Molecular Characterisation of Urine and Vaginal Samples and their Antibiotic Susceptibility Pattern Associated with Β- Lactamases Resistance Gene
Article Information
Sonia Sethi*, Rinkimishra, Vaishnavigupta, SandeepShrivastava
Dr. B.Lal Institute of Biotechnology, Malviya Industrial Area, Malviya Nagar, Jaipur,India.
*Corresponding Author:Sonia Sethi,Dr. B. Lal Institute of Biotechnology, Malviya Industrial Area, Malviya Nagar, Jaipur, India./p>
Received: 30December2022;Accepted: 07 January2023;Published: 19 January 2023
Citation:
Sonia Sethi, Rinkimishra, Vaishnavigupta, SandeepShrivastava. Molecular Characterisation of Urine and Vaginal Samples and their Antibiotic Susceptibility Pattern Associated with Β- Lactamases Resistance Gene.Archives of Nephrology and Urology 6 (2023): 01-06.
View / Download Pdf Share at FacebookAbstract
Infection or inflammation of the vagina is called vaginitis. The two most common causes of vaginitis are bacterial vaginitis and Candida vaginitis. The causative organisms may be sexually transmitted or endogenous, usually by Gardnerellavaginalis and Mycoplasma hominis in combination with other anaerobes. Urinary tract infections (UTI) refer to the presence of microbial pathogens within the urinary tract and are caused by a range of pathogens but most commonly by Escherichia coli, Klebsiellapneumoniae etc. Highly increase in antibiotic- resistant among uropathogens, including ESBL producing E.coli, AmpC β-lactamase or carbapenemase-producing Enterobacteriaceae (e.g., New Delhi metallo-β-lactamase [NDM]) are being reported among health care unit. Carbapenem-resistant Enterobacteriaceae (CRE) is a growing concern worldwide and a major challenge by the WHO 2014. The aim of this study was to determine the occurrence of CTX- M gene and TEM gene and the possible role of ESBL in resistance to antibiotic. A study was conducted on a total of 100 urine and vaginal samples from Dr. B. Lal clinical laboratory and was cultured on Nutrient Agar and MacConkey agar for the detection of etiologic agents. Bacteria isolation, identification & biochemical characterization was done. Kirby-Bauer Disk Diffusion (KBDS) method was used for detection of ESBL-producing strains. A disc of Cefotaxime (30 μg) and Clavulanic acid (30 μg) were placed at a distance of 20 mm on Muller Hilton agar plate along with Imipenem(30μg), Piperacillin/Tazobactam(10μg) and Ceftazidime(30μg). The zone of inhibition generally greater than 20mm was considered as ESBL producer. Total 14 isolates were identified as ESBL producer. In order to find out the plasmid profile of these isolates, plasmid DNA isolation was done. Isolates was found to have one or more than one plasmid gene responsible for the ESBL resistance. This finding concludes that CTX-M gene is most common gene in ESBL p
Keywords
Antibiotic sensitivity; Enterobacteriaceae; ESBL; CTX-M; TEM gene.
Antibiotic sensitivity articles; Enterobacteriaceae articles; ESBL articles; CTX-M articles; TEM gene articles
Antibiotic sensitivity articles Antibiotic sensitivity Research articles Antibiotic sensitivity review articles Antibiotic sensitivity PubMed articles Antibiotic sensitivity PubMed Central articles Antibiotic sensitivity 2023 articles Antibiotic sensitivity 2024 articles Antibiotic sensitivity Scopus articles Antibiotic sensitivity impact factor journals Antibiotic sensitivity Scopus journals Antibiotic sensitivity PubMed journals Antibiotic sensitivity medical journals Antibiotic sensitivity free journals Antibiotic sensitivity best journals Antibiotic sensitivity top journals Antibiotic sensitivity free medical journals Antibiotic sensitivity famous journals Antibiotic sensitivity Google Scholar indexed journals Enterobacteriaceae articles Enterobacteriaceae Research articles Enterobacteriaceae review articles Enterobacteriaceae PubMed articles Enterobacteriaceae PubMed Central articles Enterobacteriaceae 2023 articles Enterobacteriaceae 2024 articles Enterobacteriaceae Scopus articles Enterobacteriaceae impact factor journals Enterobacteriaceae Scopus journals Enterobacteriaceae PubMed journals Enterobacteriaceae medical journals Enterobacteriaceae free journals Enterobacteriaceae best journals Enterobacteriaceae top journals Enterobacteriaceae free medical journals Enterobacteriaceae famous journals Enterobacteriaceae Google Scholar indexed journals ESBL articles ESBL Research articles ESBL review articles ESBL PubMed articles ESBL PubMed Central articles ESBL 2023 articles ESBL 2024 articles ESBL Scopus articles ESBL impact factor journals ESBL Scopus journals ESBL PubMed journals ESBL medical journals ESBL free journals ESBL best journals ESBL top journals ESBL free medical journals ESBL famous journals ESBL Google Scholar indexed journals CTX-M articles CTX-M Research articles CTX-M review articles CTX-M PubMed articles CTX-M PubMed Central articles CTX-M 2023 articles CTX-M 2024 articles CTX-M Scopus articles CTX-M impact factor journals CTX-M Scopus journals CTX-M PubMed journals CTX-M medical journals CTX-M free journals CTX-M best journals CTX-M top journals CTX-M free medical journals CTX-M famous journals CTX-M Google Scholar indexed journals TEM gene articles TEM gene Research articles TEM gene review articles TEM gene PubMed articles TEM gene PubMed Central articles TEM gene 2023 articles TEM gene 2024 articles TEM gene Scopus articles TEM gene impact factor journals TEM gene Scopus journals TEM gene PubMed journals TEM gene medical journals TEM gene free journals TEM gene best journals TEM gene top journals TEM gene free medical journals TEM gene famous journals TEM gene Google Scholar indexed journals Piperacillin/Tazobactam articles Piperacillin/Tazobactam Research articles Piperacillin/Tazobactam review articles Piperacillin/Tazobactam PubMed articles Piperacillin/Tazobactam PubMed Central articles Piperacillin/Tazobactam 2023 articles Piperacillin/Tazobactam 2024 articles Piperacillin/Tazobactam Scopus articles Piperacillin/Tazobactam impact factor journals Piperacillin/Tazobactam Scopus journals Piperacillin/Tazobactam PubMed journals Piperacillin/Tazobactam medical journals Piperacillin/Tazobactam free journals Piperacillin/Tazobactam best journals Piperacillin/Tazobactam top journals Piperacillin/Tazobactam free medical journals Piperacillin/Tazobactam famous journals Piperacillin/Tazobactam Google Scholar indexed journals Polymerase chain reaction articles Polymerase chain reaction Research articles Polymerase chain reaction review articles Polymerase chain reaction PubMed articles Polymerase chain reaction PubMed Central articles Polymerase chain reaction 2023 articles Polymerase chain reaction 2024 articles Polymerase chain reaction Scopus articles Polymerase chain reaction impact factor journals Polymerase chain reaction Scopus journals Polymerase chain reaction PubMed journals Polymerase chain reaction medical journals Polymerase chain reaction free journals Polymerase chain reaction best journals Polymerase chain reaction top journals Polymerase chain reaction free medical journals Polymerase chain reaction famous journals Polymerase chain reaction Google Scholar indexed journals
Article Details
1. Introduction
In recent times, because of augmentation of bacterial resistance to various antibiotics, the management of irresistible diseases due to harmful pathogenic bacteria has become a challenge [1]. In medical system resistance for antibiotics has become a foremost issue and mainly in urinary tract infections. Although, UTI is curable, but nowadays becoming burdensome to manage the disease due to antibiotic resistance, especially in the Enterobacteriaceae family [2]. The traditional mechanism of resistance among the Enterobacteriaceae family is the exhibition of hydrolytic enzymes, the “β-lactamases” [3]. Increase in prevalence of β-lactamases causes complications in UTIs producing uropathogens [4]. ESBL producing organisms infections are linked with high medical expenditure, mortality and morbidity. ESBLs occur principally among Escherichia coli and Klebsiella species which are plasmid mediated and genes transfer horizontally between GNB members [5]. Out of all gram-negative bacteria that produce β- lactamases are of major concern due to their ability to spread globally and the assessable inadequate curable alternative because of multiple resistance genes and the enzymes' which are associated with resistance to various non-beta-lactam antibiotics [6]. ESBLs are Class A β lactamases that are forbidden in vitro by β-lactamase inhibitors such as clavulanic acid and cannot hydrolyse cefoxitin or cefotetan i.e. cephamycins or imipenem and meropenem i.e. carbapenems but can hydrolyze penicillin, oxyimino-cephalosporins, and monobactams [7]. Studies of the antimicrobial susceptibility of GNB revealed that there is resistance augmentation due to hyperproduction of ESBL and AmpC enzymes over time. This phenomenon was observed among E. coli, K. pneumoniae and other GNB [8].
ESBLs have different genotypes and the most common are the SHV, TEM, and CTX-M types. Other types include VEB, PER, BEL-1, BES-1, SFO-1, TLA, and IBC [9]. Detection of Beta Lactamases resistant strains proves to be a guide in treatment using antibiotics and also minimizes spreading of infections [10].
2. Materials and Methods
2.1 Grouping of the study patients
This study was conducted at Dr. B. Lal Institute of Biotechnology, Jaipur (Rajasthan). A total of 100 samples of bacterial isolates were isolated and identified from different samples i.e. Urine and vaginal processed at Dr. B. Lal Clinical Laboratory Pvt. Ltd.
2.2 Grouping of the study patients
All Mid Stream Urine samples were cultured on routine culture media by semi- quantitative method as described in World Health Organization (WHO) manual [11]. In short, 1μL of urine was inoculated on MacConkey agar plate (Hi Media Laboratories Pvt. Ltd., India) by streaking using calibrated loop, and incubated aerobically for 18-48hrs at 37°C. Samples that did not yield significant bacterial growth, those that had multiple organisms and samples with suspected contamination as per lab report, were excluded from the study. Growth of 100 colonies or more, i.e. 105 colony forming units (CFU)/mL urine, was considered as culture positive. Isolation and identification of isolates were done following their morphology in Gram's staining, cultural characteristics and biochemical properties mainly IMViC (Indole test, Methyl red test, Voges-proskauer test and Citrate test) as per the Manual of Clinical Microbiology [12].
2.3 Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing of all isolates was performed by Kirby-Bauer’s disc diffusion method and interpretation of the results was done as described in CLSI 2013[13]. Antibiotic discs (HiMedia Laboratories Pvt. Ltd., India) used were, Cefotaxime (CTX) (30μg), Imipenem (10μg), Piperacillin/Tazobactam (PTZ), and Clavulanic acid (CEC). Control strains of P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used in parallel as a part of quality control. Organisms resistant to two or more classes of antimicrobial agents were considered to be multidrug resistance (MDR). The procedure involves nutrient broth having isolated organism and after some time sterile cotton swab was dipped into the suspension. The cotton swab was revolved few times and pressed against the inside wall of the tube and inoculated on the fresh surface of a Mueller-Hinton agar (MHA) plate by streaking the swab over it. For obtaining even distribution, the swab was streaked two or three times at an angle over the MH agar. After 2minutes, antibiotic discs were applied and pressed down to ensure attachment with agar surface. The plates are then inverted and incubated aerobically at 37°C after the application of disc.
2.4 Screening of ESBL-Producing Strains
According to CLSI guidelines, strains showing zone of inhibition of ≤25mm for cefotaxime were considered for conformational test for ESBL. ESBL production among potential ESBL-producing isolates was confirmed phenotypically using combined disc method. Comparison of the zone of inhibition was made for the cefotaxime (30µg) discs alone vs. that of the ceftazidime/Clavulanic acid (CAC) (10μg) when placed 25mm apart (center to center). Isolates showing an increase in zone diameter of ≥5 mm around either of the clavulanate combined discs compared to that of the disc alone was considered ESBL producer.
2.5 Molecular detection of β-lactamase genes
All the phenotypic ESBL Escherichia coli isolates were subjected to molecular analysis for the confirmation of ESBL production. Molecular detection of Escherichia coli harboring ESBL genes (bla-CTX-M, bla-TEM, and bla-SHV) was carried out by conventional polymerase chain reaction (PCR) with PCR master mix (DreamTaq Green PCR Master Mix; Thermo Scientific) using specific primers (Table 1).
|
Target genes |
Primers used (sequence 5´-3´) |
Thermal cycling condition |
PCR product size |
|
blaCTX Forward Reverse |
CGCTTTGC GATGTGCAG ACCGCGGATATC GTTGGT |
1 min of denaturation at 95°C (1 cycle), followed by 30 cycles of amplification; each of heat denaturation at 95 °C for 45 s, primer annealing at 56.9 °C for 45 s, and DNA extension at 72 °C for 1 min then one cycle for final extension at 72°C for 10 minutes. |
753bp |
|
blaSHV Forward Reverse |
ATGCGTTATATTCGCCTGTG TGCTTTGTTGTTATTCGGGCCAA |
1 min of denaturation at 95°C (1 cycle), followed by 30 cycles of amplification; each of heat denaturation at 95 °C for 45 s, primer annealing at 56.9 °C for 45 s, and DNA extension at 72 °C for 1 min then one cycle for final extension at 72°C for 10 minutes. |
550bp |
|
blaTEM Forward Reverse |
AAACGCTGGTGAAGTA TTTGTTGTTATTCGGGCCAA |
1 min of denaturation at 95°C (1 cycle), followed by 30 cycles of amplification; each of heat denaturation at 95 °C for 45 s, primer annealing at 57.4 °C for 45 s, and DNA extension at 72 °C for 1 min then one cycle for final extension at 72°C for 10 minutes. |
822bp |
2.6 Plasmid DNA Extraction and Amplification
For plasmid DNA extraction, a single colony of each potential ESBL-producing isolates was inoculated into Luria-Bertani broth and incubated till the logarithmic state. Extraction and purification of plasmid DNA of bacteria was carried out using a commercial kit (SureSpin Plasmid Mini kit from Genetix Biotech, Asia) following manufacturer’s instructions. Purified DNA from bacterial isolates was used as a template to detect ESBL genotypes: CTX-M, TEM, and SHV β-lactamase genes. Primers for the amplification of ESBL genotypes (bla-CTX-M, bla- TEM, and bla-SHV) were purchased and are as listed in above Table 1.
Polymerase chain reaction- (PCR-) based amplification of ESBL genes was carried out as the method previously described [14]. Amplification reactions were carried out in a DNA thermal cycler (CG) with the following thermal and cycling conditions: initial denaturation at 94°C for 5 min, followed by 35 cycles at 94°C for 30 sec, 45°C for 1 min, and 72°C for 1 min, and a final extension at 72°C for 10 min. The amplified PCR products were subjected to electrophoresis on a 1.5% agarose gel in 0.5 × TBE buffer. DNA was stained with ethidium bromide (1 μg/ml), and the amplified DNA bands were visualized using a UV-transilluminator.
2.7 Statistical Analysis
Data were analyzed using IBM-SPSS version 22 (Chicago, USA, 2013). Qualitative data were expressed as number and percentage and quantitative data were expressed as mean ± standard deviation (SD). Chi square test was used to compare frequencies of qualitative data and Student's t test was used to compare means in quantitative data. P values < 0.05 were considered significant.
3. Results
A total of 100 urine and vaginal samples were collected from patients that had symptoms suggestive of UTI. The age of patients ranged from 2 - 80 years (mean 41 years). These patients were 38 males and 62 females. The percentage of women was higher than that of men.
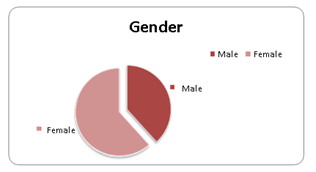
Figure 1: Gender distribution
Most of the patients (39%) reporting UTIs were between 21-40 years of age. 31 % patients were 41-80 years old, 26 % patients were above 60 years old and the lowest was 4% patient of age group less than 20.
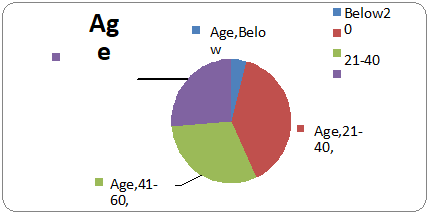
Figure 2: Age distribution
3.1 Distribution of Isolates from Clinical Specimens
One hundred clinical samples were collected from patients (62 females and 38 males) during the study period. The samples obtained were urine (n = 77) and high vaginal swabs (n = 23). Total isolates obtained was 70. Escherichia coli was isolated in the highest number from urine samples. E.coli (n=50; 71.4%) and followed by K. pneumoniae (n = 14; 20%) was a predominate isolate and the rest 8% included pseudomonas sp., Citrobactersp.andShigella sp. (Table 1 and Figure 3).
|
Identified Microorganisms |
Number of Isolates |
Percentage (%) |
|
E.coli |
50 |
71.4 |
|
Klebsiella sp. |
14 |
20 |
|
Pseudomonas sp. |
3 |
4.2 |
|
Citrobacter sp. |
2 |
2.8 |
|
Shigella |
1 |
1.4 |
Table 1: Total Number of Isolates from Clinical Specimens
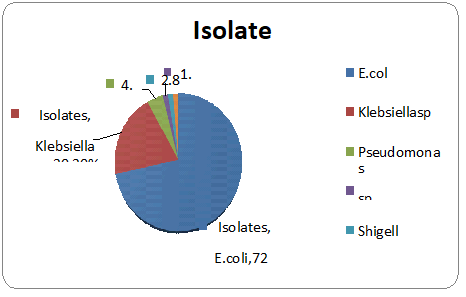
Figure 3: Percentage of patients with E. coli infection and non-E.coli infection among the isolates.
3.2 Antimicrobial Sensitivity Pattern of Uropathogenic E. coli Isolates
The antibacterial susceptibility profile was examined in all 100 uropathogenic isolates. Theresistance rate to each antibiotic was calculated as the number of resistant isolates divided by thetotal number of isolates. All ESBL-producing isolates showed 100% resistance to ampicillin, andtoallcephalosporinsincludingCefotaximeandClavulanicacid.Amongtheβ-lactam/β-lactamase inhibitor combinations 99% were resistant to amoxicillin/clavulanic acid. Out of 70isolates that were considered as potential ESBL producers, 46 (65.7%) were confirmed as ESBLproducers.
Figure 4: ESBL positive strain
3.3 Molecular analysis of ESBL genes
Among the 46 ESBL producing bacterial isolates representing E.Coli, Klebsiella, Pseudomonas, Citrobacter, were selected and characterized with PCR for genes encoding resistance. Of these, the higher resistance (n=28 ,60.8 %) was encoded by blaCTX genes. blaTEM and blaSHV genes were detected in 9 and 19.5 % of the isolates, respectively. 28 were positive for the blaCTX-M gene (60.8%), of which 19 were E. coli, 5 were Klebsiella sp., and 2 each of Citrobacter sp. and Pseudomonas sp. There was a small proportion of other gene observed in ESBL producers such as blaTEM and blaSHV genes. Out of total 46 ESBL producers, 6 were positive for the blaTEM gene (13 %), of which 3 were E.coli and 1 each of Klebseilla sp., Citrobactersp.and Pseudomonas sp. Also, 3 out of 46 ESBL producers showed presence of blaSHV gene (6.5%) in Klebseilla sp. Three (E. coli) isolate produced two ESBLs (TEM and CTX) and Klebsiella isolate produced all types of ESBLs (CTX, TEM & SHV). 37.3% of bacteria harbored multiple bla genes (≥ two genes) (Table 2). All ESBLs were resistant to cefotaxime.
|
Isolates (46) |
CTX |
TEM |
SHV |
|
E.coli |
19 |
3 |
|
|
Klebseilla sp. |
5 |
2 |
3 |
|
Pseudomonas sp. |
2 |
1 |
- |
|
Citrobacter |
2 |
- |
- |
|
Total |
28 |
6 |
3 |
Table 2: Distribution of bla gene variants in ESBL producing organisms
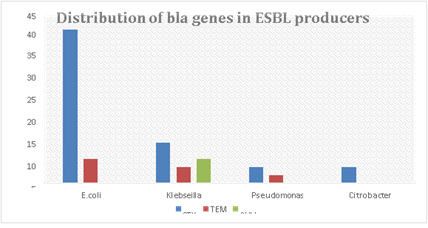
Figure 5:Distribution of bla gene variants in ESBL producing organisms
4. Discussion
In this study, we investigated the antibiotic resistance profiles and trends in MDR isolates obtained from urine and vaginal samples. Over the past two decades, there has been a significant number of infections caused by bacteria expressing extended-spectrum-β-lactamase (ESBL) and carbapenemases [15]. It has been frequently reported that UTIs occur far more frequently in girls than in boys during the first few months of life, presumably due to the shorter length of the female urethra[16].
As expected, 62% of isolates out of 100 were that of women which is 3/5th of total isolates. ESBL mediated resistance was found in 65.6% of our isolates out of 70 potential ESBL producers. This prevalence rate is much higher than other reports from India and abroad. However, the prevalence of ESBL producing strains of E.coli, pseudomonas sp. and members of enterobacteria have to be carefully monitored to prevent misuse and overuse of antibiotics because antibiotic exposure could make them more vulnerable to multidrug resistant bacteria.
The prevalence of ESBL production varies according to species, geographical areas, variations in infection control programs, different patterns of empiric antibiotic regimens and even over time. Moreover, selective pressure caused by the overuse of cephalosporins in some countries leads to the emergence of increasing rates of ESBLs production[17].
Previous report from North East India showed high rate of blaSHV (63.4%) followed by blaCTXM (60.86%) and blaTEM (54.3%). A study from Central India (Rajasthan) on 20 ESBL- producing E. coli isolates reported a high rate of blaCTXM (80%) followed by blaTEM (60%) and blaSHV (55%) [18]. Although a rapid and global spread of CTX-M-type ESBL has been observed since the 2000s, most of the ESBLs recovered in this study accordingly produced a CTX-M-type -lactamase.
The spread of ESBL genes is related to different mobile genetic elements, such as plasmid, transposons and integrons. In our study, Plasmid DNA isolation method was used for extraction of ESBL genes from ESBL producers by Plasmid DNA Extraction kit. The current limited development of novel drugs and substitutes makes the use of ARGs monitoring even more important to develop comprehensive and integrative measures for antimicrobial resistance.
Resistance of Enterobacteriaceae to third generation cephalosporins is a worldwide problem [19], which is mainly caused by ESBLs production. Production of additional β-lactamases (AmpC) also contributes to this problem, moreover, the presence of AmpC genes is often associated with multidrug resistance [20]. Previously, AmpC -β-lactamase has received less attention, but is now identified as an important cause of resistance in Enterobacteriaceae species. Global spread of β-lactamases-producing strains gives a great importance to the study of these strains in community and hospitals for reassessment of the existing treatment protocols. Although MDR rate among ESBL producers in the current study (28.5%) was lower than that reported in previous studies; (96.3%) [21] and (77.6%), there was statistically significant increase in MDR rate reported in the ESBL-producers (28.5%) than that reported in the non- ESBL-producers (1.5%) (p value = 0.04).
Out of 72 ESBL- producing isolates in the current study, 48 (99%) isolates were positive for ESBL genes, with blaCTX-M type as the most predominant. The frequency of community- acquired infections caused by blaCTX-M-producing strains have markedly increased in the last decade, that agrees with our findings, where blaCTX-M genes were detected in 254 (81.6%) of Enterobacteriaceae isolates. Our results concur with several studies on hospital and community-acquired infections, those reportedhigh prevalence of blaCTX among Enterobacteriaceae species in Egypt , Burkina Faso, Iran, Qatar and Japan. blaTEM and blaSHV-producing strains were reported previously as hospital pathogens until the late 1990s [22].
5. Conclusion
High burden of antimicrobial resistance and increased prevalence of ESBL-producing Escherichia coli associated with UTI are the major findings of this study. Diverse genotypes of ESBL E. coli along with resistance towards common antibiotics were observed. The blaCTX-M type was the pre-dominant among ESBL-producing Enterobacteriaceae, especially in combination with blaTEM enzymes. Molecular detection and identification of beta lactamases is essential for a reliable epidemiological investigation of resistance. These enzymes can be chromosomal or plasmid mediated, which may help in the distribution of antimicrobial drug resistance in health care organization. In this perspective, regular national-wide epidemiological surveillance of bacterial pathogens causing UTIs and their anti-microbial resistance would be useful in developing the treatment guidelines in our country. Further next generation sequencing (NGS) studies are essential for a more comprehensive analysis of ESBL form.
Financial support &Sponsorhip:
We are very grateful to IRSC, Dr. B. Lal Institute of Biotechnology for their kind funding support.
Conflicts of interest:
NIL
References
- Marie MA, John J, Krishnappa LG, et al. Molecular characterization of the β- lactamases in Escherichia coli and Klebsiellapneumoniae from a tertiary care hospital in Riyadh, Saudi Arabia. Microbiology and Immunology 57(12) (2013):805-810.
- Ekwealor PA, Ugwu MC, Ezeobi I. Antimicrobial evaluation of bacterial isolates from urine specimen of patients with complaints of urinary tract infections in Awka, Nigeria. International Journal of Microbiology 2 (2016):1-6.
- Parajul N, Prasad PM, Hridaya P. High rates of multidrug resistance among uropathogenic Escherichia coli in children and analyses of ESBL producers from Nepal. Antimicrobial Resistance & Infection Control 6(9)(2017):1-7.
- Elsayed TI, Ismail HA, Elgamal SA. The occurrence of multidrug resistant E. Coli which produce ESBL and cause urinary tract infections. Journal of Applied Microbiology and Biochemistry l(1)(2017):2-8.
- Chirindze LM, Zimba TF, Sekyere JO. Faecal colonization of E. coli and Klebsiella spp. producing extended-spectrum beta-lactamases and plasmid-mediated AmpC in Mozambican university students. BMC Infectious Diseases 18(1)(2018):1-8.
- Alyaman EJ, KhiyamiAM, Booq RY, et al. The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia. Annals of Clinical Microbiology and Antimicrobials 16(1)(2017): 1-13.
- Manoharan A, Sugumar M, Kumar A, et al. Phenotypic & molecular characterization of AmpC β-lactamases among Escherichia coli, Klebsiella spp. &Enterobacter spp. from five Indian medical centers. Indian Journal of Medical Research 135(3)(2012): 359-364.
- Mylvaganam H Kolstad H, Breistein RI, et al. Extended spectrum cephalosporin resistance among clinical isolates of Enterobacteriaceae in west Norway during 2006-2013; a prospective surveillance study. APMIS 125(1)(2017):52-58.
- Eftekhar F, Rastegar M, Golalipoor M, et al. Detection of extended spectrum beta-lactamases in urinary isolates ofKlebsiella pneumonia in relation to Bla SHV, Bla TEM, Bla CTX-M gene carriage. Iran J Public Health 41(2012): 127-132.
- Teethaisong Y, Eumkeb G, Chumnarnsilpa S. Phenotypic detection of AmpC β- lactamases, extended-spectrum β-lactamases and metallo-β-lactamases in Enterobacteriaceae using a resazurinmicrotitre assay with inhibitor-based methods. Journal of Medical Microbiology 65(10)(2016):1079-1087.
- Vandepitte J, Engbaek K, Rohner P, et al. Basic laboratory procedures in clinical bacteriology. 2nd ed. Geneva: World Health Organization 7 (2003).
- Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML et al.. Manual of Clinical Microbiology. 10th ed. American Society of Microbiology 5 (2011).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Twenty-thi. Clinical and Laboratory Standards Institute 10 (2013).
- Dallenne C, Da Costa A, Decre D, et al. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother 65 (2010): 490-495.
- Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis 215 (2017): S28-S36.
- Zeyaullah M, Kaul V. Prevalence of urinary tract infection and antibiotic resistance pattern in Saudi Arabia population. Global J Biol, Agricult Health Sci 4(1)(2015):206-214.
- Canto´n R, Novais A, Valverde A, et al. Prevalence and spread of extended-spectrum b-lactamase-producing Enterobacteriaceae in Europe. ClinMicrobiol Infect 14(2008): 144-153.
- Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of Extended Spectrum Beta- Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiellaspp 7(10):2173-2177.
- World Health Organization. Antimicrobial resistance: global report on surveillance: WHO; (2014).
- Jacoby G. AmpC-lactamases. ClinMicrobiol Rev 22 (2009):161-182.
- Teklu DS, Negeri AA, Legese MH, et al. Extended- spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control 8(15)(2019):39
- Chong Y, Ito Y, Kamimura T. Genetic evolution and clinical impact in extended-spectrum β- lactamase-producing Escherichia coli and Klebsiellapneumoniae. Infect Gen Evol 11(2011): 1499-1504.
