecules.
Figure 1. The connectivity map displays the 88 molecules that were associated with both DM-II and SARS-CoV-2 infection that were shown on the left and right side of the cluster of molecules, respectively. These molecules were retrieved from RNA-sequencing of islet b-cell genes using CLC Genomics Workbench 22. The functions of the molecules were represented by symbols in the legend box. Table 1 listed the full list of these molecules.
Table 1: List of molecules that overlap between DMII and SARS-CoV-2, with Entrez Gene Name and Family
|
ACE |
angiotensin I converting enzyme |
peptidase |
|
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif 2 |
peptidase |
|
ADORA2A |
adenosine A2a receptor |
G-protein coupled receptor |
|
ADRA2A |
adrenoceptor alpha 2A |
G-protein coupled receptor |
|
ADRB1 |
adrenoceptor beta 1 |
G-protein coupled receptor |
|
ADRB2 |
adrenoceptor beta 2 |
G-protein coupled receptor |
|
AGT |
angiotensinogen |
growth factor |
|
AGTR1 |
angiotensin II receptor type 1 |
G-protein coupled receptor |
|
AGTR2 |
angiotensin II receptor type 2 |
G-protein coupled receptor |
|
AKT1 |
AKT serine/threonine kinase 1 |
kinase |
|
ALB |
albumin |
transporter |
|
ALPL |
alkaline phosphatase, biomineralization associated |
phosphatase |
|
AMY2A |
amylase alpha 2A |
enzyme |
|
AMY2B |
amylase alpha 2B |
enzyme |
|
APOA2 |
apolipoprotein A2 |
transporter |
|
AR |
androgen receptor |
ligand-dependent nuclear receptor |
|
ATP10A |
ATPase phospholipid transporting 10A (putative) |
transporter |
|
C8A |
complement C8 alpha chain |
other |
|
CACNA1D |
calcium voltage-gated channel subunit alpha1 D |
ion channel |
|
CCL2 |
C-C motif chemokine ligand 2 |
cytokine |
|
CD38 |
CD38 molecule |
enzyme |
|
CDC42 |
cell division cycle 42 |
enzyme |
|
CHI3L1 |
chitinase 3 like 1 |
enzyme |
|
CLU |
clusterin |
other |
|
CPLX2 |
complexin 2 |
other |
|
CRP |
C-reactive protein |
other |
|
CXCL8 |
C-X-C motif chemokine ligand 8 |
cytokine |
|
DRD1 |
dopamine receptor D1 |
G-protein coupled receptor |
|
DRD2 |
dopamine receptor D2 |
G-protein coupled receptor |
|
EDNRA |
endothelin receptor type A |
transmembrane receptor |
|
EGR1 |
early growth response 1 |
transcription regulator |
|
F7 |
coagulation factor VII |
peptidase |
|
FOS |
Fos proto-oncogene, AP-1 transcription factor subunit |
transcription regulator |
|
FYN |
FYN proto-oncogene, Src family tyrosine kinase |
kinase |
|
GPD2 |
glycerol-3-phosphate dehydrogenase 2 |
enzyme |
|
GSS |
glutathione synthetase |
enzyme |
|
HBA1/HBA2 |
hemoglobin subunit alpha 2 |
transporter |
|
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
enzyme |
|
HSPA5 |
heat shock protein family A (Hsp70) member 5 |
enzyme |
|
HTR1A |
5-hydroxytryptamine receptor 1A |
G-protein coupled receptor |
|
IL1B |
interleukin 1 beta |
cytokine |
|
IL1RN |
interleukin 1 receptor antagonist |
cytokine |
|
IL2 |
interleukin 2 |
cytokine |
|
IL6 |
interleukin 6 |
cytokine |
|
KCNJ5 |
potassium inwardly rectifying channel subfamily J member 5 |
ion channel |
|
MAOA |
monoamine oxidase A |
enzyme |
|
MTNR1B |
melatonin receptor 1B |
G-protein coupled receptor |
|
NR3C1 |
nuclear receptor subfamily 3 group C member 1 |
ligand-dependent nuclear receptor |
|
NR3C2 |
nuclear receptor subfamily 3 group C member 2 |
ligand-dependent nuclear receptor |
|
NTRK2 |
neurotrophic receptor tyrosine kinase 2 |
kinase |
|
PCSK9 |
proprotein convertase subtilisin/kexin type 9 |
peptidase |
|
PDE11A |
phosphodiesterase 11A |
enzyme |
|
PDE3A |
phosphodiesterase 3A |
enzyme |
|
PDE3B |
phosphodiesterase 3B |
enzyme |
|
PDE4A |
phosphodiesterase 4A |
enzyme |
|
PDE4B |
phosphodiesterase 4B |
enzyme |
|
PDE4C |
phosphodiesterase 4C |
enzyme |
|
PDE4D |
phosphodiesterase 4D |
enzyme |
|
PDE5A |
phosphodiesterase 5A |
enzyme |
|
PDE7A |
phosphodiesterase 7A |
enzyme |
|
PDE7B |
phosphodiesterase 7B |
enzyme |
|
PDE8A |
phosphodiesterase 8A |
enzyme |
|
POR |
cytochrome p450 oxidoreductase |
enzyme |
|
PPARA |
peroxisome proliferator activated receptor alpha |
ligand-dependent nuclear receptor |
|
PPARG |
peroxisome proliferator activated receptor gamma |
ligand-dependent nuclear receptor |
|
PPP3CA |
protein phosphatase 3 catalytic subunit alpha |
phosphatase |
|
PTEN |
phosphatase and tensin homolog |
phosphatase |
|
PTGS1 |
prostaglandin-endoperoxide synthase 1 |
enzyme |
|
PTGS2 |
prostaglandin-endoperoxide synthase 2 |
enzyme |
|
RBP4 |
retinol binding protein 4 |
other |
|
RNF138 |
ring finger protein 138 |
enzyme |
|
SCN5A |
sodium voltage-gated channel alpha subunit 5 |
ion channel |
|
SELL |
selectin L |
transmembrane receptor |
|
SERPINC1 |
serpin family C member 1 |
enzyme |
|
SLC2A1 |
solute carrier family 2 member 1 |
transporter |
|
SLC2A2 |
solute carrier family 2 member 2 |
transporter |
|
SLC2A4 |
solute carrier family 2 member 4 |
transporter |
|
SLC5A2 |
solute carrier family 5 member 2 |
transporter |
|
SLC6A4 |
solute carrier family 6 member 4 |
transporter |
|
SLC7A11 |
solute carrier family 7 member 11 |
transporter |
|
SNCA |
synuclein alpha |
enzyme |
|
TNF |
tumor necrosis factor |
cytokine |
|
TNFSF10 |
TNF superfamily member 10 |
cytokine |
|
TNFSF11 |
TNF superfamily member 11 |
cytokine |
|
TP53 |
tumor protein p53 |
transcription regulator |
|
TUBA1A |
tubulin alpha 1a |
other |
|
TUBA1C |
tubulin alpha 1c |
other |
|
VCAM1 |
vascular cell adhesion molecule 1 |
transmembrane receptor |
|
VDR |
vitamin D receptor |
transcription regulator |
|
VEGFA |
vascular endothelial growth factor A |
growth factor |
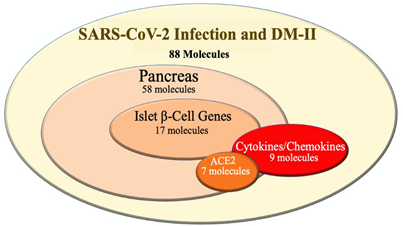
Using IPA analysis, 88 molecules were categorized based on their association with the following groups: pancreatic molecules, Ib-CG, cytokines/chemokines, and ACE2-related molecules. The Keep tool of IPA was used to identify the molecule types of the 88. Among the 88 molecules, 58, 17, and 9 molecules were identified as pancreatic, Ib-CG, and cytokines/chemokines, respectively (Figure 2). Among the 58 pancreatic molecules, 7 of them were directly linked with ACE2 receptors, including ACE, IL6, AGT, AGTR1, TNF, PTGS1, and HSPA5. The association of these 7 molecules with the pancreas, as well as a direct association with ACE2, indicated a strong relationship between the pancreas and ACE2 receptors. Next, among the 88 molecules shared between DM-II and SARS-CoV-2 infection, 17 of the 88 were identified as Ib-CG: EGR1, TP53, MTNR1B, IL1RN, FOS, EDNRA, VEGFA, PPARG, CPLX2, TNF, NR3C1, CACNA1D, SLC2A2, GPD2, RBP4, PCSK9, and ADRA2A. Lastly, a total of 9 from the 88 molecules were identified as cytokines/chemokines, (IL1B, CCL2, TNF, IL1RN, IL6, CXCL8, TNFSF11, TNFSF10, and IL2). Upon identification of all molecule types listed above, this study proceeded to investigate how these molecule types were affected by the onset of DM-II, as well as how these effects may impact the severity of SARS-CoV-2 infection.
Figure 2: The overlapping molecules between SARS-CoV-2 infection and DM-II. Total 88 molecules were identified by Pathway Explorer. Among these molecules, 58, 17, 9 and 7 molecules were associated with pancreas, islet b-cell, cytokines/chemokines and ACE2 molecules, respectively. All molecules associated with ACE2 receptor did not show a link with islet b-cell genes.
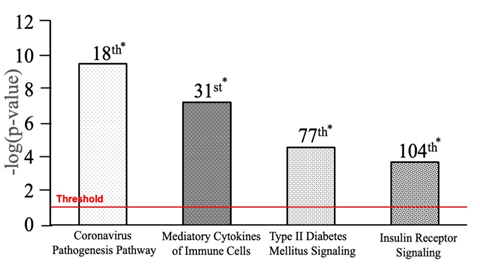
IPA’s Core Analysis revealed that a total of 264 pathways (p-value <0.05) were most associated with the 88 molecules. Among the 264 pathways, Coronavirus Pathogenesis, Mediatory Cytokines of Immune Cells, Type II Diabetes Mellitus Signaling, and Insulin Receptor Signaling pathways were ranked 18th (p-value 2.72E-10), 31st (p-value 5.16E-08), 77th (p-value 3.92E-02), and 104th in association (p-value 3.57E-02), respectively (Figure 3).
Figure 3: Top canonical pathways of DM-II and SARS-CoV-2. * = Rank. Four canonical pathways displayed in the graph were taken from 264 canonical pathways (p-value <0.05) that were associated with 88 molecules having p-value<0.05. The canonical pathway s with 18th, 31st, 77th, and 104th ranks were shown in order of increasing p values or decreasing association with the 88 molecules. All bars above the red line were significantly associated with the 88 molecules with a p-value<0.05.
Core Analysis was also conducted to identify diseases that were most associated with the list of 88 molecules. Table 2 lists the top 20 diseases with the greatest association to the 88 molecules. Infectious Diseases (p-value 2.62E-147), Endocrine System Disorders (p-value 1.72E-142), Inflammatory Response (p-value 1.69E-67), and Inflammation of Respiratory Conditions (p-value 1.57E-58) were notable diseases.
Table 2: List of the top 20 diseases and conditions*
|
Diseases/Conditions |
p-value |
|
Infection by SARS coronavirus |
2.62E-147 |
|
Non-insulin-dependent Diabetes mellitus |
1.72E-142 |
|
COVID-19 |
8.57E-141 |
|
Ischemia of heart |
2.23E-70 |
|
Stroke |
1.18E-66 |
|
Atherosclerosis |
4.41E-64 |
|
Failure of heart |
6.99E-60 |
|
Obesity |
8.06E-60 |
|
Inflammation of respiratory system component |
1.57E-58 |
|
Hypertension |
1.11E-56 |
|
Inflammation of airway |
1.11E-55 |
|
Chronic inflammatory disorder |
5.90E-54 |
|
Diabetic complication |
9.49E-54 |
|
Acute respiratory disorder |
1.92E-51 |
|
Congestive heart failure |
5.36E-51 |
|
Ischemic cardiomyopathy |
1.29E-48 |
|
Acute kidney injury |
1.51E-46 |
|
Insulin resistance |
2.09E-46 |
|
Obstruction of airway |
1.09E-45 |
|
Damage of kidney |
1.74E-44 |
|
Inflammatory Response |
1.69E-67 |
*Top 20 Diseases and Functions associated with 88 molecules that are in common with both non-insulin dependent Diabetes Mellitus and Infection by SARS Coronavirus in order of increasing p-value.
In a parallel study, the CLC Genomics Workbench 22 software was also used to identify additional genes that may be directly associated with both DM-II and SARS-CoV-2 infection. Sample Ib-CG were retrieved from study by Tang et al., 2021. This data was accessed from the GEO database with accession SRP287500. Among the five Ib-CG samples that were studied, two were treated with the SARS-CoV-2 virus, one was provided with integrated stress response inhibitor (ISRIB) treatment for infection, and the last two were given a mock treatment. The ISRIB-treated genome sample was removed from analysis due to the use of a drug that was not related to the study. Following RNA-sequence analysis of infected and mock treated sample genomes, the infected genomes were compared with the mock treated genomes through Differential Expression Between Two Groups using CLC Genomics Workbench 22. Subsequent analysis revealed that the expression of 26,057 molecules changed between infected and mock treated genome samples. From this dataset, 127 of the 26,057 molecules held a significant log fold change from mock to infected (p-value<0.05), which indicated a change in expression of these genes due to SARS-CoV-2 infection. The top five among the 127 Ib-CG with the greatest positive expression fold changes, all indicating upregulation due to positive expression fold changes, were identified as H19 imprinted maternally expressed transcript (H19), lysine-specific demethylase 5D (KDM5D), ribosomal protein S4 Y-linked 1 (RPS4Y1), DEAD-box helicase 3 Y-linked (DDX3Y), and UDP-glucuronosyltransferase 2B15 (UGT2B15) (Table 3).
Table 3: List of the top 5 islet b-cell genes (IbCG) genes with fold change
|
Symbol |
Expression Fold Change |
p-value* |
|
H19 |
105.74 |
0.0000574 |
|
KDM5D |
99.87 |
0.00181 |
|
RPS4Y1 |
2359.7 |
0.02 |
|
DDX3Y |
1229.52 |
0.03 |
|
UGT2B15 |
758.58 |
0.04 |
*p-values were identified from CLC database with the greatest log change among significant molecules that have differential expression between SARS-CoV-2 virus treated and mock treated IbCG genome samples. Molecules are listed down in order of increasing p-value.
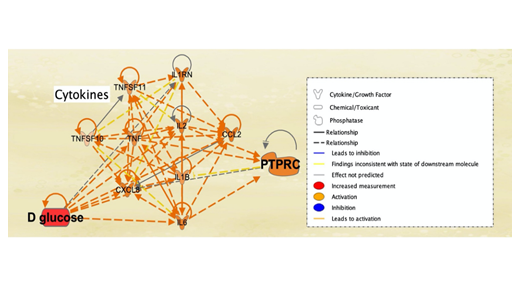
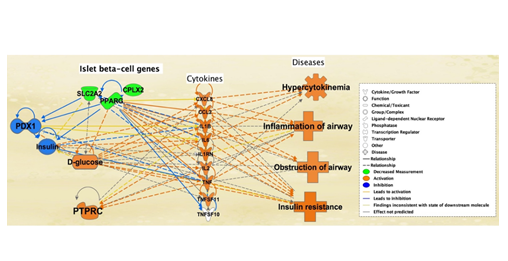
The total of 127 significantly expressed molecules from RNA-sequencing were investigated further using IPA. The dataset with the 127 genes was uploaded to IPA. Core Analysis was run on the 127 genes to demonstrate that these genes were associated with similar diseases as the 88 genes and molecules identified from the QKB: Infectious Diseases (p-value 1.91E-07), Inflammatory Response (p-value 3.42E-07), Endocrine System Disorders (p-value 7.52E-07), and Metabolic Disease (p-value 7.51E-07). Upon collection of these data, IPA was used to generate pathways of relation between the different molecule types. First, My Pathway was used to manually input the 9 cytokines that were identified from IPA’s QKB pathway. Glucose and B lymphocytes (PTPRC) were then added to this connectivity network to study potential immune responses that may arise due to a simulated increase in glucose levels. MAP was then applied to assess the effects of a simulated increase in glucose levels on immune responses and cytokines/chemokines expression. Simulated increased glucose levels, denoted by red in Figure 4, was shown to activate all 9 cytokines, as well as to activate PTPRC.
Figure 4: Effect of Increased Glucose Levels on Cytokines Associated with SARS-CoV-2 Infection and DM-II. The legend denotes red as the initial simulated upregulation of the molecule. In this pathway, glucose levels were increased (denoted in red), which resulted in the activation all 9 cytokines listed as CXCL8, ILB, IL1B, CCL2, IL2, TNF, TNFSF10, TNFSF11, IL1RN (cytokines are located in the middle, and activation due to the increased levels of glucose is denoted in orange). This further activated PTPRC (also known as B lymphocytes) expression, whose activation is also denoted in orange. This suggested increased production of autoantibodies, due to increased glucose levels. Curved orange arrows (which originate and end back to the molecule of origin) indicate that such molecules are capable of autocrine activation, following the effects of increased glucose levels.
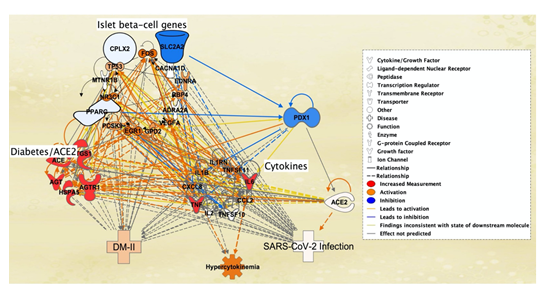
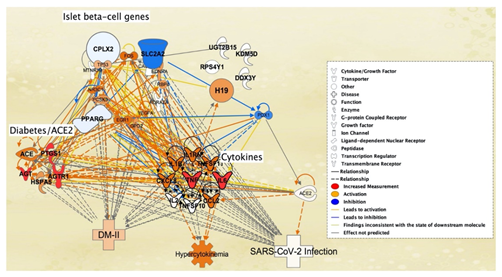
An additional pathway was created to study the potential onset of SARS-CoV-2 infection due to the expressions of intermediary molecules such as the Ib-CG, cytokines/chemokines, and pancreatic/ACE2 molecules, that were affected by DM-II (Figure 5). The 7 previously identified pancreatic molecules with direct association to ACE2 receptors and DM-II were increased in expression to imitate DM-II onset. As a result, all cytokines were activated except IL2 and TNFSF10, and DM-II, SARS-CoV-2 infection. Hypercytokinemia was also augmented due to the increased expression of the 7 molecules. In Figure 5, lines of relationship between cytokines/chemokines and ACE2 receptors indicated increased expression of 7 total cytokines. SARS-CoV-2 infection was also augmented as a result of increased ACE2 receptor expression. Expression of the 7 pancreatic and DM-II associated molecules also led to the inhibition of three Ib-CG listed as Complexin-2 (CPLX2), Solute carrier family 2 member 2 (SLC2A2), and Peroxisome Proliferator Activated Receptor Gamma (PPARg). Inhibition of these Ib-CG further inhibited the function of PDX1 (Figure 5).
Figure 5: Simulation of DM-II Using Molecular Activity Predictor (MAP) Tool. Simulation of increased levels of 7 pancreas-related diabetes/ACE2 receptor molecules, ACE, IL6, AGT, AGTR1, TNF, PTGS1 and HSPA5, (middle left) led to the induction of cytokines/chemokines - IL1B, CCL2, TNF, IL1RN, IL6, CXCL8, TNFSF11 except TNFSF10, and IL2 (middle right). Activation of three islet b-cell genes, CPLX, PPARG, SLC2A2 (top left), resulted in inhibition of PDX1 (top right). Induction of the cytokines, mainly CXCL8, CCL2 and IL6, triggered hypercytokinemia (middle bottom), ACE2 receptor expression (left), SARS-CoV-2 infection (bottom right) and DM-II (bottom left). Orange lines indicate a relationship between cytokines/chemokines and ACE2 receptor.
The 7 pancreas and DM-II associated molecules that led to the inhibition of three Ib-CG’s (SLC2A2, PPARg, and CPLX2) in Figure 5 were used to further analyze their effects on glucose levels, cytokine expression, and related diseases. Mock inhibition of SLC2A2, PPARg, and CPLX2 led to inhibition of PDX1, as well as the inhibition of insulin. This resulted in the increase in glucose levels. Increased glucose levels further resulted in the activation of all cytokines excluding TNFSF10 and PTPRC. Activation of cytokines also promoted several diseases associated with SARS-CoV-2 infection complications, including obstruction of airways, inflammation of airways, and insulin resistance with hypercytokinemia (Figure 6).
Figure 6: Effect of the simulated inhibition of islet b-cell genes on PDX1 and insulin levels is expressed in this figure. Simulated decreased measurement of the three islet b-cell molecules - SLC2A2, PPARG, and CPLX2 (top middle, in which the simulated decrease is denoted by the green color) - inhibited the PDX1 (top right, resultant inhibition being denoted by blue) gene, thereby reducing insulin levels (top right, blue). Reduction of insulin elevated glucose levels (bottom left, orange). Elevated glucose levels activated all cytokines except TNFSF10 (center). PTPRC was also activated due to inhibition of the three b-cell genes (bottom right, orange). Several additional associated diseases such as obstruction of airway, inflammation of airway, hypercytokinemia, and insulin resistance had also been activated (left, orange), suggesting that the progression of DM-II is associated with SARS-CoV-2.
Ib-CG genome samples from SARS-CoV-2 infected and mock treated patients were also studied for changes in expression levels due to SARS-CoV-2 exposure. The change in expression levels were investigated using the Differential Expression Between Two Groups on CLC Genomics Workbench 22. SARS-CoV-2 infected Ib-CG expression levels were compared with those of mock treated Ib-CG gene expression levels. As shown in Table 3, the 5 genes from the Ib-CG genome samples with greatest fold change due to SARS-CoV-2 infection were H19, KDM5D, RPS4Y1, DDX3Y, and UGTB15. The top 5 genes were added to Figure 7 on IPA to observe their interaction with the full connectivity network. Upon mock increased expression of the 7 pancreatic/ACE2 receptor associated molecules retrieved from QKB, the H19 gene was the sole molecule that had a direct relation to the cytokines TNF and IL1B. For instance, H19 was activated by the cytokine TNF. Among Ib-CG retrieved from the QKB of IPA, 3 genes SLC2A2, PPARg, and CPLX2 was also inhibited due to the onset of DM-II. However, among the genes that were retrieved from RNA-sequencing of Ib-CG genome samples, H19 was the sole activated gene (Figure 7).
Figure 7: SARS-CoV-2-induced molecules of islet b-cell genes (with greatest fold change) from the Tang et al., 2021 study - H19, KDM5D, RPS4Y1, DDX3Y and UGTB15 - were added to the pathway from Figure 5. The addition of these five islet b-cell genes are found in the top right of the pathway. After adding these five islet b-cell genes to the pathway, the expression levels of pancreatic/ACE2 receptor molecules were still increased to cause the same results as observed in figure 5. The addition of these five islet b-cell genes contributed to figure 5, as it led to the sole activation of the H19 islet b-cell gene (top right, orange).
4. Discussion
Generally, as a normal immunological process, viruses generate a mild inflammatory response after infection. However, when these infectious viruses, including SARS-CoV-2, attack patients who have pre-existing or immunosuppressive conditions (e.g., DM-II), a sudden hyperinflammatory response can be triggered in the form of hypercytokinemia. In normal circumstances, insulin acts as an anti-inflammatory molecule that promotes anti-inflammatory mechanism and directly suppresses the proinflammatory cytokines/chemokines promoting glucose homeostasis. Nonetheless, when this systemic inflammation interferes with the metabolic processes, the overall insulin sensitivity is altered by the production of reactive oxygen species, impairing glucose homeostasis/metabolism, and energy storage. Subsequently, the derailment of these metabolic processes leads to the development of insulin resistance, which in turn leads to exaggerated inflammatory response.
The pancreas harbors macrophages as resident cells in the islets at a steady state. These quiescent cells get activated in response to inflammatory signals within the pancreas. Additionally, low-grade, or chronic inflammation can cause infiltration of other immune cells into the pancreas. These cells release proinflammatory cytokines/chemokines, which interfere with insulin signaling pathways and cause metabolic disturbances, leading to onset of DM-II [34, 35]. Onset of DM-II occurs due to decreased production of insulin in the pancreas, as well as due to the dysregulation of glucose levels. These perturbations in metabolic processes exacerbate a cycle of persistent low-grade and chronic inflammation, which results from the inception of inflammation caused by immune cells. The dissemination of continuous inflammation induces proinflammatory cytokines/chemokines, reactive oxygen species, and impairs insulin signaling pathways through the functional inhibition of IRS-1, AKT/PKB and PPARg, leading to insulin resistance.
Coronavirus most likely infects the immune cells (e.g., macrophages and neutrophils), first mainly through ACE2, and possibly involving NRP-1, CD13/APN or CD26/DPP4 receptors [26, 27, 28, 29, 30]. This infection initially results in a mild inflammatory response by activating immune cells and releasing cytokines to protect other cells against the invading virus. However, influx of inflammatory cytokines, in turn, induces pancreatic dysfunction, leading to insulin resistance in the long-term, as an ancillary effect. This network-meta-analysis study investigated the molecular interactions between DM-II and SARS-CoV-2, involving pancreatic and Ib-CG, linked to ACE-2 receptor and cytokines/chemokines. These interactions were studied for their potential effects on the augmentation of SARS Coronavirus infection underlying DM-II. Through this study, it was indicated that DM-II onset inhibited the expression of three Ib-CG genes retrieved from IPA’s QKB listed as SLC2A2, PPARg, and CPLX8. The study also discovered the increased expression of RNA-sequenced gene H19, which had a positive log fold change of 105.74 (p-value 5.74E-5). Inhibition of SLC2A2, PPARg, and CPLX8 resulted in decreased expression of PDX1 and insulin. Decreased expression of insulin also further led to an increase in glucose levels. The impact of increased expression of H19 is not known; however, the increased expression of H19 by cytokine TNF in Figure 7 was in agreement with RNA-sequence results on H19, which also showed increased expression with SARS-CoV-2 infection [36]. These findings on Ib-CG gene expression changes as shown in Figure 7 suggested that DM-II may activate cytokines by maintaining high glucose levels, due to the inhibition of insulin. Additionally, Figure 7 also demonstrated that, because of DM-II onset, cytokines were also capable of changing expression levels of genes such as H19 in islet b-cells.
The increase in cytokines/chemokines levels also resulted in hypercytokinemia [37,38], noted as cytokine storm in IPA’s QKB. Increased glucose levels also contributed to the development of hypercytokinemia. This was also reported previously, in which high glucose levels lead to the production of proinflammatory cytokines [8]. As demonstrated in Figure 5, increased expression of the 7 molecules led to the subsequent increased expression of all cytokines excluding ILB1 and TNFSF10. The expression of the 7 cytokines led to activation of the ACE2 receptor, which also aligned with the findings of a prior study showing increased ACE2 receptor expression due to cytokine treatment [39]. ACE2 receptor activation resulted in the augmentation of SARS-CoV-2 infection pathology, indicating greater risk of severe infection due to DM-II. Although the ACE2 receptor is known to subsume SARS-CoV-2, other receptors such as NRP-1, CD13/APN and CD26/DPP4 may also facilitate internalization of SARS-CoV-2 into the cells during infection. The NRP-1 potentiates SARS-CoV-2 infectivity via the olfactory epithelium of the nasal cavity into the brain. Increase in NRP-1 protein expression in diabetic kidney cells implies severity of COVID-19 [28,29]. Another receptor, CD13/APN, a selective angiogenic marker, also serves as a receptor to common cold virus, which is named as “human coronavirus 299E” [40]. This angiogenic marker is expressed in non-diabetic, hypoxia-induced retinal neovascularization. However, it had recently been shown to play a role in diabetic retinal vasculature abnormalities [30]. Receptor CD26/DPP4 is expressed in epithelium as well as immune cells, and has been previously recognized to bind middle east respiratory syndrome coronavirus (MERS-CoV); however, this receptor might also capture SARS-CoV-2, and like ACE2 receptor, may also be dysregulated in diabetes [41,42]. In addition to hypercytokinemia, other complications such as obstruction of airways, inflammation of airways, and insulin resistance were also shown in Figure 6. Regarding obstruction of airways, specific complications such as asthma, chronic obstructive pulmonary disease (COPD) and emphysema may cause the obstruction of the airways due to SARS-CoV-2 infection and DM-II. Core Analysis results of the 88 molecules, retrieved from IPA’s QKB, as shown in Table 2, confirmed a close association of the 88 molecules with infectious diseases (p-value 2.62E-147), endocrine system disorders (p-value 1.72E-142), inflammatory response (p-value 1.69E-67), and respiratory disease (p-value 1.57E-58). In light of respiratory diseases, previous findings also reported that the association between the two diseases increases risk for more severe illness such as acute respiratory distress syndrome (ARDS), and increases risk of death [5]. Therefore, it was suggested that both DM-II and SARS-CoV-2 infection simultaneously increased the risk for complications.
For instance, DM-II involves the loss of Ib-CG volume in the pancreas; a study of 121 autopsies reported 63% loss of Ib-CG volume in DM-II affected subjects as opposed to healthy subjects [39]. IPA findings from this current study supported these findings with the inhibition of Ib-CG function due to onset of DM-II. Similarly, SARS-CoV-2 infection also resulted in significant changes in the expression of up to 127 Ib-CG. This might indicate that the combined effects of both diseases could result in a synergistic inhibition of Ib-CG that may hinder insulin sensitivity, and increase cytokines/chemokines production due to increased glucose levels. Further studies may, however, be needed to discover the extent of this mechanism from which SARS-CoV-2-induced pathology could be augmented, potentially by utilizing the molecules and genes which had been identified in this study. Some limitations in this study may potentially affect the findings.
These include the lack of data on the effects of the Ib-CG H19, when it is increased in expression. It is known through this study that both DM-II and SARS-CoV-2 infection both upregulate expression of H19. However, its molecular effects or interactions with other molecules and diseases in this study were not certain due to the limited information. Additionally, the islet b-cell samples which were RNA-sequenced had been sourced from a small pool of subjects of various ages and genders. It is not clear if age or gender may affect the gene expression level changes of Ib-CG. Therefore, genes that had been identified from RNA-sequence analyses such as H19 require further investigation on their expression changes with age and gender variables, as well as the effect of such expression changes on the potential augmentation of SARS-CoV-2 infection. However, genes such as SLC2A2, PPARg, and CPLX8, were drawn from IPA’s QKB, which provided sufficient information for their inhibition due to DM-II, as well as their impact on the increased expression of hypercytokinemia due to promoted cytokine production.
This network meta-analysis study established the biological mechanisms underlying the complication between DM-II and SARS-CoV-2 infection. SARS-CoV-2 infection was a node used in QKB to refer to COVID-19. The molecules and signaling pathways associated with DM-II and SARS-CoV-2 infection revealed a total 88 molecules, of which 58, 17, 9 and 7 belong to the pancreas, Ib-CG, cytokines/chemokines and ACE2 receptors, respectively. Using IPA tools, we have empirically demonstrated that COVID-19 could have initially caused infection, possibly via through ACE2 receptor actions leading to hypercytokinemia. We have also shown that DM-II could directly result in glucose dyshomeostasis, as well as decreased insulin levels in the pancreas. Decreased expression of insulin contributed indirectly to hypercytokinemia. For instance, the decrease in insulin expression resulted in increased glucose levels leading to, activation of cytokines, and the development of hypercytokinemia in patients with DM-II and COVID-19. This in-silico study, which revealed the synergistic effects of DM-II and COVID-19, has posed an innovative paradigm shift in understanding the higher morbidity rate of DM-II patients during the earlier stages of the COVID-19 pandemic. Both molecular and pathway-mediated mechanisms identified in this study may be valuable for the development of therapeutical strategies to treat DM-II patients in the course of severe inflammatory diseases with hypercytokinemia, comparable to COVID-19.
Supplementary Materials:
Not applicable.
Funding:
This study was partially sponsored by R01 DA046258 to SLChang
Acknowledgements:
The authors thank Dr. Eric Seiser from QIAGEN for providing consultation on the use of the Ingenuity Pathway Analysis (IPA) and its features.
Author Contributions:
Conceptualization, S.L.C., R.J.K., M.A.S.K., and M.K.; methodology, S.L.C., R.J.K., M.A.S.K., and M.K; software, S.L.C., R.J.K., M.A.S.K., and M.K; validation, S.L.C., M.A.S.K, and R.J.K.; formal analysis, S.L.C., R.J.K., M.A.S.K., and M.K; investigation, S.L.C., R.J.K., M.A.S.K., and M.K.; resources, S.L.C.; data curation, R.J.K, M.K..; writing—original draft preparation, S.L.C., R.J.K..; writing—review and editing, S.L.C., R.J.K., M.A.S.K., and M.K; visualization, R.J.K., M.K.; figures, R.K., M.K.; supervision, S.L.C. and M.A.S.K.; project administration, S.L.C.; funding acquisition, S.L.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Data Availability Statement: Data supporting the reported results may be accessed with a purchased license for IPA use by QIAGEN (QIAGEN Inc., Germantown, MD, USA, https://www.qiagenbioinformatics.com/products, last accessed on 23 February 2022.
Competing Interest: The authors declare no conflict of interests.
References
- Da Silva Xavier G. “The Cells of the Islets of Langerhans.” J Clin Med 7 (2018).
- Zhu Y, et al. "PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration." Stem Cell Res Ther 8 (2017): 240.
- Saini V. "Molecular mechanisms of insulin resistance in type 2 diabetes mellitus." World J Diabetes 1 (2010): 68-75.
- Slack JM. "Developmental biology of the pancreas." Development 121 (1995): 1569-1580.
- Oliveira TL, et al. "Pathophysiology of SARS-CoV-2 in Lung of Diabetic Patients." Front Physiol 11 (2020): 587013.
- Cerf ME. "Beta cell dynamics: beta cell replenishment, beta cell compensation and diabetes." Endocrine 44 (2013): 303-311.
- Cerf ME. "Beta cell dysfunction and insulin resistance." Front Endocrinol (Lausanne) 4 (2013): 37.
- Hu R, et al. "Effect of high glucose on cytokine production by human peripheral blood immune cells and type I interferon signaling in monocytes: Implications for the role of hyperglycemia in the diabetes inflammatory process and host defense against infection" Clin Immunol 195 (2018): 139-148.
- Muniangi-Muhitu H, et al. "Covid-19 and Diabetes: A Complex Bidirectional Relationship." Front Endocrinol (Lausanne) 11 (2020): 582936.
- Ibrahim S, et al. "Not so sweet and simple: impacts of SARS-CoV-2 on the beta cell" Islets 13 (2021): 66-79.
- Shirakawa J. "Pancreatic beta-cell fate in subjects with COVID-19." J Diabetes Investig 12 (2021): 2126-2128.
- Chang SL, et al. “Neuro-COVID-19: a meta-analysis of COVID-19-induced neuroinflammation and its implications.” Science Documents 1 (2020): 22-34.
- Khan B, et al. “Implications of COVID-19 in metabolic syndrome and therapeutic strategy for prevention and cure of the disease.” Science Documents 1 (2020): 35-44.
- Hojyo S, et al. "How COVID-19 induces cytokine storm with high mortality." Inflamm Regen 40 (2020): 37.
- Seo JY, et al. "Oxidative stress induced cytokine production in isolated rat pancreatic acinar cells: effects of small-molecule antioxidants." Pharmacology 64 (2002): 63-70.
- Lacy P and Stow JL. "Cytokine release from innate immune cells: association with diverse membrane trafficking pathways." Blood 118 (2011): 9-18.
- Arango Duque G. and Descoteaux A. "Macrophage cytokines: involvement in immunity and infectious diseases." Front Immunol 5 (2014): 491.
- Striz I, et al. "Cytokine networking of innate immunity cells: a potential target of therapy." Clin Sci (Lond) 126 (2014): 593-612.
- Li F. "Structure, Function, and Evolution of Coronavirus Spike Proteins" Annu Rev Virol 3 (2016): 237-261.
- Duan L, et al. "The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens." Front Immunol 11 (2020): 576622.
- Martinez-Flores D, et al. "SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants." Front Immunol 12 (2021): 701501.
- Velusamy P, et al. "SARS-CoV-2 spike protein: Site-specific breakpoints for the development of COVID-19 vaccines." J King Saud Univ Sci 33 (2021): 101648.
- Li Y, et al. "Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor." Pharmacol Res 157 (2020): 104833.
- Ashraf UM, et al. "SARS-CoV-2, ACE2 expression, and systemic organ invasion." Physiol Genomics 53 (2021): 51-60.
- Tang X, et al. "SARS-CoV-2 infection induces beta cell transdifferentiation." Cell Metab 33 (2021): 1577-1591 e1577.
- Deverakonda CKV, Meredith E, Ghosh M, Shapiro LH: Coronavirus receptors as immune
modulators J Immunol 206 (2021): 923-29. - Millet JK, Jaimes JA, Whittaker GR. Molecular diversity of coronavirus host cell entry receptors, FEMS Microbiol Rev 45 (2021): fuaa057.
- Mayi BS, et al. “The role of Neuropilin-1 in Covid-19.” PLoS Pathog 17 (2021): e1009153.
- Gudowska-Sawczuk M, et al. The role of Neuropilin-1 (NRP-1) in SARS-CoV-2 infection: Review.” J Clin Med 10 (2021): 2772.
- Hossain A, et al. Protective effects of bestatin in the retina of streptozotocin-induced diabetic mice.” Exp Eye Res 149 (2016):100-106.
- Xin Y, et al. "RNA Sequencing of Single Human Islet Cells Reveals Type 2 Diabetes Genes." Cell Metab 24 (2016): 608-615.
- Vounzoulaki E, et al. "Progression to type 2 diabetes in women with a known history of gestational diabetes: systematic review and meta-analysis." BMJ 369 (2020): m1361.
- Caradonna A, et al. "Meta-Analysis of APP Expression Modulated by SARS-CoV-2 Infection via the ACE2 Receptor." Int J Mol Sci 23 (2022).
- Evans A, et al. “The role of inflammatory cells in fostering pancreatic cancer cell growth and invasion.” Front Physiol 13 (2012): 270.
- Guo J, et al. “Immune regulation of islet homeostasis and adaptation.” Mol Cell Biol 12 (2020): 764-774.
- Arisan ED, et al. "The Prediction of miRNAs in SARS-CoV-2 Genomes: hsa-miR Databases Identify 7 Key miRs Linked to Host Responses and Virus Pathogenicity-Related KEGG Pathways Significant for Comorbidities." Viruses 12 (2020).
- Ragab D, et al. "The COVID-19 Cytokine Storm; What We Know So Far." Front Immunol 11 (2020): 1446.
- Ragab D, et al. "A case of COVID-19, with cytokine storm, treated by consecutive use of therapeutic plasma exchange followed by convalescent plasma transfusion: A case report." J Med Virol 93 (2021): 1854-1856.
- Fignani D, et al. "SARS-CoV-2 Receptor Angiotensin I-Converting Enzyme Type 2 (ACE2) Is Expressed in Human Pancreatic beta-Cells and in the Human Pancreas Microvasculature." Front Endocrinol (Lausanne) 11 (2020): 596898.
- King SB, et al. Comparative genomic analysis reveals varying levels of mammalian adaptation to coronavirus infections” PLoS Comput Biol 17 (2021): e1009560.
- Radzikowska U, et al. “Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthama, COPD, obesity, hypertension, and COVID-19 risk factors.” Allergy 75 (2020): 2829-2845.
- Valencia I, et al. “DPP4 and ACE2 in Diabetes and COVID-19: Therapeutic targets for cardiovascular complications?” 7 (2020): 1161.