Molecular Characterization of Clinical Strains of Extended-Spectrum Βeta-Lactamases-Producing Klebsiella Pneumoniae Isolated in A Tertiary Hospital in Dakar-Senegal
Article Information
Komla Mawunyo Dossouvi1, *, Bissoume Sambe Ba2, Gora Lo1, 4, Abdoulaye Cissé3, Awa Ba-Diallo1, 4, Issa Ndiaye3, Assane Dieng1, Serigne Mbaye Lo Ndiaye1, Cheikh Fall3, Alioune Tine1, Farba Karam1, Habsa Diagne-Samb1, Ousmane Sow3, Safietou Ngom-Cisse1, Halimatou Diop-Ndiaye1, 4, Gnatoulma Katawa5, Coumba Toure-Kane4, Aïssatou Gaye-Diallo1, 4, Simplice Damintoti Karou5, Souleymane Mboup1,4, Cheikh Saad Bouh Boye1, Abdoulaye Seck6, Makhtar Camara1, 4
1Bacteriology-Virology laboratory, National University Hospital Aristide Le Dantec, Dakar, Senegal
2World Health Organization WCARO, Dakar Senegal
3Pole of Microbiology, Institut Pasteur de Dakar, Senegal
4Institut de Recherche en Santé, de Surveillance Epidémiologique et de Formation (IRESSEF), Dakar, Senegal
5Ecole Supérieure des Techniques Biologiques et Alimentaires (ESTBA), Université de Lomé, Lomé, Togo
6Medical Analysis laboratory, Institut Pasteur de Dakar, Senegal
*Corresponding author: Komla Mawunyo Dossouvi, Bacteriology-Virology laboratory, National University Hospital Aristide Le Dantec, Dakar, Senegal
Received: 15 December 2022; Accepted: 22 December 2022; Published: 06 January 2022
Citation: Komla Mawunyo Dossouvi, Bissoume Sambe Ba, Gora Lo, Abdoulaye Cissé, Awa Ba- Diallo, Issa Ndiaye, Assane Dieng, Serigne Mbaye Lo Ndiaye, Cheikh Fall, Alioune Tine, Farba Karam, Habsa Diagne-Samb, Ousmane Sow, Safietou Ngom-Cisse, Halimatou Diop-Ndiaye, Gnatoulma Katawa, Coumba Toure-Kane, Aïssatou Gaye- Diallo, Simplice Damintoti Karou, Souleymane Mboup, Cheikh Saad Bouh Boye, Abdoulaye Seck, Makhtar Camara. Molecular Characterization of Clinical Strains of Extended-Spectrum Βeta- Lactamases-Producing Klebsiella Pneumoniae Isolated in A Tertiary Hospital in Dakar-Senegal. Archives of Microbiology and Immunology 7 (2023): 01-09.
View / Download Pdf Share at FacebookAbstract
Klebsiella pneumoniae (K. pneumoniae) isolates are often multidrug-resistant (MDR) and clones producing extended-spectrum beta-lactamases (ESBL) are increasingly reported all over the world. This study aimed to determine drug-susceptibility profiles and characterize ESBL genes in clinical strains of K. pneumoniae in an university teaching hospital of Dakar. Sixty-six ESBL-producing K. pneumoniae strains were selected for this study and subjected to the Kirby-Bauer disk diffusion method and standard polymerase chain reaction (PCR) for the screening of major ESBL genes (blaCTX-M, blaCTX-M-9, blaCTX-M-15, blaCTX-M-25, blaOXA-1, blaTEM, blaSHV).
All isolates were resistant to ampicillin, ticarcillin, amoxicillin/clavulanic acid combination, cefalotin, cefotaxime, ceftazidime and cefepime, while 60% (40/66) of them were resistant to fosfomycin. Amikacin, ertapenem and imipenem remained effective with respective sensitivity rate of 77.3% (51/66), 75.8% (50/66) and 100% (66/66). blaCTX-M (64/66; 97%) with its variant blaCTX-M-15 (63/66; 95.5%) were the most prevalent ESBL genes; followed by blaTEM (58/66; 87.9%), blaOXA-1 (47/66; 71.2%) and blaSHV (31/66; 47%). Sixty-four isolates out of 66 (97%) of isolates carried at least 2 ESBL genes and the most prevalent ESBL gene combinations were (blaCTX-M-15 + blaOXA-1 + blaTEM) (24/66; 36.4%) and (blaCTX-M-15 + blaTEM + blaOXA-1 + blaSHV) (18/66; 27.3%).
Cephalosporins, quinolones and aminoglycosides (except amikacin) would no l
Keywords
Klebsiella pneumoniae; Extended Spectrum beta-lactamase; tertiary hospital; Dakar-Senegal
Klebsiella pneumoniae articles; Extended Spectrum beta-lactamase articles; tertiary hospital articles; Dakar-Senegal articles
Article Details
1. Introduction
By 2030, around 24 million people, especially in low-income countries, will live in extreme poverty due to antimicrobial resistance (AMR) and around 28.3 million European people can fall into poverty. Furthermore, by 2050, around 3.8% of the world annual gross domestic product (GDP) will be lost due to AMR, while low-income countries will lose up to 5.6% of their GDP and AMR could lead 10 million people to death [1-3]. K. pneumoniae, an ESKAPE pathogen group member, is one of the main and widespread bacteria showing uncommon drug resistance patterns in the world [4-6].
pneumoniae, a human gastrointestinal tract commensal Enterobacteriaceae is an opportunistic bacterium, also implicated in about 20% of enterobacterial infections and frequently implicated in hospital-acquired infections [7-9]. In community, K. pneumoniae is often involved in serious extra-intestinal infections such as urinary tract infections (UTI) and pneumonia, while in hospitals it generally causes septicemia through secondary infections of wounds, catheters and probes [4, 10]. Moreover, carriage of K. pneumoniae among hospitalized patients can be up to 77% in the stool, 19% in the pharynx and 42% on the hands [11].
The most common resistance mechanism in MDR K. pneumoniae is the production of extended spectrum beta-lactamases (ESBL) and carbapenemases [4, 10]. ESBL hydrolyze penicillins, cephalosporins (except cephamycins) and monobactams. However, they are inactive on beta-lactamase inhibitors (BLI) and carbapenems [12, 13]. At least 13 ESBL families have been reported worldwide including CTX-M, SHV-type ESBL, TEM-type ESBL, OXA-type ESBL, IRT, CMT, GES, PER, VEB, BEL, TLA, SFO and OXY. However, the most active and prevalent ESBL families are CTX-M, SHV-type ESBL, TEM-type ESBL and OXA-type ESBL. SHV-type ESBL and TEM-type ESBL are known as “old” ESBLs while CTX-M are known as “new” ESBLs. Furthermore, OXA-1 which is not originally an ESBL-type oxacillinases, can be added to ESBLs because of its ability to hydrolyze cefepime [14, 15].
ESBL genes are frequently located on the bacterial chromosome or on mobile genetic supports (plasmids, integrons and transposons). These mobile genetic carriers are strongly involved in intra- and interspecific bacterial genes exchanges and are therefore powerful catalysts for the global spread of multidrug resistant bacteria. ESBL-producing strains are often resistant to aminoglycosides and quinolones as mostly supported by the same mobile genetic elements [16-20].
The screening and study of ESBL K. pneumoniae strains is therefore critical in order to improve therapeutic management of patients, prevent and contain epidemics and collect useful epidemiological data that will help public health authorities to plan short-, medium- and long-term activities. To our knowledge, few studies focused on circulating clinical strains of ESBL-producing K. pneumoniae in Senegal and in West Africa globally. This study therefore aimed to describe antibiotics resistance profiles and to characterize ESBL genes in clinical K. pneumoniae strains isolated in Aristide le Dantec University Teaching Hospital (HALD), one of the major tertiary facilities in Dakar, Senegal. Additionally, we compared community-acquired (CA) to hospital-acquired (HA) isolates, and uropathogenic K. pneumoniae (UPKP) to No-Uropathogenic K. pneumoniae isolates (No-UPKP).
2. Materials and methods
2.1 Bacterial isolates
This study focused on 66 ESBL-producing K. pneumoniae strains isolated from different biological samples, including urines, pus, sputum, bronchial fluid and vaginal secretions in routine diagnosis activities of HALD bacteriology laboratory from January 01st, 2018 to December 31, 2020. Isolation and identification were done on Eosin Methylene Blue (EMB) agar (Merck KGaA, Darmstadt, Germany) and Api 20E for Enterobacteriaceae (bioMérieux, Lyon / France), respectively. Majority of isolates was from urine specimen (n = 42, 63.63%). 54 (81.8%) isolates were community-acquired whereas 12 (18.2%) were hospital-acquired strains. Bacterial isolates were stored at -80 °C in brain heart infusion broth with 15% glycerol until the study begins.
2.2 Antibiotic susceptibility testing
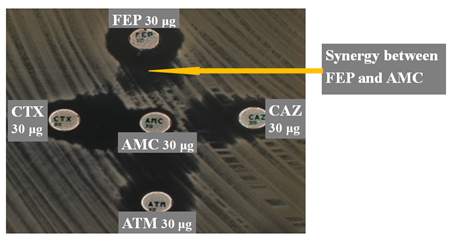
We used Kirby-Bauer disc diffusion method for the antibiotic susceptibility testing and results were interpreted according to the guidelines of the antibiogram committee of the French society of microbiology (CA-SFM, 2022). Briefly, bacterial suspensions were prepared at 0.5 McFarland and inoculated onto Mueller-Hinton agar for an overnight incubation at 37°C. These following antibiotic disks were tested: ampicillin (AMP, 10 μg), ticarcillin (TIC, 75 μg), amoxicillin-clavulanic acid (AMC, 20/10 μg), cefalotin (CEF, 30 μg), cefoxitin (FOX, 30 μg), cefotaxime (CTA, 30 μg), ceftazidime (CAZ, 30 μg), cefepime (CEP, 30 μg), aztreonam (AZT, 30 μg), imipenem (IMP, 10 μg), ertapenem (ERT, 10 μg), nalidixic acid (NAL, 30 μg ), ciprofloxacin (CIP, 5 μg), gentamicin (GEN, 10 μg), amikacin (AMI, 30 μg), fosfomycin (FOS, 50 μg), tetracycline (TET, 30 μg) and sulfamethoxazole-trimethoprim (TRS, 1.25 μg / 23.75 μg). Escherichia coli ATCC 25922 was used for quality control. ESBL production was detected by double-disk synergy test using AMC disc surrounded at a radius of 30 mm by CEP, CAZ, CTA and AZT (figure 1).
2.3 DNA Extraction
We used mechanical thermal lysis method to extract bacterial DNA. Briefly, a well-separated bacterial colony was dispersed in a tube containing 1ml of sterile distilled water, vortexed, boiled for 15 minutes at 100°C and centrifuged at 13,200 rpm for 10 min. The supernatant was carefully recovered, aliquoted and stored at -20°C until used. Invitrogen™ Qubit™ 3 Fluorometer, (Thermo Fisher Scientific Inc, Strasbourg / France) was used for the DNA quantification.
2.4 ESBL genes amplification
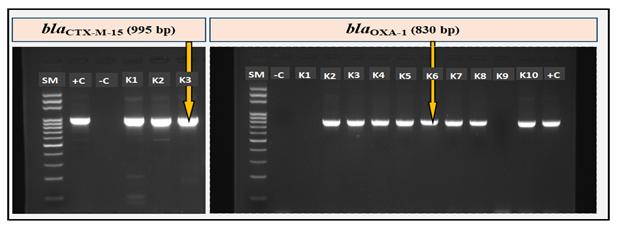
Each DNA sample was subjected to a simplex end-point PCR (on Thermocycler 2720, Applied Biosystems, Lincoln Centre Drive, Foster City, California 94404, USA). Specific primers (table 1) were used to amplify ESBL genes (blaCTX-M, blaCTX-M-9, blaCTX-M-15, blaCTX-M-25, blaOXA-1, blaTEM, blaSHV). Each reaction included positive and negative controls. Reaction volume of each PCR reaction was 20 μl (2.5 μl of DNA + 17.5 μl Master Mix FIREPol, Tartu / Estonia) and amplification of ESBL genes was according to the program below: initial denaturation at 95°C for 3 min, 35 PCR cycles (denaturation: 94°C, 30 sec; annealing; extension: 72°C, 60 sec) and a final elongation at 72°C for 7 min. Each amplicon (10 μl) was separated on 2% agarose gel in 1X TAE buffer for 35 min at 135 volts and the amplified fragment detected using a GelDoc imager (BioRad, Hercules, California / USA).
2.5 Statistical analysis
Statistical analyses were performed with Microsoft-Excel. The statistic test used is the Chi-square at 5% and 10% risk thresholds. p-values ??are obtained from the proportion comparison test and the level of significance for all statistical tests was set at p < 0.05 for the strong evidence of difference and 0.05 < p < 0.1 for moderate evidence of difference [33, 34, 37].
Table 1. Oligonucleotide primers sequence used for PCR to detect ESBL genes
|
Target genes |
Sequences genes |
Sizes (bp) |
Annealing Temp (°C) |
References |
|
blaCTX-M |
F: 5’ - ATGTGCAGYACCAGTAARGTKATGGC - 3’ |
592 |
55 |
[35] |
|
R: 5’ - TGGGTRAARTARGTSACCAGAAYSAGCGG - 3’ |
||||
|
blaCTX-M-9 |
F: 5’ - GTGACAAAGAGAGTGCAACGG - 3’ |
856 |
55 |
[35] |
|
R: 5’ - ATGATTCTCGCCGCTGAAGCC - 3’ |
||||
|
blaCTX-M-15 |
F: 5’ - CACACGTGGAATTTAGGGACT - 3’ |
995 |
50 |
[35] |
|
R: 5’ - GCCGTCTAAGGCGATAAACA - 3’ |
||||
|
blaCTX-M-25 |
F: 5’ - GCACGATGACATTCGGG - 3’ |
327 |
52 |
[35] |
|
R: 5’ - AACCCACGATGTGGGTAGC - 3’ |
||||
|
blaOXA-1 |
F: 5’ - ATGAAAAACACAATACATATC - 3’ |
830 |
56 |
[36] |
|
R: 5’ - AATTTAGTGTGTTTAGAATGG - 3’ |
||||
|
blaTEM |
F: 5’ - TTGGGTGCACGAGTGGGTTA - 3’ |
506 |
55 |
[35] |
|
R: 5’ - TAATTGTTGCCGGGAAGCTA - 3’ |
||||
|
blaSHV |
F: 5’ - TCGGGCCGCGTAGGCATGAT - 3’ |
628 |
52 |
[35] |
|
R: 5’ - AGCAGGGCGACAATCCCGCG - 3’ |
3. Results
3.1 Antibiotic susceptibility testing
Based on resistance profiles observed, all 66 strains of this study are MDR. Antibiotic resistance profiles are compiled in the table 2. All strains (100%) were resistant to ampicillin (AMP), ticarcillin (TIC), Amoxicillin-clavulanic acid (AMC), cefalotin (CEF), cefotaxim (CTA), ceftazidime (CAZ) and cefepime (CEP). Strains were almost resistant to sulfamethoxazole-trimethoprim (TRS) and tetracycline (TET), respectively 65/66 (98.5%) and 57/66 (86.4%) and are highly resistant to most used antibiotics such as ciprofloxacin (CIP) and gentamicin (GEN), respectively 51 (77.3%) and 45 (68.2%). Forty strains out of 66 (60.6%) were resistant to fosfomycin (FOS). Amikacin, ertapenem and imipenem remained effective with respective sensitivity rate of 77.3% (51/66), 75.8% (50/66) and 100% (66/66) (table 2). CA were significantly more resistant to FOX (p = 0.07) and to sulfamethoxazole-trimethoprim (p = 0.03) than HA isolates. Based on resistance profiles, no significant difference was observed comparing UPKP to No-UPKP (table 2)
Table 2: Antibiotics resistance rate of total strains, CA and HA, UPKP and no-UPKP.
|
Antibiotics |
Total strains |
Pathogenicity |
Origin |
|||||
|
Class |
codes |
N (%) |
UPKP |
No-UPKP |
p |
CA |
HA |
p |
|
N (%) |
N (%) |
N (%) |
N (%) |
|||||
|
Beta-lactams |
AMP |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
12 (100) |
1 |
|
TIC |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
12 (100) |
1 |
|
|
AMC |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
12 (100) |
1 |
|
|
CEF |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
12 (100) |
1 |
|
|
FOX |
26 (39.4) |
16 (38.1) |
10 (41.7) |
0.8 |
24 (44.4) |
2 (16.7) |
0.07* |
|
|
CTA |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
24 (100) |
1 |
|
|
CAZ |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
24 (100) |
1 |
|
|
CEP |
66 (100) |
42 (100) |
24 (100) |
1 |
54 (100) |
24 (100) |
1 |
|
|
AZT |
57 (86.4) |
38 (90.5) |
19 (79.2) |
0.19 |
47 (87) |
10 (83.3) |
0.73 |
|
|
IMP |
0 |
0 |
0 |
- |
0 |
0 |
- |
|
|
ERT |
10 (15.2) |
6 (14.3) |
4 (16.7) |
0.79 |
9 (16.7) |
1 (8.3) |
0.47 |
|
|
Quinolones and Fluoroquinolones |
NAL |
39 (59.1) |
25 (59.5) |
14 (58.3) |
0.92 |
32 (59.3) |
7 (58.3) |
0.95 |
|
CIP |
51 (77.3) |
34 (81) |
17 (70.8) |
0.34 |
43 (79.2) |
8 (66.7) |
0.33 |
|
|
Aminoglycosides |
GEN |
45 (68.2) |
27 (64.3) |
18 (75) |
0.36 |
35 (64.8) |
10 (83.3) |
0.21 |
|
AMI |
15 (22.7) |
9 (21.4) |
6 (25) |
0.73 |
11 (20.4) |
4 (33.3) |
0.33 |
|
|
Phosphonic acid |
FOS |
40 (60.6) |
25 (59.5) |
15 (62.5) |
0.82 |
34 (63) |
6 (50) |
0.4 |
|
Cyclines |
TET |
57 (86.4) |
36 (85.7) |
21 (87.5) |
0.83 |
46 (85.2) |
11 (91.7) |
0.55 |
|
Antifolates |
TRS |
65 (98.5) |
41 (97.6) |
24 (100) |
0.44 |
54 (100) |
11 (91.7) |
0.03** |
UPKP, Uropathogenic K. pneumoniae; No-UPKP, K. pneumoniae isolated from pus, sputum, bronchial fluid and vaginal secretions; CA, Community-acquired; HA, Hospital-acquired; AMP, ampicillin; TIC, ticarcillin; AMC, Amoxicillin-clavulanic acid; CEF, cefalotin; FOX, cefoxitin; CTA, cefotaxim; CAZ, ceftazidime; CEP, cefepime; AZT, aztreonam; IMP, imipenem; ERT, Ertapenem; NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin; AMI, amikacin; FOS, fosfomycin; TET, tetracycline; TRS, sulphamethoxazole-trimethoprim; **, strong evidence of difference (p < 0.05); *, moderate evidence of difference (0.05 < p < 0.1).
Twenty-two strains out of 66 (31.8%) shared identical resistance profiles. These isolates with identical drug resistance profiles were generally uropathogenic and community-acquired (Table 3). These identical resistance patterns ranged from 13 to 16 antibiotics.
Table 3. Identical drug resistance profiles in tested isolates.
|
Drugs |
Total Strains N (%) |
Pathogenicity |
Origin |
||
|
UPKP N (%) |
No-UPKP N (%) |
CA N (%) |
HA (%) |
||
|
AMP, TIC, AMC, CEF, FOX, CTA, CAZ, CEP, AZT, ERT, NAL, CIP, GEN, FOS, TET, TRS |
3 (4.5) |
3 (7.1) |
0 |
3 (5.6) |
0 |
|
AMP, TIC, AMC, CEF, FOX, CTA, CAZ, CEP, AZT, NAL, CIP, GEN, AMI, FOS, TET, TRS |
2 (3) |
1 (2.4) |
1 (4.2) |
2 (3.7) |
0 |
|
AMP, TIC, AMC, CEF, FOX, CTA, CAZ, CEP, AZT, NAL, CIP, GEN, AMI, TET, TRS |
3 (4.5) |
2 (4.8) |
1 (4.2) |
2 (3.7) |
1 (8.3) |
|
AMP, TIC, AMC, CEF, CTA, CAZ, CEP, AZT, NAL, CIP, GEN, AMI, TET, TRS |
2 (3) |
2 (4.8) |
0 |
1 (1.9) |
1 (8.3) |
|
AMP, TIC, AMC, CEF, CTA, CAZ, CEP, AZT, NAL, CIP, GEN, FOS, TET, TRS |
4 (6.1) |
2 (4.8) |
2 (8.3) |
3 (5.6) |
1 (8.3) |
|
AMP, TIC, AMC, CEF, CTA, CAZ, CEP, AZT, NAL, CIP, FOS, TET, TRS |
3 (4.5) |
2 (4.8) |
1 (4.2) |
3 (5.6) |
0 |
|
AMP, TIC, AMC, CEF, CTA, CAZ, CEP, AZT, NAL, CIP, GEN, TET, TRS |
4 |
4 |
0 |
3 |
1 |
UPKP, Uropathogenic K. pneumoniae; No-UPKP, K. pneumoniae isolated from pus, sputum, bronchial fluid and vaginal secretions; CA, Community-acquired; HA, Hospital-acquired; AMP, ampicillin; TIC, ticarcillin; AMC, Amoxicillin-clavulanic acid; CEF, cefalotin; FOX, cefoxitin; CTA, cefotaxim; CAZ, ceftazidime; CEP, cefepime; AZT, aztreonam; IMP, imipenem; ERT, Ertapenem; NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin; AMI, amikacin; FOS, fosfomycin; TET, tetracycline; TRS, sulphamethoxazole-trimethoprim; N, total isolates number
3.2 ESBL genes
Of the 7 ESBL genes screened during this study, blaCTX-M (64/66; 97%) and its variant blaCTX-M-15 (63/66; 95.5%) were the most prevalent, followed by blaTEM (58/66; 87.9%), blaOXA-1 (47/66; 71.2%) and blaSHV (31/66; 47%). None strain carried blaCTX-M-9 or blaCTX-M-25 (table 4 and figure 2). All HA (12/12; 100%) carried blaCTX-M-15. We noticed that UPKP carried significantly more blaOXA-1 than No-UPKP (p = 0.08) (table 4). For the prevalence of blaCTX-M-15, blaTEM and blaSHV, no significant difference was noted when comparing UPKP to No-UPKP and CA to HA (table 4).
In terms of accumulation of ESBL genes, (65/66; 98.5%) of isolates carried at least 2 ESBL genes out of the 7 sought and (01/66; 1.5%) strain did not carry any of the 7 genes sought. The combination (blaTEM + blaSHV) genes was detected in only one strain (01/66; 1.5%). The most prevalent ESBL gene combinations were (blaCTX-M-15 + blaOXA-1 + blaTEM) (24/66; 36.4%) and (blaCTX-M-15 + blaTEM + blaOXA-1 + blaSHV) (18/66; 27.3%) (table 5).
Table 4: Prevalences of ESBL genes in total strains, CA and HA, UPKP and No- UPKP.
|
ESBL |
Total strains |
Pathogenicity |
Origin |
|||||
|
Family |
Genes |
N (%) |
UPKP |
No- UPKP |
p |
CA |
HA |
p |
|
N (%) |
N (%) |
N (%) |
N (%) |
|||||
|
Cefotaximase-Munich |
blaCTX-M |
64 (97) |
41 (97.6) |
23 (95.8) |
0.85 |
50 (96.2) |
12 (100) |
0.55 |
|
blaCTX-M-9 |
0 |
0 |
0 |
- |
0 |
0 |
- |
|
|
blaCTX-M-15 |
63 (95.5) |
41 (97.6) |
22 (91.7) |
0.26 |
51 (94.4) |
12 (100) |
0.4 |
|
|
blaCTX-M-25 |
0 |
0 |
0 |
- |
0 |
0 |
- |
|
|
Oxacillinase |
blaOXA-1 |
47 (71.2) |
33 (78.5) |
14 (58.3) |
0.08* |
40 (74.1) |
7 (58.3) |
0.27 |
|
Temoneira |
blaTEM |
58 (87.9) |
36 (85.7) |
22 (91.7) |
0.48 |
48 (88.9) |
10 (83.3) |
0.59 |
|
Sulfhydryl variable |
blaSHV |
31 (47) |
19 (45.2) |
12 (50) |
0.71 |
25 (46.3) |
6 (50) |
0.82 |
UPKP, Uropathogenic K. pneumoniae; No-UPKP, K. pneumoniae isolated from pus, sputum, bronchial fluid and vaginal secretions; CA, community-acquired; HA, hospital-acquired; %, percentage; N, number of isolates; **, strong evidence of difference (p < 0.05); *, moderate evidence of difference (0.05 < p < 0.1).
Table 5: Prevalence of ESBL genes combinations in total strains, CA and HA, UPKP and No-UPKP.
|
Combination of ESBL genes |
Total strains |
Pathogenicity |
Origin |
||||
|
N (%) |
UPKP |
No-UPKP |
p |
CA |
HA |
p |
|
|
N (%) |
N (%) |
N (%) |
N (%) |
||||
|
blaCTX-M-15 + blaTEM + blaOXA-1 + blaSHV |
18 (27.3) |
12 (28.6) |
6 (25) |
0.75 |
15 (27.8) |
3 (25) |
0.84 |
|
blaCTX-M-15 + blaOXA-1 + blaTEM |
24 (36.4) |
17 (40.8) |
7 (29.2) |
0.35 |
20 (37) |
4 (33.3) |
0.81 |
|
blaCTX-M-15 + blaTEM + blaSHV |
7 (10.6) |
3 (7.1) |
4 (16.7) |
0.22 |
5 (9.3) |
2 (16.7) |
0.45 |
|
blaCTX-M-15 + blaOXA-1 + blaSHV |
3 (4.5) |
3 (7.1) |
0 |
0.18 |
3 (5.6) |
0 |
0.4 |
|
blaCTX-M-15 + blaTEM |
7 (10.6) |
4 (9.5) |
3 (12.5) |
0.7 |
6 (11.1) |
1 (8.3) |
0.77 |
|
blaCTX-M-15 + blaSHV |
2 (3) |
1 (2.4) |
1 (4.2) |
0.68 |
1 (1.9) |
1 (8.3) |
0.24 |
|
blaCTX-M-15 + blaOXA-1 |
1 (1.5) |
1 (2.4) |
0 |
0.44 |
1 (1.9) |
0 |
0.63 |
|
blaTEM + blaOXA-1 |
1 (1.5) |
0 |
1 (4.2) |
0.18 |
1 (1.9) |
0 |
0.63 |
|
blaTEM + blaSHV |
1 (1.5) |
0 |
1 (4.2) |
0.18 |
1 (1.9) |
0 |
0.63 |
UPKP, Uropathogenic K. pneumoniae; No-UPKP, K. pneumoniae isolated from pus, sputum, bronchial fluid and vaginal secretions; CA, community-acquired; HA, hospital-acquired %, percentage; N, number of isolates; **, strong evidence of difference (p < 0.05); *, moderate evidence of difference (0.05 < p < 0.1).
4. Discussion
4.1 Antibiotic susceptibility testing
High resistance rates (68.2% to 100%) reported during our study for AMP, TIC, AMC, CEF, CTA, CAZ, CEP, TRS, TET, CIP and GEN are very suggestive as CIP, GEN, 3rd and 4th generation cephalosporins are widely prescribed as empirical therapies to treat community-acquired and hospital-acquired bacterial infections. It is therefore necessary to expand research on the susceptibility profiles of Enterobacteriaceae to these first-line antibiotics. This could lead to modification or adjustment of empirical antibiotic therapies in Senegal.
[28, 29, 31, 32] mentioned prevalence of resistance to CAZ (50% and 67%); CEP (38% and 75%); CIP (83%, 67% and 65%); GEN (42%, 38% and 8%); AMI (12% and 0%); TRS (58% et 50%) and TET (42%) in Enterobacteriaceae ESBL-producing isolates. By comparing theirs results with those obtained during our present study, we noted that our isolates were generally more resistant. Right now, we do not have the exact reason why clinical K. pneumoniae from Dakar seem to be more resistant than those isolated in other countries. However, self-medication and the easy access to antibiotics without medical prescription could be among main causes. 60.6% of the strains in our study were resistant to fosfomycin. This is alarming as until now, fosfomycin was an alternative and capital molecule to treat UTIs caused by MDR Enterobacteriaceae without resorting to carbapenems. Clinicians must thererfore properly control fosfomycin prescription and researchers must conduct large-scale studies on the resistance profiles of Enterobacteriaceae to this antibiotic. It is also necessary to investigate resistance mechanisms involved in fosfomycin resistance in Dakar. It would be worrying in case resistance to fosfomycin was mediated by genes located on mobile genetic elements, as these mobile genetic elements are major carriers of transmission of inter- and intraspecific drug resistance. These studies could help to quickly contain Enterobacteriaceae resistance to fosfomycin and to continue to use it as an alternative treatment to carbapenems.
To “protect” carbapenems (by delaying their use), it is therefore important to study the susceptibility profiles of our 66 K. pneumoniae strains to alternative antibiotics such as tigecycline, temocillin, ceftazidime-avibactam and ceftolozane-tazobactam. Only 15.2% of the strains in this study were resistant to Amikacin. While waiting for larger-scale studies on the efficacy of amikacin on MDR K. pneumoniae in Senegal, amikacin would constitute an alternative molecule for the treatment of MDR K. pneumoniae infections. Nevertheless, clinicians should carefully control the prescription of amikacin to delay the emergence of amikacin-resistant bacterial clones. In this study, all CA strains were resistant to TRS, and CA were significantly more resistant to TRS than HA (p = 0.03). This might be due to the fact that in our area, TRS is a widely consumed drug in community. TRS selection pressure may have caused high carriage of TRS-resistant K. pneumoniae in the community. In addition, it is necessary to investigate the resistance mechanisms, involved genes and their genetic supports.
4.2 ESBL genes
Since the appearance of CTX-M-type ESBLs in 1990 [21], they have been reported all over the world and have become the major ESBL, relegating SHV, TEM and OXA-types ESBLs. The results obtained during our study follow this trend. Indeed, blaCTX-M-15, gene encoding CTX-M-15, which confers a high level of resistance to cefotaxime, ceftriaxone, ceftazidime and aztreonam [22], was present in 95.5% of isolates. blaCTX-M-15 was present in 97% of clinical isolates in Spain [23]; in 91% of hospital-acquired isolates in Portugal and in 84% of isolates in Ethiopia [24, 25]. [26] had previously mentioned a high prevalence of blaCTX-M-15 (96.9%) in clinical K. pneumoniae strains isolated in another university teaching hospital in Dakar. Paradoxically, [27] and [28] had reported respectively an absence of blaCTX-M-15 in 70 strains of K. pneumoniae in Uganda and a low prevalence of 30%. blaCTX-M-9 and blaCTX-M-25 were not detected during our study. This seems to confirm claims that CTX-M-9 and CTX-M-25 are minor variants of CTX-M. [26] had also mentioned absence of blaCTX-M-9 and blaCTX-M-25 in Dakar.
During this study, prevalence of “old” beta-lactamases (TEM and SHV-type) genes were respectively 87.9% and 47%. Prevalence of blaTEM of 78.1% in Senegal [26], 40% in China [28], and 3.8% in Uganda had been recently reported. Moreover, [28] had mentioned a predominance of blaSHV of 60%. Some variants of blaTEM and blaSHV encode penicillinases while others code for ESBLs. We need Therefore to sequence these blaTEM and blaSHV genes to find accurate variants present in our isolates as well as their real impact on the ESBL phenotypes observed in isolates.
The presence of blaOXA-1 (known to strongly hydrolyze cefepime) in 71.2% of isolates strains seems to be one main reason for the resistance of all isolates to cefepime. Our isolates seem to carry more blaOXA-1 than isolates reported during other similar studies. Indeed, the highest prevalence of blaOXA-1 in Sudan, Ghana and Dakar was 30% [26, 29, 30]. The logical continuation of this study will be the whole genome sequencing (WGS). This will provide additional informations and deepen this study. Indeed, WGS will detect other ESBL genes different from those sought in our study. WGS will also specify TEM and SHV variants responsible for ESBL production in the isolate that carried only (blaTEM + blaSHV) combination. The fact that the strains of our study cumulated on average 4 out of 7 ESBL genes sought could be the cause of their resistance to all penicillins and cephalosporins classes. This high accumulation of ESBL genes per strain combined with resistance to multiple antibiotic families is suggestive of the carriage of mobile genetic elements (plasmids, transposons and integrons). WGS could confirm and deepen our assertion. Finally, we will also check bacterial clones within this studied bacterial population. This will provide more epidemiological data and will contribute to initiate preventive and curative measures, especially in hospital settings.
5. Conclusion
We note that amikacin and carbapenems would still be effective in the treatment of MDR ESBL-producing K. pneumoniae infections, and blaCTX-M-15 seems to be predominant ESBL gene in clinical K. pneumoniae strains isolated in Dakar. Furthermore, uropathogenic strains carried significantly more blaOXA-1 than No-uropathogenic strains. We recommend to avoid monotherapy and to prohibit C3G, C4G and fluoroquinolones as empirical treatment of UTI in Dakar-Senegal. More than half of our strains were resistant to fosfomycin. We therefore recommend an in-depth and larger-scale study in order to assess the true extent of Enterobacteriaceae fosfomycin resistance in Dakar. These actions may help preserve the efficacy of this precious molecule. Whole-genome sequencing could provide additional and in-depth informations. Such studies will help to initiate or improve the public health policy of limiting MDR bacteria spread, especially in hospital settings.
Author Contributions
Conceptualization: Makhtar Camara, Bissoume Sambe Ba, Komla Mawunyo Dossouvi
Samples collection: Assane Dieng, Serigne Mbaye Lo Ndiaye, Alioune Tine, Farba Karam, Habsa Diagne Samb
Methodology: Makhtar Camara, Bissoume Sambe Ba, Komla Mawunyo Dossouvi
Lab works: Komla Mawunyo Dossouvi, Abdoulaye Cissé, Issa Ndiaye, Ousmane Sow
Data curation: Komla Mawunyo Dossouvi, Abdoulaye Cissé, Issa Ndiaye, Ousmane Sow
Formal analysis: Komla Mawunyo Dossouvi, Bissoume Sambe Ba, Cheikh Fall
Supervision: Makhtar Camara, Bissoume Sambe Ba, Cheikh Fall
Original draft writing: Komla Mawunyo Dossouvi
Review and editing: Makhtar Camara, Bissoume Sambe Ba, Gnatoulma Katawa, Simplice Damintoti Karou, Cheikh Fall
Project administration: Abdoulaye Seck, Gora Lo, Awa Ba-Diallo, Safietou Ngom-Cisse, Halimatou Diop-Ndiaye, Coumba Toure-Kane, Aïssatou Gaye-Diallo, Souleymane Mboup, Cheikh Saad Bouh Boye
Senior project administrator: Makhtar Camara
Ethical research approval
Our study has received the Ethical Research approval of the Research Ethics Committee (CER) of Cheikh Anta Diop University (UCAD) under the reference CER/UCAD/AD/MSN/051/2020.
Conflict of interests
The authors have not declared any conflict of interests.
Acknowledgements
Authors thank Abdoul Aziz Wane, Brice Léon Mosso, Amadou Mactar Gueye, Sélom Amétépé and El-Hadj Ali Niang for their assistance.
References
- Antimicrobial resistance and the United Nations sustainable development cooperation framework: Guidance for United Nations country teams (2021).
- N-environment programme, U N. Summary for Policymakers—Environmental Dimensions of Antimicrobial Resistance. UNEP - UN Environment Programme (2022).
- Murray CJ, Ikuta KS & Sharara F. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399 (2022): 629-655.
- De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, et al. Antimicrobial Resistance in ESKAPE Pathogens. Clin Microbiol Rev 33 (2020): e00181-19.
- Navon-Venezia S, Kondratyeva K & Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41 (2017): 252-275.
- Wyres KL & Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol 45 (2018) :131-139.
- Sarr H, Diop A, Diallo F, Niang AA, Dièye B, Diagne R, Lo S, et al. Prévalence des entérobactéries productrices de bêta-lactamases à spectre élargie et résistance à l’imipenème au laboratoire de Bactériologie-virologie du CHNU/Fann—Dakar (2021).
- Camara M, Mane MT, Ba-Diallo A, Dieng A, Diop-Ndiaye H, Karam F, et al. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae clinical isolates in a Senegalese teaching hospital: A cross sectional study. Afr J Microbiol Res, 11 (2017): 1600-1605.
- Odari R & Dawadi P. Prevalence of Multidrug-Resistant Klebsiella pneumoniae Clinical Isolates in Nepal. J Trop Med (2022): 5309350.
- Choby JE, Howard-Anderson J & Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med 287 (2020): 283-300.
- Effah CY, Sun T, Liu S & Wu Y. Klebsiella pneumoniae: An increasing threat to public health. Ann Clin Microbiol Antimicrob 19 (2020): 1.
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14 (2001): 933-951.
- Ghafourian S, Sadeghifard N, Soheili S & Sekawi Z. Extended Spectrum Beta-lactamases: Definition, Classification and Epidemiology. Curr Issues Mol Biol 17 (2015): 11-22.
- Castanheira M, Simner PJ, & Bradford PA. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob Resist 3 (2021): dlab092.
- Paterson DL & Bonomo RA. Extended-Spectrum β-Lactamases: A Clinical Update. Clin Microbiol Rev 18 (2005): 657-686.
- Juraschek K, Malekzadah J, Malorny B, Käsbohrer A, Schwarz S, Meemken D, et al. Characterization of qnrB-carrying plasmids from ESBL- and non-ESBL-producing Escherichia coli. BMC Genomics 23 (2022): 365.
- Latifi B, Tajbakhsh S, Ahadi, L, & Yousefi, F. Coexistence of aminoglycoside resistance genes in CTX-M-producing isolates of Klebsiella pneumoniae in Bushehr province, Iran. Iranian J Microbiol 13 (2021):161-170.
- Carr VR, Shkoporov A, Hill C, Mullany P & Moyes DL. Probing the Mobilome: Discoveries in the Dynamic Microbiome. Trends Microbiol 29 (2021): 158-170.
- Che Y, Yang Y, Xu X, Brinda K, Polz MF, Hanage WP & Zhang T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. PNAS, 118 (2021): e2008731118.
- Partridge SR, Kwong SM, Firth N & Jensen SO. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev 31(2018): e00088-17.
- Bauernfeind A, Grimm H & Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18 (1990): 294-298.
- Poirel L, Gniadkowski M, & Nordmann P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J Antimicrob Chemother 50 (2002): 1031-1034.
- Xercavins M, Jiménez E, Padilla E, Riera M, Freixas N, Boix-Palop et al. High clonal diversity of ESBL-producing Klebsiella pneumoniae isolates from clinical samples in a non-outbreak situation. A cohort study. Antimicrob Resist Infect Control 9 (2020): 5.
- Carvalho I, Chenouf NS, Carvalho JA, Castro AP, Silva V, Capita R, et al. Multidrug-resistant Klebsiella pneumoniae harboring extended spectrum β-lactamase encoding genes isolated from human septicemias. PLoS ONE 16 (2021): e0250525.
- Sewunet T, Asrat D, Woldeamanuel Y, Ny S, Westerlund F, Aseffa A, et al. High prevalence of blaCTX-M-15 and nosocomial transmission of hypervirulent epidemic clones of Klebsiella pneumoniae at a tertiary hospital in Ethiopia. JAC Antimicrob Resist 3 (2021): dlab001.
- Ngom B, Diagne R, Dia M, Ka R, Diallo F, Dieye B, et al. Molecular typing of extended spectrum β-lactamase producing klebsiella pneumoniae strains isolated in the university hospital center of Dakar. J Bacteriol Mycol: Open Access 7 (2019): 22-24.
- Mbyemeire H, Ssekatawa K, Kato CD & Wampande EM. Molecular characterization and distribution of cephalosporin resistance determinants in Escherichia coli and Klebsiella pneumoniae isolated from patients attending Kampala International University Teaching Hospital in Bushenyi, Western Uganda. Alexandria J Med 57 (2021): 205-214.
- Hao Y, Jiang Y, Ishaq HM, Liu W, Zhao H, Wang M, et al. Molecular Characterization of Klebsiella pneumoniae Isolated from Sputum in a Tertiary Hospital in Xinxiang, China. Infect Drug Resist 15 (2022): 3829-3839.
- Altayb HN, Elbadawi HS, Alzahrani FA, Baothman O, Kazmi I, Nadeem MS, et al. Co-Occurrence of β-Lactam and Aminoglycoside Resistance Determinants among Clinical and Environmental Isolates of Klebsiella pneumoniae and Escherichia coli: A Genomic Approach. Pharmaceuticals 15 (2022): 1011.
- Pankok F, Taudien S, Dekker D, Thye T, Oppong K, Wiafe Akenten C, et al. Epidemiology of Plasmids in Escherichia coli and Klebsiella pneumoniae with Acquired Extended Spectrum Beta-Lactamase Genes Isolated from Chronic Wounds in Ghana. Antibiotics 11 (2022): 689.
- Taraghian A, Esfahani BN, Moghim S, & Fazeli H. Characterization of Hypervirulent Extended-Spectrum beta-Lactamase-Producing Klebsiella pneumoniae Among Urinary Tract Infections: The First Report from Iran. Infect Drug Resist 13 (2020): 3103-3111.
- Sivaraman GK, Sudha S, Muneeb KH, Shome B, Holmes M, & Cole J. Molecular assessment of antimicrobial resistance and virulence in multi drug resistant ESBL-producing Escherichia coli and Klebsiella pneumoniae from food fishes, Assam, India. Microb Pathog 149 (2020): 104581.
- Dahiru T. P - value, a true test of statistical significance? a cautionary note. Ann Ib Postgrad Med 6 (2008): 21-26.
- Nahm FS. What the P values really tell us. Korean J Pain 30 (2017): 241-242.
- Gundran R, Cardenio P, Villanueva M, Sison F, Benigno C, Kreausukon K, et al. Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended- spectrum β- lactamase- producing E. coli isolates from broiler farms in the Philippines. BMC Vet Res 15 (2019): 227.
- Weill F-X, Guesnier F, Guibert V, Timinouni M, Demartin M, Polomack L, et al. Multidrug Resistance in Salmonella enterica Serotype Typhimurium from Humans in France (1993 to 2003). J Clin Microbiol 44 (2006): 700-708.
- Cave & Supakorn C. What is a P-value? Biosci (2016).