Molecular Detection of Multi-drug Resistant Tuberculosis in Clinical Isolates from Two Urban Centres in Malawi
Article Information
Tionge Daston Sikwese1, Takondwa Msosa1, Hussein Hassan Twabi1, Samuel Dzunda2, David Chaima1, Billy Banda2, Yusuf Kanamazina2, Mirriam Nyenje2, Alinune Musopole3, Marriott Nliwasa1, Victor Ndhlovu3*
1Helse Nord TB Initiative, Kamuzu University of Health Sciences, Blantyre, Malawi
2Public Health Institute of Malawi, National TB Reference Laboratory, Lilongwe, Malawi
3Department of Biomedical Sciences, Kamuzu University of Health Sciences, Blantyre, Malawi
*Corresponding author : Dr. Victor Ndhlovu, Department of Biomedical Sciences, Kamuzu University of Health Sciences, Blantyre, Malawi.
Received: 28 April 2025; Accepted: 05 May 2025; Published: 22 September 2025
Citation: Tionge Sikwese, Takondwa Msosa, Hussein Twabi, Samuel Dzunda, David Chaima, Billy Banda, Yusuf Kanamazina, Mirriam Nyenje, Alinune Musopole, Marriott Nliwasa, Victor Ndhlovu. Molecular Detection of Multi-drug Resistant Tuberculosis in Clinical Isolates from Two Urban Centres in Malawi. Archives of Clinical and Biomedical Research. 9 (2025): 376-382.
View / Download Pdf Share at FacebookAbstract
Introduction: Suboptimal chemotherapy allows Mycobacterium tuberculosis to develop drug resistance owing to development of resistant mutants in the mycobacterial population. Early diagnosis of TB and identification of drugresistance is of particular importance in human immunodeficiency virus (HIV)-infected individuals, as delay of therapy and subsequent development of drug-resistant TB can be devastating due to compromised immune systems.
Methodology: We conducted a cross-sectional evaluation study using presumptive M. tuberculosis positive clinical isolates at two urban sites in Malawi (Blantyre and Lilongwe) to assess the presence of mutant genes on first and second line TB drugs using Line Probe Assay (LPA) and the gold standard drug susceptibility test (DST).
Results: For the Lilongwe site, the incidence of MDR-TB by Line Probe Assay (LPA) was found to be 14.06% (95% CI: 8%-20%) whereas that for Rif mono-resistance was 6.25% (95% CI: 2%-10%). Contrastingly, MDRTB by DST was 23.44% (95 CI:16% - 21%) while mono-resistance was 6.25% (95% CI:2% -10). There was a substantial agreement on the detection of MDR-TB (kappa statistic was 0.75 with 95% CI of 0.62-0.88). Blantyre site, at 9.5% confidence interval, the point estimate for MDR-TB was 0% while for INH mono-resistance TB was 3.3%.
Conclusion: There is high incidence of MDR-TB among patients whose samples are sent to the Lilongwe site than previously thought. A short turnaround time to diagnosis, and the ability to simultaneously detect rifampicin and isoniazid resistance, makes LPA a reliable tool for the early detection of multidrug-resistant tuberculosis.
Keywords
Line Probe Assay (LPA); Phenotypic DST; Multi-drug resistant tuberculosis
Line Probe Assay (LPA) articles; Phenotypic DST articles; Multi-drug resistant tuberculosis articles
Article Details
1. Introduction
Tuberculosis (TB) remains a global public health emergency, which, until coronavirus (COVID-19), was the leading cause of death from a single infectious agent, Mycobacterium tuberculosis (Mtb), ranking above HIV/ AIDS [1]. Globally, there were 1.4 million TB deaths among HIV-negative people and an additional 187,000 among HIV-positive people in 2021 alone [1]. Suboptimal chemotherapy enables M. tuberculosis to develop drug resistance through the emergence of resistant mutants, which are introduced into the mycobacterial population [2,3]. Mono-resistant M. tuberculosis is when an isolate is resistant to only one drug, whereas multi-drug resistant (MDR) M. tuberculosis refers to the in-vitro resistance to at least rifampicin (RIF) and isoniazid (INH), the two main first-line anti-TB drugs. A majority of RIF-resistant M. tuberculosis isolates possess genetic mutations within the 81bp core region of the rpoB gene (Rifampicin resistance determining region RRDR) [4-6], whereas INH resistance is mediated via mutations within the katG gene or the promoter region of the inhA gene [7,8].
Multi-drug-resistant TB is an emerging issue in Malawi, and results from the national TB prevalence survey completed in 2014 showed MDR-TB prevalence at 0.4% among new patients and 4.8 % in previously treated TB patient populations, respectively [9].
Early diagnosis of TB and identification of drug resistance is of particular significance in human immunodeficiency virus (HIV)-infected individuals, as delay of therapy and the resulting drug-resistant TB can be devastating, especially for compromised immune systems.
Currently, phenotypic drug susceptibility testing (DST) remains the gold standard for the detection of resistance among TB patients. However, the method suffers from a long culture period of ~ 3-6 weeks to obtain results [10]. Additionally, critical drug concentrations for second-line drugs have not been completely established. The development of a new, rapid DST that is user-friendly and inexpensive is thus an urgent priority for determining the susceptibility to second-line drugs [11]. A rapid molecular test known as Geno Type MTBDR plus (Hain, Life Science) is a Line Probe Assay (LPA), which has been approved by WHO since 2008 for the diagnosis of MDR-TB [12], uses Polymerase Chain Reaction (PCR) and reverse hybridization methods for the rapid detection of mutations associated with drug resistance [13]. They are designed to identify Mycobacterium Tuberculosis Complex (MTBC) and simultaneously detect mutations in the rpoB, katG, and inhA genes associated with drug resistance [14,15].
Patients infected with MDR-TB strains have particularly poor treatment outcomes and are more likely to remain sources of infection for longer periods of time [8,16,17]. This study was conducted to investigate the emerging cases of MDR-TB from two urban centres in Malawi (Blantyre and Lilongwe) and evaluate the performance of LPA against the standard DST. Monitoring circulating MDR-TB strains remains a critical part of any TB control program and a key factor to reduce and contain the spread of these resistant strains. Understanding the mechanisms of action of anti-TB drugs and the development of drug resistance will allow us to identify new drug targets and better ways to detect drug resistance [5]. To date and to the best of our knowledge, few studies in Malawi have focused on the burden of MDR-TB using molecular-based techniques [18,19]. Additionally, such studies are limited as they have focused only on particular geographical regions of the country and may not be representative of national data.
2. Methodology
2.1 Ethical consideration
Ethical clearance was obtained from the College of Medicine Research Ethics Committee (Certificate number COMREC: P.09/21/3418).
2.2 Study setting and population
This was a cross-sectional evaluation study using pre-existing M. tuberculosis positive clinical isolates from Kamuzu University of Health Sciences (KUHeS)/Malawi Liverpool Welcome Trust (MLW) TB laboratory and the TB national Reference Laboratory, complete with patient data. The study was approved by the College of Medicine Research Ethics Committee (Comrec). All the isolates originating from KUHeS/MLW TB laboratory belonged to a previous study (Exact TB study), with inclusion criteria: adults aged ≥ 18 years, having TB symptoms (cough of any duration, fever, night sweats or weight loss), able to submit sputum, willing and able to provide informed consent. Exclusion criteria: Presence of clinical danger signs (unable to walk unaided, confused or agitated, breathless when speaking or at rest), already taking TB treatment, or taken TB treatment within the last 60 days, unable to submit sputum and refuses offer of sputum induction, previously been enrolled in the trial), which was conducted at ART clinics in Bangwe Health Centre and Queen Elizabeth Central Hospital in Blantyre, Malawi. These health centres represent high-density suburbs of Blantyre with an estimated TB prevalence of 1,014 per 100,000. The KUHeS/MLW TB laboratory receives samples from Blantyre and surrounding areas, while the National TB reference laboratory in Lilongwe receives samples from all the regions of Malawi, which are suspected to be MDR-TB (These are patients who are already on TB treatment).
2.3 Study procedures
The isolates were retrieved from frozen state using the unique patient identification number (PID) and generated laboratory number, allowed to thaw at 37°C before being inoculated into tubes of Middlebrook 7H9 broth base supplemented with oleic acid, albumin, dextrose, and catalase (OADC) and an antibiotic mixture of polymyxin B, amphotericin B, nalidixic acid, trimethoprimandazlocillin (PANTA). The tubes were loaded in a BACTEC MGIT 96 °C machine, and 37 °C monitored weekly for growth of the culture.
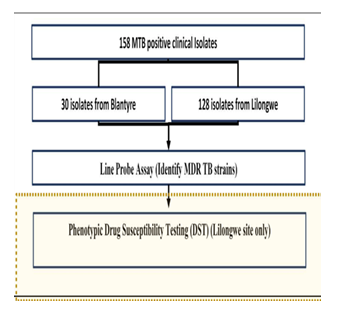
Genotype MTBDRplus VER 2.0-line probe assay and Phenotypic DST Laboratory methods were used to detect MDR-TB in clinical M. tuberculosis positive clinical Isolates. Figure 1 below shows the simplified procedural flow diagram.
Line probe assay
The GenoType MTBDRplus VER 2.0 in-vitro assay, which uses DNA strips, was carried out in accordance with the guidelines provided by the manufacturer (Hain Lifescience, Nehren, Germany).
DNA isolation, multiplex polymerase chain reaction (PCR) amplification, and reverse hybridization were the procedures for the LPA assay. The BSL-3 lab extracted mycobacterial DNA in accordance with the manufacturer's instructions. To summarize, a 500 ml aliquot of MGIT sample was centrifuged for 15 minutes at 10,000Xg, the pellet was re-suspended in 100 ml of sterile distilled water, and the supernatant was disposed of. After that, the suspension was heat-killed on a heat block for 40 minutes at 85°C. After 15 minutes of sonication, the material was centrifuged for 10 minutes at 13,000 × g. A 5 ml aliquot of the DNA sample was used for PCR with the master mixture for amplification composing of 35 ml primer nucleotide mixture (provided with kit), 5 ml of 10XPCR buffer with 15 mM MgCl2, 2 ml of 25 mM MgCl2, 0.2 ml of HotStarTaq DNA polymerase (Hain Lifescience, Nehren, Germany), 3 ml nuclease free molecular grade water and 5 ml of DNA sample in a final volume of 50 ml. Ten cycles of denaturation at 95°C for 30 seconds and 58°C for 2 minutes comprised the amplification process, which included 15 minutes of denaturation at 95°C. 20 cycles at 95°C for 25 seconds, 53°C for 40 seconds, and 70°C for 40 seconds were then performed, followed by an 8-minute final extension at 70°C. The twincubator, an automated device, was used to perform hybridization. Strips were taken out, fixed on paper, and the results were analyzed following hybridization and washing. Twenty-one of the 27 reaction zones on the MTBDRplus assay strip are mutation probes, and the remaining six are control probes used to confirm the test protocols. A conjugate control and amplification control (AC), an M. tuberculosis complex-specific control (TUB), an rpoB locus AC, a katG locus AC, and an inhA locus AC were among the six control probes. Specific probes cover locations in katG and inhA for INH resistance, whereas the probes cover the rpoB gene for RIF resistance detection. The sample under test is assumed to be resistant to the specific antibiotic if at least one of the wild-type bands is absent or if bands indicating a mutation in each drug resistance-related gene are present. The sample is sensitive to the appropriate antibiotic when all of the wild-type probes of a gene stain positively and no discernible mutation is found in the area under investigation.
DST Testing
Drug susceptibility testing was performed on baseline isolates and samples from patients suspected of treatment failure or relapse to identify the presence of resistance to any of the TB drugs under study. Drug susceptibility testing was performed as previously described [20]. Briefly, the M. tuberculosis confirmed culture was added to a liquid medium (BACTEC MGIT 960, Becton Dickinson) with 1 % proportion test of susceptibility to isoniazid 0.2 μg/ml, rifampin 1 μg/ml, streptomycin 2 μg/ml, ethambutol 5 μg/ml, kanamycin 5 μg/ml, and ofloxacin 4 μg/ml. For one to be classified as MDR-TB infected, they had to be resistant to isoniazid and rifampicin (whether or not they had resistance to other anti-TB drugs). Each batch consisted of one H37RV sensitivity strain, which was run for quality control purposes.
2.4 Statistical analysis
To determine associations with the primary outcome: MDR-TB, univariate and multivariate logistic regression were employed. A cut-off p-value of 0.05 was used to determine the strength of associations. Chi-squared test was used to compare the distributions of test results for LPA and DST. All the results were presented in tables. Agreements between the LPA method and the reference method (DST) were assessed using Kappa statistics. Cohen’s kappa values were interpreted as follows: from 0.00 to 0.20 indicating slight agreement; from 0.21 to 0.40 suggesting fair agreement; from 0.41 to 0.6 showing moderate agreement; from 0.61 to 0.80 indicating substantial agreement; and above 0.8 showing excellent agreement [21]. Only available data was considered in the analysis of the different variables (only complete cases were considered, depending on the variables being analysed. R statistical language was used to analyse the data [22].
3. Results
A total of 128 M. tuberculosis positive samples from the Lilongwe site were run on molecular Genotype MTBDRplus VER 2.0 LPA and compared with phenotypic DST, whereas for the Blantyre site, only LPA was conducted. The median age of the participants was 35 (IQR: 29.0 – 70.0) for the Lilongwe site, while for the Blantyre site, the median age was 21 (IQR: 11.0 – 38.25)
Overall, the incidence of MDR-TB by Genotype MTBDRplus VER 2.0 LPA was found to be 18/128 (14.06%), whereas that by DST was 30/128 (23.44%). RIF Mono-resistance (rpoB gene mutation) was found to occur at 8/128 (6.25%) for LPA and 8/128 (6.25%) for DST, while a large number of samples were found to be non-resistant to all drugs tested, with 102/128 (79.69%) for LPA and 90/128 (70.31%) for DST. The distributions of the test results under LPA and DST did not differ significantly (Chi-squared test: df = 4, p-value = 0.1991) (Table 1).
|
Category |
Incidence by LPA n (%) |
Incidence by DST n (%) |
|
|
MDR-TB |
18 (14.06%) |
30 (23.44%) |
|
|
Mono-resistance |
8 (6.25%) |
8 (6.25%) |
|
|
No resistance |
102 (79.68%) |
90 (70.31%) |
|
|
Total (n) |
128 |
128 |
|
Table 1: Comparison of the incidence of MDR-TB by LPA vs DST.
Next, we cross-tabulated the results from the two tests in order to determine the agreement between the two tests, with the row being DST and the column the Genotype MTBDRplus VER 2.0 LPA. We found significant agreement between Genotype MTBDRplus VER 2.0 LPA and DST in detecting MDR-TB (Table 2). A kappa statistic of 0.75 (95% CI: 0.62-0.88) was obtained.
|
Genotype MTBDRplus VER 2.0 LPA |
||||
|
DST |
MDR |
Mono-resistance |
No resistance |
|
|
MDR |
18 |
4 |
8 |
|
|
Mono-resistance |
0 |
4 |
4 |
|
|
No resistance |
0 |
0 |
90 |
|
Table 2: Agreement of the incidence of MDR-TB by LPA vs DST.
Using MTBDRplus VER 2.0 LPA (compared to DST) at the Lilongwe site, the sensitivity, specificity, positive predicting value, and negative predicting value for detection of mono-resistance were 50%, 97%, 50%, and 97% respectively; and for detection of multi-drug resistance the sensitivity, specificity, positive predicting value, and negative predicting value were 62%, 100%, 100% and 90% respectively (Table 3).
To determine the incidence of MDR-TB at the Lilongwe site only, a kappa statistical analysis was conducted and found to be 67.08% with a 95% confidence interval of 51.74% - 82.42%. Using MTBDRplus VER 2.0 LPA (compared to DST) at the Lilongwe site, the sensitivity, specificity, positive predicting value, and negative predicting value for detection of RIF resistance were 66%, 100%, 100%, and 87% respectively; for detection of INH resistance the sensitivity, specificity, positive predicting value, and negative predicting value were 17%, 100%, 100% and 80% respectively; while for multi-drug resistance the sensitivity, specificity, positive predicting value, and negative predicting value were 100% each (Table 3).
|
Drug |
Sensitivity (95% CI) |
Specificity (95% CI) |
Positive Predicting Value (95% CI) |
Negative Predicting Value (95% CI) |
Agreement |
Cohen’s Kappa (p-value) |
|
Mono |
50% (16%- 84%) |
97% (92%-99%) |
50% (16%-84%) |
97% (92%-99%) |
94% |
0.467 (0.0052) |
|
Multi |
62% (42%-79%) |
100% (96%-100%) |
100% (81%-100%) |
90% (83%-95%) |
91% |
0.717 (0) |
Table 3: Performance characteristics of MTBDRplus VER 2.0 LPA when compared to the DST method in the detection of mono-resistance (RIF resistance) and multi-drug resistance.
To determine associations with the primary outcome: MDR-TB, univariate and multivariate logistic regression were employed, and a cut-off p-value of 0.05 was used to determine the strength of associations. There was no significant difference in the likelihood of developing MDR-TB between those less than 36 years old and those older. The likelihood of developing MDR-TB was lower for males compared to females (odds ratio was 0.23, and 95% CI was 0.076-0.646). No significant difference was observed in the likelihood of developing MDR-TB between participants with smear grades of scanty, 1+, 2+, and 3+, and those with negative smear grades (Table 4).
|
Variable |
Stratum |
Frequency (%) |
COR (95% CI) |
P-value |
AOR (95% CI) |
P-value |
|
Age |
< 36 years |
56 (60.2%) |
1 |
1 |
||
|
>= 36 years |
37 (39.8%) |
0.74 (0.28, 1.9) |
0.5397 |
0.95 (0.33, 2.74) |
0.9311 |
|
|
Sex |
Female |
42 (42%) |
1 |
1 |
||
|
Male |
58 (58%) |
0.28 (0.1, 0.69) |
0.00739 |
0.27 (0.09, 0.74) |
0.0125 |
|
|
Smear grade |
Negative |
34 (36. 6%) |
1 |
1 |
||
|
Scanty |
10 (10.8%) |
1.91 (0.38, 9.57) |
0.4184 |
2.52 (0.46, 14.45) |
0.2833 |
|
|
1+ |
17 (18.3%) |
0.64 (0.15, 2.34) |
0.5105 |
0.91 (0.2, 3.84) |
0.8983 |
|
|
2+ |
18 (19.4%) |
0.44 (0.09, 1.73) |
0.2685 |
0.56 (0.1, 2.4) |
0.4526 |
|
|
3+ |
14 (15.1%) |
0.76 (0.18, 2.89) |
0.6996 |
0.95 (0.21, 3.96) |
0.9478 |
Table 4: Logistic regression analysis by age, sex, and smear grade.
4. Discussion
This is the first study to describe the burden of MDR-TB based on Genotype MTBDRplus VER 2.0 LPA and phenotypic DST results in Malawi. Although other studies have reported DR-TB in Malawi, the data were only based on INH mono-resistance [19] or limited to only a single urban hospital [23]. Additionally, this is the first laboratory-based study to combine MDR-TB data from two different regions of the country.
Both LPA and Cartridge-Based Nucleic Acid Amplification Test (NAAT) have been endorsed by the World Health Organization (WHO), but there is no clarity regarding the superiority of one over the other [13]. This notwithstanding, studies elsewhere have demonstrated that LPAs can significantly reduce the time to treatment initiation for MDR-TB patients [13,24]. In the current study, for both Lilongwe and Blantyre sites, all samples, 128 (100%) and 30 (100%), respectively, had a positive TUB band on LPA, suggesting they were all members of M. tuberculosis complex. The sensitivity of LPAs in detecting M. tuberculosis complex has been questioned in some studies owing to the fact that it has been unable to detect the TUB band correctly in up to 6% of smear-positive M. tuberculosis samples [13].
Out of 128 samples from Lilongwe, LPA was able to detect 14.06% as MDR-TB compared to 23.44% by DST, representing 61% concordance. These results are inconsistent with previous studies [14] where LPA was able to detect 66 out of the 68 (97%) DST MDR-TB isolates correctly. RIF mono-resistance was found to occur at 6.06% and 7.07% for Genotype MTBDRplus VER 2.0 LPA and DST, respectively (Table 1), whereas no INH mono-resistance was detected. The high rates of RIF mono-resistance, which is usually rare, could be attributed to the fact that over 70% of TB cases in Malawi were found in HIV infected individuals at the height of the pandemic [25,26]. In the current study, we were unable to conduct a comparative analysis of HIV status owing to a small sample size. Interestingly, the WHO policy is that LPAs may not completely replace mycobacterial culture and DST [13], but studies continue to suggest the increasing importance of LPAs, as it has been shown to significantly reduce time to treatment initiation for MDR-TB patients. The sensitivities of LPA compared to DST for the detection of RIF, INH, and MDR-TB were 66%, 17% and 100% respectively, whereas the specificities were 100% throughout (Table 3). In our results, the sensitivity of LPA for RIF and INH resistance was lower compared to other studies [13,14,27]. Although INH sensitivity was detected at a lower level, other studies have suggested a correlation with RIF resistance [27]. Additionally, other studies in India [28] and Germany [15] have reported very high sensitivities and specificities for INH resistance detection by LPA. Our failure to detect INH mono-resistance could be in part because 10% to 25% of INH-resistant strains are thought to have mutations outside the katG and inhA loci [12,29]. Several other factors have been proposed for low-sensitivity LPA results, including reagent quality, preparation, and presence of PCR inhibitors [14]. Overall, our results suggest that Genotype MTBDRplus VER 2.0 LPA may be as good as DST at detecting MDR-TB, consistent with previous studies that have reported higher sensitivities and specificities of LPA [12].
At the Blantyre site (KUHeS/MLW TB laboratory), only LPA was conducted on a total number of 30 M. tuberculosis positive samples [30]. We could not conduct DST owing to inadequate infrastructure. LPA was able to detect 1/30 (3.3%) as being INH mono-resistant, in which none were either RIF or MDR-TB. The study population had HIV status as a variable, but due to small sample size, we did not calculate the association between HIV and the development of MDR-TB owing to the absence of any MDR-TB. At a 95% confidence interval, the point estimate for MDR-TB was 0% while for mono-resistance TB was 3.3%.
In this study, the majority of the suspected (52.5%) and lab-confirmed (56%) MDR-TB patients were from the age group between 15 and 45years, in concordance with the national data. The high frequencies of MDR-TB among young age groups may indicate the possibility of propagation of MDR-TB in the community because of the higher mobility of youth.
Our results show that there is a positive association between MDR-TB and Sex (Table 4). The likelihood of males developing MDR-TB was lower compared to females, with an odds ratio of 28%(95% CI is 1% - 78%). However, this may be attributed to the fact that most males do not present to the hospital when they get ill and may not truly represent what obtains in the community.
In conclusion, this study has demonstrated that MTBDRplus VER 2.0 LPA can perform better at detecting the incidence of MDR-TB. Combining DST and LPA results would remain the best option in detecting MDR TB among patients.
This study was limited by the fact that we were unable to conduct a complete comparison of the two methods for the Blantyre isolates. Additionally, using secondary data meant we were unable to collect some social-demographic information, such as occupation, HIV status (especially at the National TB Reference Laboratory), previous TB treatment, residence, and antibiotic history. Therefore, the final multivariable logistic regression model was not properly adjusted for due to this missing information, which could lead to significant residual confounding. Additionally, using a more robust method would improve the accuracy of these results.
5. Conclusion
There is a high incidence of MDR-TB among patients whose samples are sent to Lilongwe National TB Reference Laboratory, which otherwise remain undetected. Larger studies covering the three geopolitical regions of the country need to be undertaken. With performance characteristics compared to other conventional methods of detecting MDR-TB, a short turnaround time to diagnosis, and the ability to simultaneously detect rifampicin and isoniazid resistance, LPA is an excellent and reliable tool for the early detection of multidrug-resistant tuberculosis. LPA should be implemented in all regions of Malawi for the detection of MDR-TB in addition to DST.
Acknowledgement
We would like to acknowledge the Helse Nord Tuberculosis Initiative (HNTI) for funding this study.
Conflict of interest
Authors declare no conflict.
Authors’ contributions
VN, MN, and TS conceived the study. VN, TS, DC, and SD performed the investigations. MN, TK, TS, HT, and AM analyzed the data. VN, MN, TS, TK, AM, MN, BB, and YK wrote the report. MN, VN, and TS oversaw the research. All authors contributed to the study design and reviewed the report.
References
- World Health Organization. Global tuberculosis report 2022. Geneva: WHO (2022).
- Joshi JM. Tuberculosis chemotherapy in the 21st century: back to the basics. Lung India 28 (2011): 193-200.
- Dookie N, Rambaran S, Padayatchi N, et al. Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother 73 (2018): 1138-1151.
- Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79 (1998): 3-29.
- Palomino JC. Molecular detection, identification, and drug resistance detection in Mycobacterium tuberculosis. FEMS Immunol Med Microbiol 56 (2009): 103-111.
- Drobniewski FA, Wilson SM. The rapid diagnosis of isoniazid and rifampicin resistance in Mycobacterium tuberculosis: a molecular story. J Med Microbiol 47 (1998): 189-196.
- da Silva PEA, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66 (2011): 1417-1430.
- Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 13 (2009): 1320-1330.
- United States Agency for International Development. Malawi tuberculosis fact sheet. USAID (2016).
- World Health Organization. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Geneva: WHO (n.d.).
- Theron G, Peter J, Richardson M, et al. The diagnostic accuracy of the GenoType MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev (2014): CD010705.
- Meaza A, Kebede A, Yaregal Z, et al. Evaluation of genotype MTBDRplus VER 2.0 line probe assay for the detection of MDR-TB in smear-positive and negative sputum samples. BMC Infect Dis 17 (2017): 280.
- Desikan P, Panwalkar N, Mirza SB, et al. Line probe assay for detection of Mycobacterium tuberculosis complex: an experience from Central India. Indian J Med Res 145 (2017): 70-73.
- Yadav RN, Singh BK, Sharma SK, et al. Comparative evaluation of GenoType MTBDRplus line probe assay with solid culture method in early diagnosis of multidrug-resistant tuberculosis at a tertiary care centre in India. PLoS One 8 (2013): e72036.
- Hillemann D, Rusch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 45 (2007): 2635-2640.
- Seung KJ, Keshavjee S, Rich ML. Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb Perspect Med 5 (2015): a017863.
- Dalton T, Cegielski P, Akksilp S, et al. Prevalence of and risk factors for resistance to second-line drugs in people with multidrug-resistant tuberculosis in eight countries: a prospective cohort study. Lancet 380 (2012): 1406-1417.
- Ndhlovu V, Kiran A, Sloan D, et al. Genetic diversity of Mycobacterium tuberculosis clinical isolates in Blantyre, Malawi. Heliyon 5 (2019): e02638.
- Sobkowiak B, Glynn JR, Houben RMGJ, et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics 19 (2018): 613.
- Centers for Disease Control and Prevention. Drug susceptibility testing: laboratory information. Atlanta: CDC (n.d.).
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 33 (1977): 159-174.
- R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing (2022).
- Kumwenda GP, Chipungu G, Sloan DJ, et al. The occurrence and frequency of genomic mutations that mediate isoniazid and rifampicin resistance in Mycobacterium tuberculosis isolates from untreated pulmonary tuberculosis cases in urban Blantyre, Malawi. Malawi Med J 30 (2018): 1-5.
- Singla N, Satyanarayana S, Sachdeva KS, et al. Impact of introducing the line probe assay on time to treatment initiation of MDR-TB in Delhi, India. PLoS One 9 (2014): e102989.
- van Lettow M, Chan AK, Ginsburg AS, et al. Timing and uptake of ART during treatment for active tuberculosis in HIV co-infected adults in Malawi. Public Health Action 1 (2011): 6-12.
- Singano V, Kip E, Ching’ani W, et al. Tuberculosis treatment outcomes among prisoners and the general population in Zomba, Malawi. BMC Public Health 20 (2020): 700.
- Nathavitharana RR, Cudahy PGT, Schumacher SG, et al. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. Eur Respir J 49 (2017): 1601075.
- Raizada N, Sachdeva KS, Chauhan DS, et al. A multi-site validation in India of the line probe assay for the rapid diagnosis of multidrug-resistant tuberculosis directly from sputum specimens. PLoS One 9 (2014): e88626.
- de Abreu Maschmann R, Spies FS, de Souza Nunes L, et al. Performance of the GenoType MTBDRplus assay directly on sputum specimens from Brazilian patients with tuberculosis treatment failure or relapse. J Clin Microbiol 51 (2013): 1606-1608.
- Barnard M, Parsons L, Miotto P, et al. Molecular detection of drug-resistant tuberculosis by line probe assay: laboratory manual for resource-limited settings. Geneva: WHO (2012).