Molecular Epidemiology of Human Adenovirus in Asturias (2011–2023)
Article Information
Marta E. Alvarez-Argüelles1, 2, José María González Alba1, Susana Rojo-Alba1, 2, José A. Boga1, 2, Zulema Pérez-Martínez1, 2, Costanza Gómez-deOña2, Cristina Ochoa-Varela1, 2, Mercedes Rodríguez-Pérez1, 2 and Santiago Melón García*, 1, 2
1Unit of Virology, Microbiology Department, Hospital Universitario Central de Asturias, Oviedo, Spain
2Health Research Institute of the Principality of Asturias (ISPA), Oviedo, Spain
*Corresponding author: Santiago Melón García, Unit of Virology, Microbiology Department, Hospital Universitario Central de Asturias, Oviedo, Spain
Received: 21 August 2023 Accepted: 28 August 2023 Published: 07 September 2023
Citation: Marta E. Alvarez-Argüelles, José María González Alba, Susana Rojo-Alba, José A. Boga, Zulema Pérez-Martínez, Costanza Gómez-deOña, Cristina Ochoa-Varela, Mercedes Rodríguez- Pérez and Santiago Melón García. Molecular Epidemiology of Human Adenovirus in Asturias (2011–2023). Archives of Microbiology and Immunology. 7 (2023): 198-212.
View / Download Pdf Share at FacebookAbstract
Human Adenoviruses (HAdV) are implicated in multiples pathologies causing mild to severe disease. The predominant genotypes detected in association with disease differ among different countries or regions, and change over time. In Spain and elsewhere little is known about the molecular epidemiology of HAdV. From a total of 250 HAdV, members of five species were present: A (1), B (126), C (87), D (27) and E (9). The most found genotypes were B3 (119), C2 (48), C1 (29) and D8 (26). Genotypes E4, C5, C6, B7, A31, B35 and D56 were also detected. HAdV diversity increases over the years until the B3 genotype displaces all other types in 2016 and 2023. HAdV detected in Asturias were similar to those already described in other countries, no new local genotype was observed. Genotypes 1-7 were more frequent in children under 15 years of age, while types 8-56 were more frequent in the elderly. Multiple HAdV introductions must have occurred given that only small transmission clades can be inferred. The diversity of the epidemic increased with the years until it disappeared one year periodically. The COVID-19 epidemic accelerated the loss of diversity suggesting that interventions during the pandemic were able to reduce HAdV transmission.
Keywords
Human Adenovirus, Molecular epidemiology pattern, HAdV Genotypes
Human Adenovirus articles, Molecular epidemiology pattern articles, HAdV Genotypes articles
Article Details
1. Background
Human Adenoviruses (HAdV) are a leading pathogen of clinical diseases, such as gastroenteritis, conjunctivitis, respiratory illnesses, hemorrhagic cystitis, and systemic infections. HAdV have been categorized into seven species (A-G) [1-3] and have more than 113 types. Types were initially distinguished by serotyping the neutralization loops of the hexon capsid protein (types 1-51) and later by sequencing all three major capsid proteins: penton, hexon, and fiber (types 52-105) [4,5]. HAdV infection can occur sporadically, endemically, or epidemically and often is influenced by HAdV species and type. Transmission of novel strains between countries or across continents and replacement of dominant serotypes by new strains may occur. A full understanding of the epidemiology of HAdV is important to help clinicians identify infections early and take preventive measures [6]. The search for HAdV in clinical specimens has been increasing in recent years, however few hospitals routinely carry out HAdV culture and typing tests. Here, we conducted a 13-year study detecting, isolating, and genotyping HAdV in clinical samples in Asturias from 2011, to gain a better understanding of HAdV infection through long-term investigation. The aim of this study are to enrich the poor data of epidemiological molecular studies on HAdV in Spain, to support the benefit of molecular surveillance as a tool to determine the epidemiology of viruses circulating in each community, and to underline the need for the design and support of similar long-term studies.
2. Materials and methods
2.1 Samples and patients
From January 2011 to February 2023, 179,081 samples (respiratory samples of all types, stool, blood, tears, corneal scrapings, biopsies, etc.) for diagnosis of HAdV among other pathogens were processed (Table 1). In 250 samples, belong same number of patients hexon gene were sequenced. These specimens were chosen at random among the positive samples detected by a real time PCR, with cycle threshold lower than 25, specially in young and old people, and with apparent clinical significance. Of them, 188 (75%) were nasal/pharyngeal swabs, 37 (15%) were conjunctival and the remaining 25 (10%) were from various sources (blood, cutaneous, endomyocardial biopsy, genital ulcer, pleural fluid, sputum, stool, urine, vaginal and wound). These samples belonged to 134 (53.6%) male and 116 (46.4%) female. In age groups, 58 (23%) were younger than 2 years, 87 (35%) between 2-4 years, 50 (20%) between 5-14 years, 44 (17.6%) were between 15-68 and 11 (4.4%) older. Demographic data (age and sex) from those patients and type of sample are in
2.2 Virus detection and sequencing
Nucleic acids were extracted/purified by using an automated nucleic acid purifier Magnapure 96 (Roche Diagnostics, Mannheim, Germany/Switzerland) following manufacturer´s instructions. Adenoviral genome was detected by a real time (RT)-PCR according to Lu et al [32]. For characterization, a fragment of the hexon gene were amplified by a nested-PCR using primers and protocol described by Lu et al. [32] PCR products were analyzed by agarose gel electrophoresis, extracted by using Montage DNA Gel Extraction Kit (Millipore, USA) and sequenced with Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) using inner primers.
2.3 Phylogenetic reconstructions
The genotyping analysis of HAdV was used to characterize which types circulated in Asturias and to investigate whether phylogenetic clusters occurred in the study period. Nucleotide sequences were translated and aligned using the MUSCLE algorithm implemented in MEGA. Sequences (until 862 nucleotides) generated in this work have been deposited in GenBank with the following accession numbers: OQ680228-OQ680477. For type characterization, phylogenetic trees were constructed using ModelFinder, tree reconstruction and ultrafast bootstrap (1000 replicates) with IQ-TREE 2.1.3. HAdV complete genomes (1090) available in the NCBI Database www.ncbi.nlm.nih.gov; accessed on December 2022) (accession numbers are in Supplementary File S2) were used as reference. The best-fit nucleotide substitution model GTR+F+I+G4 was identified according to Bayesian information criterion. Bootstrap values were estimated using the SH test and ultrafast bootstrap. Diversity (D = 1 − ∑f2) of HAdV genotypes in Asturias was analyzed over time, a measure of variability that takes into account the frequencies (f) of all types. To investigate the possible transmission clades that occurred in Asturias. One sequence of each type identified used for the Basic Local Alignment Search Tool (BLAST) in January 2023 and the same phylogenetic analysis was performed for each type independently using the downloaded sequences.Transmission clusters were defined as viral sequences circulating in Asturias grouped into a single and well supported monophyletic clade with >80% bootstrap values.
2.4 Statistical analysis
Statistical tests (Chi-square, Fisher’s, t-Student’s, etc.) were performed using GraphPad InStat version 3.00 for Windows 95 (GraphPad Software, San Diego, CA).
3. Results
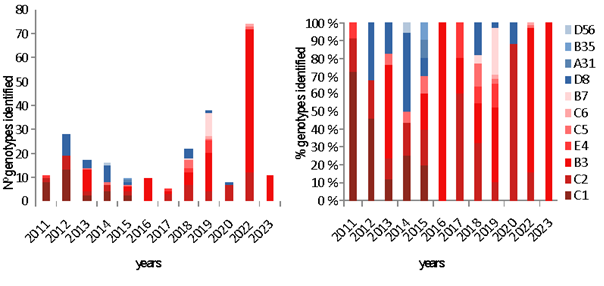
In the study period (January 2011 to February 2023), HAdV was detected in 15035 (9.2%) samples. Table 1 and Fig 1 show the annual distribution of HAdV types. In 2021 no samples were sequenced and in 2023 they were only sequenced up to February.
Table 1: Samples processed incidence and distribution of HAdV types from 2011 to 2023.
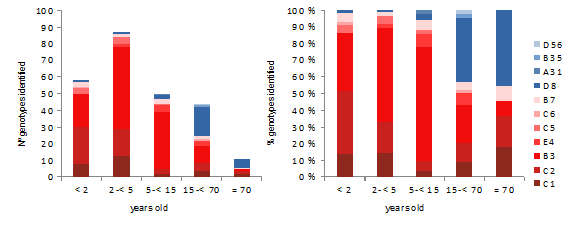
During the period studied, five species (11genotypes) of HAdV were identified: A (1- 0,4%;genotype 31), B (126 -50,4%; genotypes 3, 7, 35), C (87 -34,8%; genotypes 1, 2, 5, 6), D (27 -10.8%, genotypes 8, 56) and E (9 -3.6%; genotype 4). HAdV types according age and sex, and most common genotype are showed in Table 2 by year. Distribution of HAdV genotypes by age is showed in Fig 2.
Table 2: Distribution of HAdv genotypes by sex and age, and most common genotype by year
|
HAdv |
Sex |
Age (years) |
|
||
|
Genotype |
Male/Female |
<2 |
02-Apr |
May-14 |
≥15 |
|
A31 |
1/- |
1 |
|||
|
B3 |
65/49 |
20 |
49 |
34 |
11 |
|
B7 |
05-Jun |
3 |
2 |
3 |
3 |
|
B35 |
1/- |
1 |
|||
|
C1 |
Dec-17 |
8 |
13 |
2 |
6 |
|
C2 |
28/20 |
22 |
16 |
3 |
7 |
|
C5 |
04-Apr |
3 |
4 |
1 |
|
|
C6 |
2/- |
1 |
1 |
||
|
D8 |
Oct-16 |
1 |
1 |
2 |
22 |
|
D56 |
1/- |
1 |
|||
|
E4 |
05-Apr |
2 |
4 |
3 |
|
|
Most common genotype by year |
|||||
|
D8-2012 |
03-Jun |
1 |
8 |
||
|
B3-2013 |
06-Mar |
2 |
5 |
1 |
1 |
|
D8-2014 |
04-Mar |
7 |
|||
|
B3-2016 |
02-Aug |
1 |
3 |
2 |
4 |
|
C1-2018 |
01-Aug |
4 |
1 |
4 |
|
|
B3-2019 |
08-Aug |
2 |
8 |
4 |
2 |
|
C2-2020 |
04-Mar |
7 |
|||
|
B3-2022 |
37/23 |
7 |
29 |
22 |
2 |
|
B3-2023 |
06-May |
2 |
3 |
5 |
1 |
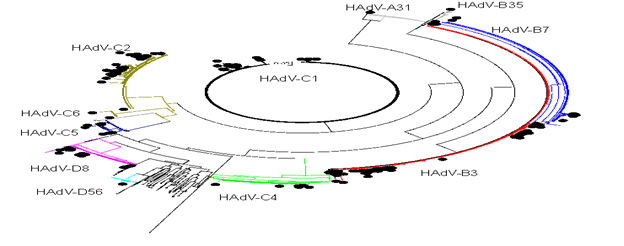
Genotypes 1 to 7 were found in 190 (97.4%) children less than 15 years of age and in 31 (56%) olders. Moreover, genotypes 8 to 56 were found in 5 (2,6%) children under 15 years of age, in 19 (43%) patients between 15 and 70 years and in 5 (45%) olders. (p<0.0001) The most common B3 genotype was found in 103 (90.4%) of children under 15 years old, and C1/C2 in 59 (76.6%) of children under 4 years old. In opposite, D8 was detected in 22 (84.6%) of people older than 14 years old. The phylogenetic genotyping of the HAdVs studied is shown in Fig 3.
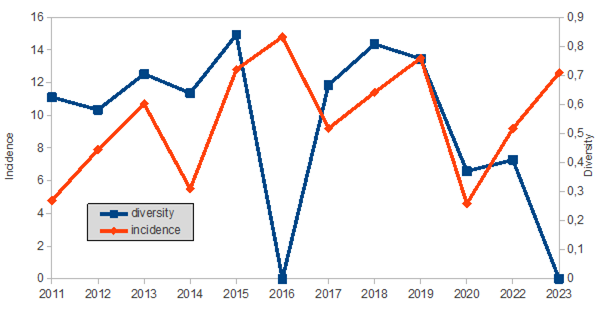
On the other hand, and Fig 4 shows the annual diversity and incidence of the HAdV genotypes throughout the time of the study.
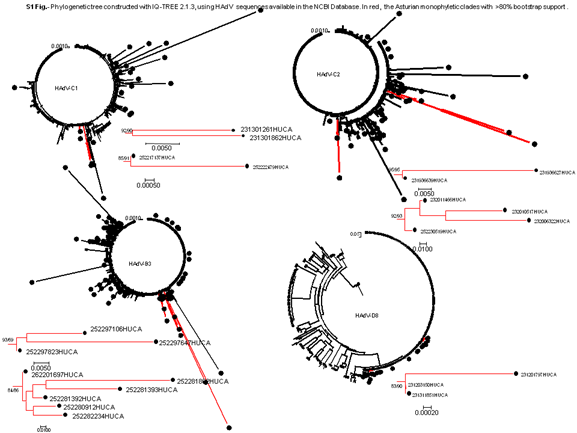
Analyzing the HAdV genotypes circulating, seven well-supported monophyletic clade (bootstrap support of >80%) were identified by phylogeny (Table 3, Supplementary Fig. S1)
Table 3: Characteristics of the samples belonging to the monophyletic cluster found in Asturias
4. Discussion
Human Adenoviruses, implicated in various pathologies, have seen an increase in the search due to their importance. For example, in our laboratory we have gone from processing around 3000 specimens per year to 3000 samples per month. In them, around 10% HAdV was detected. Adenovirus outbreaks are more likely to cause serious disease in infants and immune-compromised hosts, as well as in adults with respiratory or cardiac issues [7,8], but infect anyone and can alter biochemical markers [9]. The predominant HAdV types detected in association with clinical presentation differ between different geographic regions and may shift over time according to the year of surveillance. Thus, monitoring the molecular epidemiology of HAdV in different sites and over the years is of high importance [6,10,11].
HAdV as one of the causative agents of severe acute respiratory infection can involve all ages, however, their infections occur more frequently in children younger than 5 years old [12], In this study HAdV mainly was sequenced in children under 15 years of age in up to 78% of cases, and half of them occur in children under 4 years of age. In this paediatric age group (children under 15 years of age ) genotypes 1 to 7 accounted for almost 90% of the cases according to other studies [13]. In older patients, these genotypes accounted for half of the cases. And in patients older than 70 years, it almost reached 60%, indicating that these genotypes increase with age. In total, eleven different HAdV genotypes were found throughout the study. However, diversity usually increases from year to year. Even the majority one-year genotypes changed in subsequent years, at least before the SARS-Cov-2 pandemic. After 2020, HAdv-B3 became predominant two years in a row.
Genotype B3 and, in general, those belonging to species B were the most frequently detected in our sampling, although there are years in which it was not detected. HAdV-B causes more severe respiratory tract infections than other genotypes and is more likely to result in hospitalization. For example, HAdV-B3 is considered to be related to severe and complicated pneumonia [11]. Our findings are consistent with the global predominance of B3 (28% before the COVID-19 pandemic and 71% after) as the most common genotype implicated in reportable adenoviral infections in children and adults [14]. This was the most identified genotype every 3 years and is the predominant one in the current year.
Other HAdV-B genotype found, but not in a high rate, was B7. Several reports linking HAdV-B7 with more severe acute respiratory disease [13,15-17], and in South America has been a predominant strain [7]. In this study, HAdV-B7 was only found just before the onset of the COVID-19 epidemic, and stopped circulating. Finally, one HAdV-B35 was found in an urine of a man of 50 years old. This genotype has been described and associated with hemorrhagic cystitis [18], as in our case. Other HAdV that are associated with symptomatic respiratory infections worldwide include species C (types 1, 2, and 5) and E (type 4) [10,19-21]. Infections with HAdV-C often are endemic, mild, and most commonly seen in young children, but are important in immunocompromised patients [19,22].
In this work, HAdV-C was the second most frequent species, accounting for one in three of the cases, and basically in children younger than) 5 years, specially HAdV-C2 and HAdV-C1. HAdV-C2 was the first mainly in patients under 2 years of age and the second in patients between 2 and 5 years of age. Others HAdV-C types found was C5 and C6, both related to acute respiratory illnesses [23]. Before SARS-Cov-2 pandemic, HAdV-C species were the most indentified in America [24], but not in our region, that followed the incidence reported in United Kingdom of 12% for C1 and 19% for C2 [25]. With respect to the HAdV-E specie, E4 is associated with respiratory infections but mostly among military recruits and at a low frequency in the general population [26].HAdV-E4 was reported with sporadic circulation, but increased in many countries in recent years [27]. In contrast to other types, HAdv-E4 was found in people older than two years. In this context, in people older than 15 years, the most frequently detected HAdV genotype was HAdV-D8. HAdV-D species cause keratoconjunctivitis, and the most common cause of adenoviral keratoconjunctivitis has been associated to type 8 [28]. In our study all of D8 genotypes were isolated from conjunctival swabs (smears) except one. In addition to the HAdV-D genotypes, we found a case of D56, also associated with conjunctivitis. D56 has been reported in cases of epidemic keratoconjunctivitis as well as in cases of male urethritis with concurrent conjunctivitis [29].
Finally, only one case was found in our study of HAdV-A31 in a stool of a male of 8 year old. A31 is associated with gastroenteritis in children, circulates worldwide and has a high seroprevalence [30]. In the only study found carried out in Spain (Madrid) between the years 2005-2013 where 154 sequences were included, the most frequent genotypes were B3 (24%), C6 (21%), C5 (20%) and C2 (19%) [31]. The distribution of the genotypes is different from that of our region. As previously mentioned, the appearance of the COVID-19 epidemic accelerated the loss of diversity. Before the SARS-Cov-2 pandemic, the five species with the majority of C but predominance of type B3 were found. After 2020, the three main species (B, C, D) were maintained with a clear dominance also of type B3. Analyzing the occurrence of the different types in each year, of the eleven HAdV characterized, only four (C1, C2, B3 and D8) were found more frequently in each year of study. And if the distribution of genotypes over time is observed, the diversity seems to show a pattern every six years, increasing with the years until decreasing due to the dominance of the B3 genotype. Establishing transmission chains in viral infections is a challenge: infections are usually mild or asymptomatic and viruses may persist in the environment for long periods. In this case, no major transmission clades were identified. Only one C2 variant and one D8 variant were detected that could be transmitted in more than one year. This suggests that multiple HAdV introductions must have occurred and the dissemination of HAdV infections is controlled among immunocompetent hosts. Furthermore, the results do not fully prove transmission, as the genetic diversity of circulating HAdV remains obscure without sufficient sampling of circulating strains and sequencing of short fragments. Globally, C1, C2, B3, E4, C5, C6, B7, B14, and B55 were the main genotypes in causing outbreaks [7]. In our study, phylogenetically related (possible outbreaks) viral sequences were identified in genotypes C1, C2, B3 and D8; the first three cases related to individuals younger than 7 years and D8 to individuals older than 15 years.
Our study is limited to a geographic region. Due to the small sample size, the results may not accurately reflect the diversity circulating in this area; for the same reason we did not analyse whether HAdV types show any seasonality. In addition, molecular characterization based on more than one gene is currently advised to better identify recombination phenomena and new types, but sequencing was done before methods based on more than one gene became available. The construction of these data sets is limited due to the lack of sequence submission to GenBank. Despite the small number of sequences, they represent 1% of all those available in GenBank.
5. Conclusions
In summary, up to 5 species and 11 HAdV genotypes were found in the last decade in Asturias. Diversity followed a pattern decreasing every 6 years. The COVID-19 epidemic accelerated the loss of diversity suggesting that interventions during the pandemic were able to reduce HAdV transmission. Genotypes 1-7 were associated with children and the rest with older individuals. Each type of HAdV has been associated mainly with a specific presentation of the disease and in our sampling we have found that the associations already described are fulfilled in all cases. In Spain and elsewhere, there are a limited number of studies in regards to the incidence and molecular epidemiology of HAdV cases. Due to the involvement of HAdV in several pathologies, and the relationship with certain ages, as demonstrated in this study, the implementation of a laboratory-based surveillance system that includes genotyping should be considered.
Acknowledgement
Project partially funded by Grupin IDI/2021/000033
References
- Madisch I, Harste G, Pommer H, Heim A. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classi?cation and taxonomy. J. Virol 79 (2005): 15265-15276
- Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, et al. Molecular evolution of human adenoviruses. Sci. Rep 3 (2013): 1812.
- Greber UF. Adenoviruses - infection, pathogenesis and therapy. FEBS Lett 594 (2020): 1818-1827.
- Aoki K, Benkö M, Davison AJ, Echavarria M, Erdman DD, Harrach B, et al. Members of the Adenovirus Research Community. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology J Virol 85 (2011): 5703-4.
- Seto D, Chodosh J, Brister JR, Jones MS. Members of the Adenovirus Research Community. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol 85 (2011): 5701-2.
- Shen K, Wang Y, Li P, Su X. Clinical features, treatment and outcomes of an outbreak of type 7 adenovirus pneumonia in centralized residence young adults. J Clin Virol 154 (2022): 105244.
- Lynch JP 3rd, Kajon AE. Adenovirus: Epidemiology, Global Spread of Novel Serotypes, and Advances in Treatment and Prevention. Semin Respir Crit Care Med 37 (2016): 586-602.
- Kumthip K, Khamrin P, Ushijima H, Maneekarn N. Enteric and non-enteric adenoviruses associated with acute gastroenteritis in pediatric patients in Thailand, 2011 to 2017. PLoS One 14 (2019): e0220263.
- Gómez de Oña C, Alvarez-Argüelles M E, Rojo-Alba S, Casares H, Arroyo M, Rodríguez J, et al. Alterations in biochemical markers in adenovirus infection. Translational pediatrics 10 (2021): 1248-1258.
- Zadheidar S, Yavarian J, Heydarifard Z, Nejati A, Sadeghi K, Ghavami N, et al.. Molecular epidemiology of human adenoviruses in children with and without respiratory symptoms: Preliminary findings from a case-control study. BMC Pediatr 22 (2022): 583.
- Shen K, Wang Y, Li P, Su X. Clinical features, treatment and outcomes of an outbreak of type 7 adenovirus pneumonia in centralized residence young adults. J Clin Virol 154 (2022): 105244.
- Shafiei-Jandaghi NZ, Yavarian J, Malekshahi SS, Naseri M, Shadab A, Ghavami N, et al. Identification of adenovirus species in Iranian pediatric population with severe acute respiratory infections. Future Virol 14 (2019): 577-83
- Xie L, Zhang B, Xiao N, Zhang F, Zhao X, Liu Q, et al. Epidemiology of human adenovirus infection in children hospitalized with lower respiratory tract infections in Hunan, China. J Med Virol 91 (2019): 392-400.
- Wang YF, Shen FC, Wang SL, Kuo PH, Tsai HP, Liu CC, et al. Molecular Epidemiology and Clinical Manifestations of Adenovirus Respiratory Infections in Taiwanese Children. Medicine (Baltimore) 95 (2016): e3577.
- Scott MK, Chommanard C, Lu X, Appelgate D, Grenz L, Schneider E, et al. Human Adenovirus Associated with Severe Respiratory Infection, Oregon, USA, 2013-2014. Emerg Infect Dis 22 (2016): 1044-51.
- Barnadas C, Schmidt DJ, Fischer TK, Fonager J. Molecular epidemiology of human adenovirus infections in Denmark, 2011-2016. J Clin Virol 104 (2018): 16-22.
- Callaway Z, Kim SH, Kim JY, Kim DW, Kim C-K. Adenovirus infection with serious pulmonary sequelae in Korean children. Clin Respir J 5 (2011): 92-8.
- Hofland CA, Eron LJ, Washecka RM. Hemorrhagic adenovirus cystitis after renal transplantation. Transplant Proc 36 (2004): 3025-7.
- Lee J, Choi EH, Lee HJ. Comprehensive serotyping and epidemiology of human adenovirus isolated from the respiratory tract of Korean children over 17 consecutive years (1991-2007). J MedVirol 82 (2010): 624-631.
- Heim A, Ebnet C, Harste G, Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 70 (2003): 228-39.
- Yeung R, Eshaghi A, Lombos E, et al. Characterization of culture positive adenovirus serotypes from respiratory specimens in Toronto, Ontario, Canada: September 2007-June (2008).
- Wu X, Zhang J, Lan W, Quan L, Ou J, Zhao W, et al. Molecular Typing and Rapid Identification of Human Adenoviruses Associated With Respiratory Diseases Using Universal PCR and Sequencing Primers for the Three Major Capsid Genes: Penton Base, Hexon, and Fiber. Front Microbiol 12 (2022): 911694.
- Probst V, Datyner EK, Haddadin Z, Rankin DA, Hamdan L, Rahman HK, et al. Human adenovirus species in children with acute respiratory illnesses. J Clin Virol 134 (2021): 104716.
- Centers for Disease Control and Prevention Adenoviruses | Surveillance Data for National Adenovirus Type Reporting System (NATRS) (2022).
- Cooper RJ, Hallett R, Tullo AB, Klapper PE. The epidemiology of adenovirus infections in Greater Manchester, UK 1982-96. Epidemiol Infect 125 (2000): 333-345.
- Marcone DN, Culasso ACA, Reyes N, Kajon A, Viale D, Campos RH, et al. Genotypes and phylogenetic analysis of adenovirus in children with respiratory infection in Buenos Aires, Argentina (2000-2018). PLoS One 16 (2021): e0248191.
- Coleman KK, Wong CC, Jayakumar J, Nguyen TT, Wong AWL, Yadana S, et al. Adenoviral infections in Singapore: should new antiviral therapies and vaccines be adopted? J Infect Dis 221 (2020): 566-77.
- Seo JW, Lee SK, Hong IH, Choi SH, Lee JY, Kim HS, et al. Molecular Epidemiology of Adenoviral Keratoconjunctivitis in Korea. Ann Lab Med 42 (2022): 683-687.
- Otto WR, Lamson DM, Gonzalez G, Weinberg GA, Pecora ND, Fisher BT, et al. Fatal Neonatal Sepsis Associated with Human Adenovirus Type 56 Infection: Genomic Analysis of Three Recent Cases Detected in the United States. Viruses 13 (2021): 1105.
- Götting J, Baier C, Panagiota V, Maecker-Kolhoff B, Dhingra A, Heim A. High genetic stability of co-circulating human adenovirus type 31 lineages over 59 years. Virus Evol 8 (2022): veac067.
- Calvo C, García-García ML, Sanchez-Dehesa R, Román C, Tabares A, Pozo F, et al. Eight Year Prospective Study of Adenoviruses Infections in Hospitalized Children. Comparison with Other Respiratory Viruses. PLoS One 10 (2015): e0132162.
- Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol 151 (2006): 1587-602.
Supplementary File: S2
|
Sequence_ID |
isolate_source |
Collection_Date |
Age |
Sex |
Genotype |
|
231112888HUCA |
pharyngeal exudate |
29-07-11 |
1 |
Female |
1 |
|
231113147HUCA |
blood |
04-08-11 |
0 |
Male |
2 |
|
231117445HUCA |
conjunctival exudate |
18-10-11 |
32 |
Female |
4 |
|
231117854HUCA |
urine |
24-10-11 |
28 |
Female |
2 |
|
231200908HUCA |
pharyngeal exudate |
16-01-12 |
2 |
Male |
2 |
|
231201797HUCA |
conjunctival exudate |
26-01-12 |
78 |
Male |
8 |
|
231201959HUCA |
pharyngeal exudate |
30-01-12 |
1 |
Male |
1 |
|
231202275HUCA |
conjunctival exudate |
02-02-12 |
32 |
Female |
8 |
|
231202664HUCA |
pharyngeal exudate |
07-02-12 |
3 |
Female |
2 |
|
231202709HUCA |
conjunctival exudate |
08-02-12 |
64 |
Female |
8 |
|
231202726HUCA |
conjunctival exudate |
08-02-12 |
54 |
Female |
8 |
|
231202819HUCA |
conjunctival exudate |
09-02-12 |
56 |
Female |
8 |
|
231202919HUCA |
conjunctival exudate |
10-02-12 |
59 |
Female |
8 |
|
231203150HUCA |
conjunctival exudate |
14-02-12 |
39 |
Male |
8 |
|
231204488HUCA |
nasal wash |
01-03-12 |
39 |
Male |
2 |
|
231206599HUCA |
pharyngeal exudate |
28-03-12 |
1 |
Female |
2 |
|
231209922HUCA |
conjunctival exudate |
14-05-12 |
11 |
Female |
8 |
|
231212324HUCA |
nasal wash |
14-06-12 |
15 |
Female |
2 |
|
231212745HUCA |
pharyngeal exudate |
21-06-12 |
100 |
Male |
1 |
|
231219650HUCA |
pharyngeal exudate |
21-10-12 |
2 |
Female |
2 |
|
231224240HUCA |
conjunctival exudate |
27-12-12 |
64 |
Male |
8 |
|
231300484HUCA |
conjunctival exudate |
09-01-13 |
26 |
Female |
8 |
|
231301050HUCA |
aspirate |
17-01-13 |
1 |
Male |
2 |
|
231301261HUCA |
nasal exudate |
19-01-13 |
3 |
Male |
1 |
|
231301637HUCA |
pharyngeal exudate |
23-01-13 |
4 |
Male |
3 |
|
231301861HUCA |
pharyngeal exudate |
26-01-13 |
1 |
Female |
1 |
|
231301862HUCA |
pharyngeal exudate |
26-01-13 |
2 |
Male |
1 |
|
231303015HUCA |
nasopharyngeal exudate |
12-02-13 |
0 |
Female |
5 |
|
231304103HUCA |
pharyngeal exudate |
27-02-13 |
35 |
Female |
3 |
|
231304802HUCA |
pharyngeal exudate |
08-03-13 |
4 |
Male |
3 |
|
231305133HUCA |
pharyngeal exudate |
13-03-13 |
3 |
Male |
1 |
|
231305178HUCA |
pharyngeal exudate |
14-03-13 |
8 |
Female |
2 |
|
231305188HUCA |
pharyngeal exudate |
14-03-13 |
3 |
Female |
3 |
|
231311851HUCA |
conjunctival exudate |
13-06-13 |
100 |
Female |
8 |
|
231312106HUCA |
pharyngeal exudate |
17-06-13 |
4 |
Female |
3 |
|
231312326HUCA |
pharyngeal exudate |
20-06-13 |
7 |
Male |
3 |
|
231312737HUCA |
nasal exudate |
26-06-13 |
0 |
Male |
3 |
|
231313679HUCA |
conjunctival exudate |
11-07-13 |
40 |
Female |
8 |
|
231313817HUCA |
pharyngeal exudate |
13-07-13 |
2 |
Male |
3 |
|
231314692HUCA |
nasopharyngeal exudate |
29-07-13 |
0 |
Male |
3 |
|
231402626HUCA |
conjunctival exudate |
10-03-14 |
89 |
Male |
8 |
|
231402723HUCA |
conjunctival exudate |
10-03-14 |
73 |
Female |
8 |
|
231403901HUCA |
conjunctival exudate |
10-03-14 |
19 |
Male |
8 |
|
231405710HUCA |
conjunctival exudate |
10-03-14 |
51 |
Female |
8 |
|
231405872HUCA |
conjunctival exudate |
19-03-14 |
67 |
Male |
8 |
|
231406074HUCA |
conjunctival exudate |
19-03-14 |
36 |
Male |
8 |
|
231407089HUCA |
pharyngeal exudate |
16-03-14 |
2 |
Male |
2 |
|
231407615HUCA |
conjunctival exudate |
22-03-14 |
0 |
Male |
2 |
|
231407717HUCA |
pharyngeal exudate |
24-03-14 |
1 |
Male |
2 |
|
231408083HUCA |
conjunctival exudate |
28-03-14 |
63 |
Female |
8 |
|
231422013HUCA |
conjunctival exudate |
18-11-14 |
49 |
Male |
56 |
|
231422873HUCA |
pharyngeal exudate |
01-12-14 |
3 |
Male |
5 |
|
231422955HUCA |
pharyngeal exudate |
02-12-14 |
1 |
Female |
1 |
|
231500133HUCA |
stool |
02-01-15 |
0 |
Male |
5 |
|
231506651HUCA |
pharyngeal exudate |
23-03-15 |
3 |
Male |
1 |
|
231507200HUCA |
urine |
30-03-15 |
61 |
Male |
2 |
|
231507521HUCA |
pharyngeal exudate |
05-04-15 |
0 |
Female |
2 |
|
231508624HUCA |
pharyngeal exudate |
16-04-15 |
3 |
Male |
3 |
|
231517460HUCA |
nasopharyngeal exudate |
19-08-15 |
1 |
Female |
3 |
|
231519497HUCA |
pharyngeal exudate |
20-09-15 |
0 |
Female |
8 |
|
231519946HUCA |
stool |
27-09-15 |
8 |
Male |
31 |
|
231521960HUCA |
urine |
22-10-15 |
64 |
Male |
35 |
|
231607316HUCA |
wound exudate |
08-03-16 |
17 |
Female |
3 |
|
231608400HUCA |
pharyngeal exudate |
17-03-16 |
1 |
Female |
3 |
|
231610616HUCA |
pharyngeal exudate |
10-04-16 |
5 |
Female |
3 |
|
231611442HUCA |
wound exudate |
19-04-16 |
28 |
Male |
3 |
|
231612761HUCA |
pleural fluid |
04-05-16 |
84 |
Male |
3 |
|
231612996HUCA |
stool |
06-05-16 |
3 |
Female |
3 |
|
231613523HUCA |
urine |
12-05-16 |
8 |
Female |
3 |
|
231613812HUCA |
stool |
16-05-16 |
3 |
Female |
3 |
|
231613862HUCA |
pharyngeal exudate |
17-05-16 |
3 |
Female |
3 |
|
231614112HUCA |
conjunctival exudate |
19-05-16 |
32 |
Female |
3 |
|
231711597HUCA |
stool |
09-04-17 |
76 |
Male |
2 |
|
231712928HUCA |
pharyngeal exudate |
24-04-17 |
1 |
Male |
2 |
|
231713057HUCA |
pharyngeal exudate |
26-04-17 |
2 |
Female |
2 |
|
231718312HUCA |
pharyngeal exudate |
22-06-17 |
0 |
Male |
3 |
|
231719290HUCA |
conjunctival exudate |
05-07-17 |
24 |
Female |
4 |
|
231720177HUCA |
nasopharyngeal exudate |
17-07-17 |
1 |
Male |
1 |
|
231802985HUCA |
conjunctival exudate |
18-01-18 |
31 |
Male |
8 |
|
231810967HUCA |
nasopharyngeal exudate |
26-03-18 |
33 |
Female |
3 |
|
231812092HUCA |
conjunctival exudate |
07-04-18 |
2 |
Female |
8 |
|
231812098HUCA |
nasopharyngeal exudate |
08-04-18 |
4 |
Female |
1 |
|
231814699HUCA |
sputum |
02-05-18 |
72 |
Male |
2 |
|
231815448HUCA |
pharyngeal exudate |
09-05-18 |
2 |
Female |
5 |
|
231815465HUCA |
blood |
10-05-18 |
84 |
Female |
1 |
|
231816976HUCA |
conjunctival exudate |
25-05-18 |
35 |
Male |
8 |
|
231817051HUCA |
pharyngeal exudate |
26-05-18 |
0 |
Male |
2 |
|
231817497HUCA |
blood |
31-05-18 |
54 |
Female |
1 |
|
231817542HUCA |
pharyngeal exudate |
31-05-18 |
2 |
Male |
2 |
|
231817634HUCA |
pharyngeal exudate |
31-05-18 |
0 |
Male |
2 |
|
231817907HUCA |
urine |
04-06-18 |
54 |
Female |
1 |
|
231818170HUCA |
nasopharyngeal exudate |
06-06-18 |
2 |
Male |
2 |
|
231818202HUCA |
nasopharyngeal exudate |
06-06-18 |
6 |
Female |
5 |
|
231818217HUCA |
blood |
07-06-18 |
54 |
Female |
1 |
|
231818308HUCA |
nasopharyngeal exudate |
07-06-18 |
2 |
Male |
5 |
|
231818705HUCA |
nasopharyngeal exudate |
13-06-18 |
2 |
Male |
2 |
|
231818892HUCA |
pharyngeal exudate |
14-06-18 |
9 |
Male |
4 |
|
231818968HUCA |
pharyngeal exudate |
15-06-18 |
11 |
Female |
1 |
|
231825910HUCA |
nasopharyngeal exudate |
12-09-18 |
0 |
Male |
3 |
|
231828826HUCA |
conjunctival exudate |
15-10-18 |
70 |
Female |
8 |
|
231828839HUCA |
nasopharyngeal exudate |
15-10-18 |
65 |
Female |
4 |
|
231833746HUCA |
nasopharyngeal exudate |
29-11-18 |
2 |
Female |
1 |
|
231834056HUCA |
nasopharyngeal exudate |
03-12-18 |
2 |
Male |
1 |
|
231834363HUCA |
conjunctival exudate |
05-12-18 |
1 |
Male |
3 |
|
231834369HUCA |
pharyngeal exudate |
05-12-18 |
3 |
Female |
1 |
|
231834370HUCA |
pharyngeal exudate |
05-12-18 |
1 |
Male |
3 |
|
231835154HUCA |
nasopharyngeal exudate |
13-12-18 |
0 |
Male |
2 |
|
231836966HUCA |
nasopharyngeal exudate |
29-12-18 |
16 |
Female |
7 |
|
231837001HUCA |
nasopharyngeal exudate |
29-12-18 |
1 |
Male |
3 |
|
231904615HUCA |
pharyngeal exudate |
29-01-19 |
3 |
Male |
3 |
|
231906748HUCA |
nasopharyngeal exudate |
08-02-19 |
4 |
Female |
3 |
|
231909530HUCA |
pharyngeal exudate |
23-02-19 |
0 |
Male |
7 |
|
231909842HUCA |
conjunctival exudate |
25-02-19 |
3 |
Female |
3 |
|
231909848HUCA |
nasopharyngeal exudate |
26-02-19 |
3 |
Male |
3 |
|
231911751HUCA |
nasopharyngeal exudate |
08-03-19 |
1 |
Male |
7 |
|
231912457HUCA |
pharyngeal exudate |
13-03-19 |
1 |
Male |
1 |
|
231913746HUCA |
nasopharyngeal exudate |
21-03-19 |
0 |
Male |
3 |
|
231914121HUCA |
conjunctival exudate |
25-03-19 |
4 |
Female |
3 |
|
231914124HUCA |
nasopharyngeal exudate |
25-03-19 |
5 |
Female |
7 |
|
231914915HUCA |
pharyngeal exudate |
30-03-19 |
5 |
Female |
7 |
|
231915364HUCA |
nasopharyngeal exudate |
03-04-19 |
3 |
Female |
7 |
|
231915591HUCA |
pharyngeal exudate |
04-04-19 |
3 |
Male |
3 |
|
231915681HUCA |
pharyngeal exudate |
04-04-19 |
3 |
Female |
3 |
|
231917121HUCA |
vaginal |
05-06-19 |
58 |
Female |
3 |
|
231918578HUCA |
conjunctival exudate |
02-05-19 |
47 |
Male |
7 |
|
231920562HUCA |
pharyngeal exudate |
05-06-19 |
37 |
Female |
3 |
|
231922132HUCA |
conjunctival exudate |
31-05-19 |
12 |
Male |
8 |
|
231922134HUCA |
nasopharyngeal exudate |
31-05-19 |
3 |
Male |
4 |
|
231922135HUCA |
pharyngeal exudate |
31-05-19 |
3 |
Male |
3 |
|
231922136HUCA |
pharyngeal exudate |
31-05-19 |
0 |
Female |
7 |
|
231922142HUCA |
pharyngeal exudate |
01-06-19 |
8 |
Male |
3 |
|
231922179HUCA |
pharyngeal exudate |
02-06-19 |
2 |
Female |
1 |
|
231922783HUCA |
nasopharyngeal exudate |
06-06-19 |
3 |
Male |
7 |
|
231922961HUCA |
pharyngeal exudate |
07-06-19 |
9 |
Male |
7 |
|
231922976HUCA |
pharyngeal exudate |
08-06-19 |
4 |
Female |
4 |
|
231923660HUCA |
nasopharyngeal exudate |
14-06-19 |
1 |
Female |
2 |
|
231924319HUCA |
nasal exudate |
20-06-19 |
7 |
Male |
3 |
|
231924526HUCA |
pharyngeal exudate |
22-06-19 |
5 |
Female |
3 |
|
231924846HUCA |
pharyngeal exudate |
25-06-19 |
2 |
Female |
1 |
|
231925749HUCA |
nasopharyngeal exudate |
04-07-19 |
8 |
Male |
3 |
|
231926141HUCA |
nasopharyngeal exudate |
09-07-19 |
8 |
Male |
4 |
|
231926142HUCA |
conjunctival exudate |
09-07-19 |
8 |
Male |
4 |
|
231926170HUCA |
nasopharyngeal exudate |
09-07-19 |
10 |
Male |
4 |
|
231929561HUCA |
nasopharyngeal exudate |
20-08-19 |
1 |
Female |
5 |
|
231929685HUCA |
pharyngeal exudate |
21-08-19 |
1 |
Female |
3 |
|
231932486HUCA |
genital ulcer exudate |
19-09-19 |
73 |
Female |
7 |
|
231936627HUCA |
nasal exudate |
25-10-19 |
1 |
Male |
2 |
|
231936639HUCA |
endomyocardial biopsy |
25-10-19 |
43 |
Male |
2 |
|
231937414HUCA |
pharyngeal exudate |
03-11-19 |
4 |
Female |
2 |
|
231941472HUCA |
nasopharyngeal exudate |
03-12-19 |
52 |
Male |
6 |
|
232003159HUCA |
conjunctival exudate |
13-01-20 |
28 |
Female |
8 |
|
232006322HUCA |
nasopharyngeal exudate |
23-01-20 |
0 |
Male |
2 |
|
232009890HUCA |
pharyngeal exudate |
06-02-20 |
0 |
Female |
2 |
|
232010517HUCA |
pharyngeal exudate |
10-02-20 |
1 |
Female |
2 |
|
232010665HUCA |
pharyngeal exudate |
10-02-20 |
1 |
Male |
2 |
|
232011434HUCA |
pharyngeal exudate |
13-02-20 |
0 |
Male |
2 |
|
232011466HUCA |
pharyngeal exudate |
13-02-20 |
0 |
Male |
2 |
|
232015270HUCA |
cutaneous |
05-03-20 |
65 |
Female |
1 |
|
232015566HUCA |
nasopharyngeal exudate |
06-03-20 |
0 |
Female |
2 |
|
232302445HUCA |
conjunctival exudate |
18-01-23 |
0 |
Male |
3 |
|
232303183HUCA |
nasopharyngeal exudate |
20-01-23 |
10 |
Male |
3 |
|
232305783HUCA |
conjunctival exudate |
31-01-23 |
33 |
Female |
3 |
|
232306242HUCA |
pharyngeal exudate |
02-02-23 |
6 |
Female |
3 |
|
232306407HUCA |
nasopharyngeal exudate |
02-02-23 |
1 |
Male |
3 |
|
232306611HUCA |
nasopharyngeal exudate |
03-02-23 |
5 |
Male |
3 |
|
232306613HUCA |
pharyngeal exudate |
03-02-23 |
6 |
Female |
3 |
|
232306614HUCA |
pharyngeal exudate |
03-02-23 |
2 |
Female |
3 |
|
232306797HUCA |
pharyngeal exudate |
04-02-23 |
3 |
Male |
3 |
|
232306799HUCA |
pharyngeal exudate |
04-02-23 |
4 |
Female |
3 |
|
232307689HUCA |
nasopharyngeal exudate |
08-02-23 |
10 |
Male |
3 |
|
242242022HUCA |
pharyngeal exudate |
20-02-22 |
3 |
Female |
2 |
|
242248319HUCA |
nasopharyngeal exudate |
26-02-22 |
1 |
Male |
6 |
|
242256621HUCA |
pharyngeal exudate |
07-03-22 |
2 |
Female |
2 |
|
242266224HUCA |
nasopharyngeal exudate |
18-03-22 |
2 |
Male |
2 |
|
252209368HUCA |
nasopharyngeal exudate |
10-05-22 |
0 |
Male |
2 |
|
252216986HUCA |
pharyngeal exudate |
19-05-22 |
2 |
Female |
1 |
|
252217137HUCA |
nasopharyngeal exudate |
20-05-22 |
1 |
Male |
1 |
|
252217536HUCA |
pharyngeal exudate |
20-05-22 |
2 |
Female |
3 |
|
252222479HUCA |
pharyngeal exudate |
29-05-22 |
0 |
Male |
1 |
|
252227760HUCA |
pharyngeal exudate |
07-06-22 |
4 |
Male |
5 |
|
252227935HUCA |
nasopharyngeal exudate |
08-06-22 |
9 |
Female |
3 |
|
252228809HUCA |
nasopharyngeal exudate |
09-06-22 |
2 |
Male |
1 |
|
252229369HUCA |
pharyngeal exudate |
11-06-22 |
2 |
Male |
2 |
|
252230519HUCA |
nasopharyngeal exudate |
13-06-22 |
2 |
Female |
2 |
|
252273752HUCA |
nasopharyngeal exudate |
12-09-22 |
1 |
Male |
3 |
|
252273768HUCA |
nasopharyngeal exudate |
12-09-22 |
2 |
Female |
1 |
|
252277437HUCA |
nasopharyngeal exudate |
27-09-22 |
9 |
Female |
1 |
|
252279435HUCA |
pharyngeal exudate |
05-10-22 |
2 |
Female |
2 |
|
252280912HUCA |
nasopharyngeal exudate |
10-10-22 |
2 |
Male |
3 |
|
252281383HUCA |
nasopharyngeal exudate |
12-10-22 |
5 |
Male |
3 |
|
252281392HUCA |
pharyngeal exudate |
12-10-22 |
3 |
Female |
3 |
|
252281393HUCA |
pharyngeal exudate |
12-10-22 |
3 |
Male |
3 |
|
252281600HUCA |
pharyngeal exudate |
13-10-22 |
2 |
Female |
3 |
|
252281862HUCA |
conjunctival exudate |
14-10-22 |
50 |
Female |
3 |
|
252282209HUCA |
pharyngeal exudate |
15-10-22 |
1 |
Female |
2 |
|
252282234HUCA |
pharyngeal exudate |
15-10-22 |
4 |
Male |
3 |
|
252282277HUCA |
pharyngeal exudate |
15-10-22 |
2 |
Female |
3 |
|
252282296HUCA |
pharyngeal exudate |
16-10-22 |
6 |
Female |
2 |
|
252282304HUCA |
pharyngeal exudate |
16-10-22 |
0 |
Male |
2 |
|
252285381HUCA |
nasopharyngeal exudate |
25-10-22 |
4 |
Male |
3 |
|
252285553HUCA |
nasopharyngeal exudate |
26-10-22 |
5 |
Male |
2 |
|
2522911786HUCA |
nasopharyngeal exudate |
15-11-22 |
3 |
Female |
3 |
|
252293430HUCA |
pharyngeal exudate |
20-11-22 |
2 |
Male |
3 |
|
252293434HUCA |
nasopharyngeal exudate |
20-11-22 |
2 |
Female |
3 |
|
252296009HUCA |
nasopharyngeal exudate |
28-11-22 |
5 |
Male |
3 |
|
252297106HUCA |
nasopharyngeal exudate |
01-12-22 |
6 |
Female |
3 |
|
252297286HUCA |
nasopharyngeal exudate |
01-12-22 |
5 |
Male |
3 |
|
252297296HUCA |
pharyngeal exudate |
29-11-22 |
12 |
Male |
3 |
|
252297570HUCA |
pharyngeal exudate |
02-12-22 |
8 |
Female |
3 |
|
252297645HUCA |
pharyngeal exudate |
02-12-22 |
2 |
Female |
3 |
|
252297647HUCA |
nasopharyngeal exudate |
02-12-22 |
3 |
Male |
3 |
|
252297664HUCA |
pharyngeal exudate |
02-12-22 |
2 |
Male |
3 |
|
252297823HUCA |
nasopharyngeal exudate |
02-12-22 |
0 |
Male |
3 |
|
252297854HUCA |
nasopharyngeal exudate |
03-12-22 |
2 |
Female |
2 |
|
252297856HUCA |
pharyngeal exudate |
03-12-22 |
3 |
Female |
3 |
|
252297857HUCA |
pharyngeal exudate |
03-12-22 |
2 |
Male |
3 |
|
252298546HUCA |
nasopharyngeal exudate |
06-12-22 |
1 |
Female |
3 |
|
252299395HUCA |
pharyngeal exudate |
10-12-22 |
3 |
Male |
3 |
|
252299397HUCA |
pharyngeal exudate |
10-12-22 |
6 |
Male |
3 |
|
252299398HUCA |
pharyngeal exudate |
10-12-22 |
6 |
Male |
3 |
|
252299400HUCA |
pharyngeal exudate |
10-12-22 |
3 |
Male |
3 |
|
252299401HUCA |
pharyngeal exudate |
10-12-22 |
2 |
Male |
3 |
|
252299412HUCA |
pharyngeal exudate |
10-12-22 |
2 |
Female |
3 |
|
252299525HUCA |
pharyngeal exudate |
11-12-22 |
8 |
Female |
3 |
|
252299526HUCA |
pharyngeal exudate |
11-12-22 |
7 |
Female |
3 |
|
252299560HUCA |
nasopharyngeal exudate |
11-12-22 |
6 |
Male |
3 |
|
252299958HUCA |
nasopharyngeal exudate |
11-12-22 |
7 |
Male |
3 |
|
262201243HUCA |
nasopharyngeal exudate |
16-12-22 |
4 |
Male |
3 |
|
262201281HUCA |
nasopharyngeal exudate |
15-12-22 |
5 |
Male |
3 |
|
262201497HUCA |
pharyngeal exudate |
17-12-22 |
1 |
Male |
3 |
|
262201556HUCA |
pharyngeal exudate |
17-12-22 |
43 |
Female |
3 |
|
262201566HUCA |
pharyngeal exudate |
17-12-22 |
3 |
Male |
3 |
|
262201653HUCA |
pharyngeal exudate |
18-12-22 |
2 |
Male |
3 |
|
262201683HUCA |
pharyngeal exudate |
18-12-22 |
6 |
Male |
3 |
|
262201697HUCA |
nasopharyngeal exudate |
18-12-22 |
8 |
Male |
3 |
|
262201712HUCA |
pharyngeal exudate |
18-12-22 |
3 |
Female |
3 |
|
262203151HUCA |
pharyngeal exudate |
22-12-22 |
4 |
Male |
3 |
|
262203157HUCA |
nasopharyngeal exudate |
22-12-22 |
1 |
Male |
3 |
|
262203726HUCA |
pharyngeal exudate |
25-12-22 |
1 |
Female |
3 |
|
262203746HUCA |
nasopharyngeal exudate |
26-12-22 |
8 |
Female |
3 |
|
262203747HUCA |
nasopharyngeal exudate |
26-12-22 |
5 |
Male |
3 |
|
262203756HUCA |
pharyngeal exudate |
26-12-22 |
7 |
Female |
3 |
|
262203757HUCA |
pharyngeal exudate |
26-12-22 |
2 |
Male |
3 |
|
262203790HUCA |
pharyngeal exudate |
26-12-22 |
5 |
Male |
3 |
|
262204068HUCA |
pharyngeal exudate |
27-12-22 |
2 |
Male |
3 |
|
262204069HUCA |
nasopharyngeal exudate |
27-12-22 |
3 |
Female |
3 |
|
262204174HUCA |
pharyngeal exudate |
27-12-22 |
3 |
Female |
3 |
|
262204224HUCA |
pharyngeal exudate |
27-12-22 |
7 |
Male |
3 |
|
262204522HUCA |
pharyngeal exudate |
29-12-22 |
0 |
Male |
3 |
|
262204698HUCA |
nasopharyngeal exudate |
29-12-22 |
8 |
Male |
3 |