Mitochondrial Genome Screening Identified 26 Novel Variants in Children with Nonsyndromic Congenital Hearing Impairment
Article Information
Hema Bindu L1, Shehnaz Sultana1, Penagaluru Pardhanandana Reddy2, 3*
1Institute of Genetics and Hospital for Genetic Diseases, Osmania University, Hyderabad, Telangana, India
2MAA Research Foundation, Hyderabad, Telangana, India
3Bhagwan Mahavir Medical Research Centre, Hyderabad, Telangana, India
*Corresponding author: Penagaluru Pardhanandana Reddy, Research Director, Bhagwan Mahavir Medical Research Centre, Hyderabad, Telangana, India
Received: 09 March 2022; Accepted: 24 March 2022; Published: 13 April 2022
Citation:
Hema Bindu L, Shehnaz Sultana, Penagaluru Pardhanandana Reddy. Mitochondrial Genome Screening Identified 26 Novel Variants in Children with Nonsyndromic Congenital Hearing Impairment. Archives of Internal Medicine Research 5 (2022): 114-145.
View / Download Pdf Share at FacebookAbstract
Background: Mitochondrial DNA (mtDNA) mutations may be responsible for the pathogenesis of maternally inherited hearing loss in both nonsyndromic and syndromic condition. Several mitochondrial genes, including genes coding for rRNA, tRNA, and respiratory chain complex subunits and protein coding genes play significant role in nonsyndromic deafness.
Materials and Methods: 175 children with congenital hearing impairment and 92 normal subjects were screened for 13 mitochondrial genes comprising of two small ribosomal genes (12S rRNA and 16S rRNA), 7 tRNA genes (tRNA Val, tRNA Leu (UUR), tRNA Ile, tRNA Gln, tRNA Met, tRNA Ser (UCN) and tRNA Asp) and 4 protein coding genes (NADH dehydrogenase 1, NADH dehydrogenase 2, Cytochrome Oxidase I and Cytochrome Oxidase II) genes using specific sets of overlapping oligonucleotide primers for amplification.
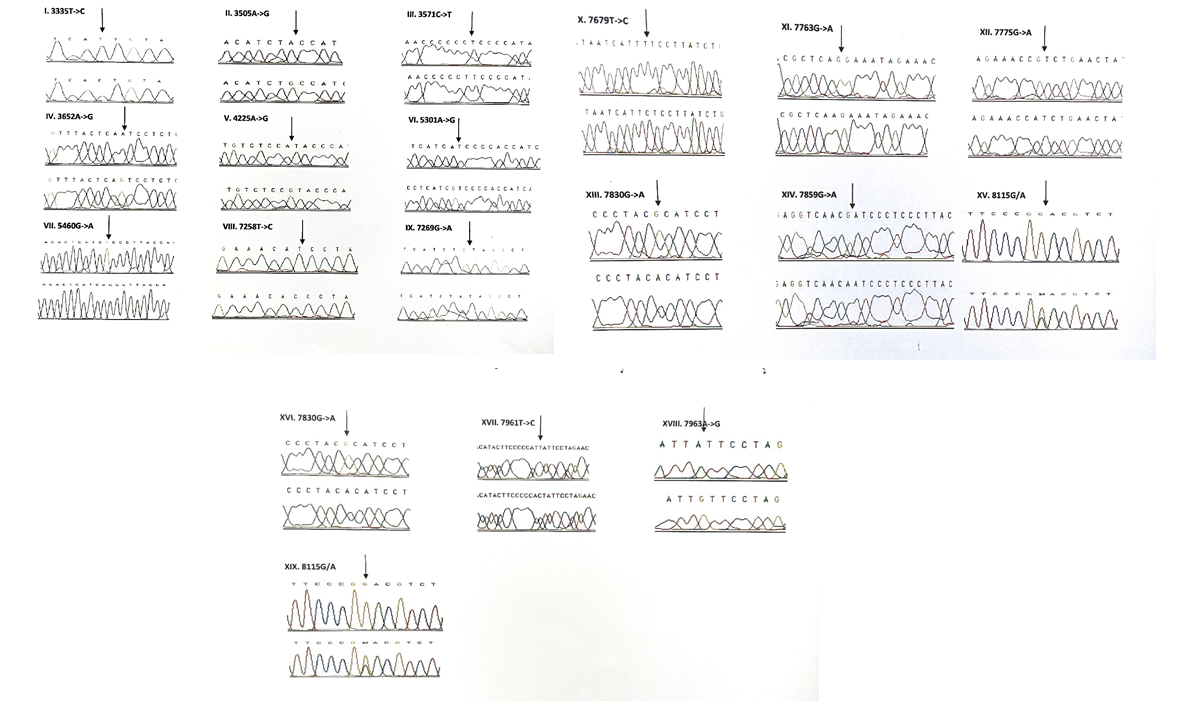
Results: A total of 26 novel variations were observed in the present study. 8 of them were in the protein coding ND1 gene, 2 in ND2 gene, 6 in ribosomal 12S rRNA, 1 in ribosomal 16S rRNA, 5 in COII and 4 in COI genes. Three variants, each belonging to ND1 (3456T/G), COI (6140C/A) and COII (8115G/A) genes were found to be heteroplasmic. Out of 26, 8 variants were observed to be transversions and 11 as transitions. Out of 19 novel variants of protein coding genes, 3 missense (A3652G, G7830A, 8115G/A) mutations and 16 silent mutations were observed.
Conclusion: This study demonstrated that various mitochondrial genes including protein-coding genes might be responsible for nonsyndromic deafness, providing new insights on the molecular bases of this pathology.
Keywords
Congenital; Nonsyndromic; Hearing Impairment; Mitochondrial Genome; Novel Variants
Congenital articles; Nonsyndromic articles; Hearing Impairment articles; Mitochondrial Genome articles; Novel Variants articles
Article Details
1. Introduction
Hearing impairment (HI) is a disabling congenital disease [1] with a global prevalence of 1.3 per 1,000 population [2] and accounts for about 1 per 1,000 live births [3]. Approximately 50% of the congenital hearing impairment cases have a genetic etiology. 70% of the cases are nonsyndromic [4] and among them almost 80% of the cases are inherited in autosomal recessive (AR) mode [5]. Out of the 30,000-50,000 human genes, 1%, i.e., 300-500 genes, are predicted to be essential for hearing [6]. Hearing impairment has been recognized as part of the clinical presentation in more than 300 genetic syndromes. Around 120 genes have been identified in hearing loss, 80 for syndromic and 41 for nonsyndromic [7]. The first genetic defect causing nonsyndromic sensorineural hearing loss (NSSNHL) detected was a mitochondrial mutation [8].
Mitochondria are the intracellular organelles found in eukaryotic organisms and produce energy in the form of ATP, which is synthesized by oxido-reduction reactions (respiratory chain) coupled to ATP synth-esis (ATP synthase). This crucial mechanism for cellular energy production is called oxidative phosphorylation, carried out by enzymatic complexes constituted by polypeptides, that are encoded by nuclear DNA and mitochondrial DNA. Thus, the ATP produced is used by cells for a number of metabolic reactions such as biochemical synthesis and the maintaining of cell function. Defect in oxidative phosphorylation can cause cellulardysfun-ction, resulting in pathologies known as mitochon-drial cytopathies. Mitochondrial cytopathies have their origin in mutations both in the mtDNA and in nuclear DNA. Numerous mtDNA point mutations and rearrangements (deletions, insertions, duplica-ions, depletion) of mitochondrial DNA are identified to be the source of some of these pathologies [9]. In the pathology of hearing loss, the tissue affected most probably is the cochlear hair cells, which are important for hearing functions since they are responsible for maintaining the ionic gradients necessary for sound signal transduction. Cochlear hair cells are affected by mitochondrial dysfunction as they are highly metabolically active and rich in mitochondria [10].
Mitochondrial DNA (mtDNA) mutations may be responsible for the pathogenesis of maternally inherited hearing loss. Whole mtDNA sequencing can detect pathogenic mutations. Several mitocho-ndrial genes, including genes coding for rRNA, tRNA, and respiratory chain complex subunits, can be significant for nonsyndromic deafness, and constant research helps in identifing novel mitocho-ndrial mutations responsible for both nonsyndromic deafness, as well as syndromic. Though nonsyn-dromic hearing loss can be caused due to different mutations in different mtDNA genes, at present-screening of the patients is typically concentrated on the ribosomal RNA 12S gene (MT-RNR1) and the transfer RNA Serine (UCN) gene (MT-TS1), overseeing other mitochondrial genes, which might affect the detection of novel mtDNA mutations involved in the pathology ofnonsyndromic hearing loss. In the present study, the contribution of 13 mitochondrial genes and their mutations has been assessed in 175 children with congenital hearing impairment and 92 control subjects in a South Indian population.
2. Materials and Methods
2.1 Study subjects
The present study was carried out in 175 children in the age group of 3-14 years with congenital neurosensory nonsyndromic hearing impairment attending Government ENT hospital, to identify the mitochondrial DNA mutations. 92 healthy subjects within the same age group were selected as controls. Children with post lingual or acquired hearing loss, and who had undergone cochlear implantationwere excluded from the study. The children were clinically examined by an ENT specialist and important charac-teristic features were recorded. An audiological evaluation was carried out by an audiologist. The severity of hearing impairment was assessed by Pure Tone Audiometry (ELKON 3 in 3, Pure tone audiometer, ISO 1964-Standard, with TDH 39 headphones and OTICON-SR 80 bone conduction receiver), Impedance Audiometry (AN22T with built in perimeters having ipsilateral and contralateral reflex testing facility), Brain Stem Evoked Response Audiometry (BERA) and Otoacoustic Emissions (OAE) (Madsen Capedla make with facility from TEOAEs and DPOAEs with CPU band software). Audiometric test was applied to each ear at frequencies of 0.5, 1.0, 2.0, 4.0 and 8.0 kHz. An audiological scientist conducted the threshold testing based on which the deafness was categorized into various types. Detailed information on epidemio-logical, clinical and genetic aspects was collected using a standard questionnaire. The study was carried out in accordance with the ethical standards and the same was approved by the Ethical Committee for Biomedical Research of Institute of Genetics and Hospital for Genetic Diseases, Ameerpet, Osmania University, Hyderabad. Written, informed consent was obtained from the parents or guardians of all the children who have participated in the study.
2.2 Mutational screening of mitochondrial genes
5ml of peripheral blood sample in EDTA was collected aseptically from each study subject and genomic DNA was isolated. Initially, a pilot study was carried out on 36 children from 18 families with congenital deafness. The mitochondrial DNA of all the 36 samples was amplified using the 24 sets of overlapping primers for amplifying the complete mitochondrial genome described by Mark et al [11]. A significant number of variants were observed in the small rRNA genes and 4 protein coding genes. Based on these results, only 13 mitochondrial genes compri-sing two small ribosomal genes (12S rRNA and 16S rRNA), 7 tRNA genes (tRNA Val, tRNA Leu (UUR), tRNA Ile, tRNA Gln, tRNA Met, tRNA Ser (UCN) and tRNA Asp) and 4 protein coding genes (NADH dehydrogenase 1, NADH dehydrogenase 2, Cyto-chrome Oxidase I and Cytochrome Oxidase II) genes were further screened in all the remaining cases. The specific sets of overlapping oligonucleotide primers used for amplifying these 13 genes in all 175 cases and 92 control subjects as shown in Table1. Polymerase chain reaction (PCR) of each sample was performed and amplified products were quantified using 2% agarose gel electrophoresis.
2.3 Cycle sequencing of the PCR product
PCR was repeated using 1µl of PCR product as a template in a 5.0 µl reaction volume. The reaction mixture consists of 2pM (0.05µl) of primer (forward), 1.8 µl of BigDye Terminator ready reaction kit (Perkin Elmer) and 2.15 µl of double distilled water to adjust the volume to 5.0 µl. These PCR reactions were carried out using 96-well plates (Applied Biosystems) in a GeneAmp 9600 thermal cycler (Perkin Elmer) employing the following conditions: 30 cycles at 96°C for 10 seconds, 55°C for 5 seconds and 60°C for 4 minutes.
2.4 Purification of PCR products after cycle sequencing
The extended PCR products were precipitated by adding 25 µl of 3M sodium acetate and ethanol in the ratio 1: 25. The plate was centrifuged at 24°C at 4000rpm for 20 minutes. The supernatant was discarded without disturbing the pellet. 100 µl of 80% alcohol was added to the plate and centrifuged at 24°C at 4000rpm for 12 minutes. Supernatant was removed and the plate was dried free of alcohol. Purified samples were dissolved in 10 µl of 50% Hi-Di formamide and analyzed in an ABI 3730 automated DNA analyzer (Perkin Elmer). The raw sequence data was analyzed and carefully edited using the sequence analysis software. The resultant edited sequences were aligned and compared with the revised Cambridge Reference Sequences (rCRS) [12] (GenBank accession number: NC_012920) using the Auto Assembler software. All the variant sites com-
pared to the reference sequence were recorded.
2.5 Insilco analysis
All the mitochondrial variants were checked from the mitochondrial genome databases, Mitomap (www. mitomap.org) and mtDB (www.genpat.uu.se/mtDB). The functional implications of the mitochondrial variants were examined using the Java based tool, Mitoanalyzer (www.cstl.nist.gov/biotech/strbase/ mitoanalyzer.html). The secondary structural altera-tion due to the missense mutations of mitochondrial genes was investigated using the GOR4 protein tool of the Biology Workbench of the San Diego Supercomputer Centre (http://workbench.sdsc.edu). The functional implications due to the variants were checked using the expasy tools PMut, SIFT and Polyphen. The amino acid conservation across species of the protein subunits was checked by comparing the protein sequences of various species.
2.6 Statistical analysis
Genotype frequencies, allele frequencies and odds ratio (OR) with an associated 95% confidence interval (CI) of all genes were calculated in patients and controls to estimate the relative risk of various mutant alleles. P-value < 0.05 was considered statistically significant and all P-values were based upon two-tailed tests.
3. Results
The Sequence analysis of the mitochondrial DNA in 175 children with congenital non-syndromic deafness resulted in a total of 106 variants in 13 mitochondrial genes. 45 variants were observed in the ribosomal genes, 3 in tRNA genes and 58 in 4 protein coding genes (ND1, ND2, COI and COII) respectively. The results of the study are presented in Tables 2-8 and Figures 1-5.
3.1 Ribosomal RNA gene mutations
3.1.1 Frequency of 12S rRNA (MTRNR1) gene mutations: A total of 29 variants were observed in 12S rRNA gene. The frequency of each mutant allele for 29 variants and their statistical significance after comparison with the controls is presented in Table 2.6 (T720C, G811A, C960A, A1053G, A1453G and G1462T) out of the 29 mutations were found to be novel and were not reported elsewhere. None of the novel mutations, except the G to T transition at the nucleotide position 1462 was observed in the control group. The T to C transition at nucleotide position 720 and A to G transition at nucleotide 1453 were observed in one patient with a mutant allele frequency of 0.57%. Homoplasmic G to A transition at nucleotide position 811 and C to A transversion at nucleotide position 960 were reported in 2 (1.14%) patients. Three patients each with an allele frequency of 1.71% were found harboringhomoplasmic A to G transition at 1053 nucleotide position and G to T transversion at 1462 nucleotide position.
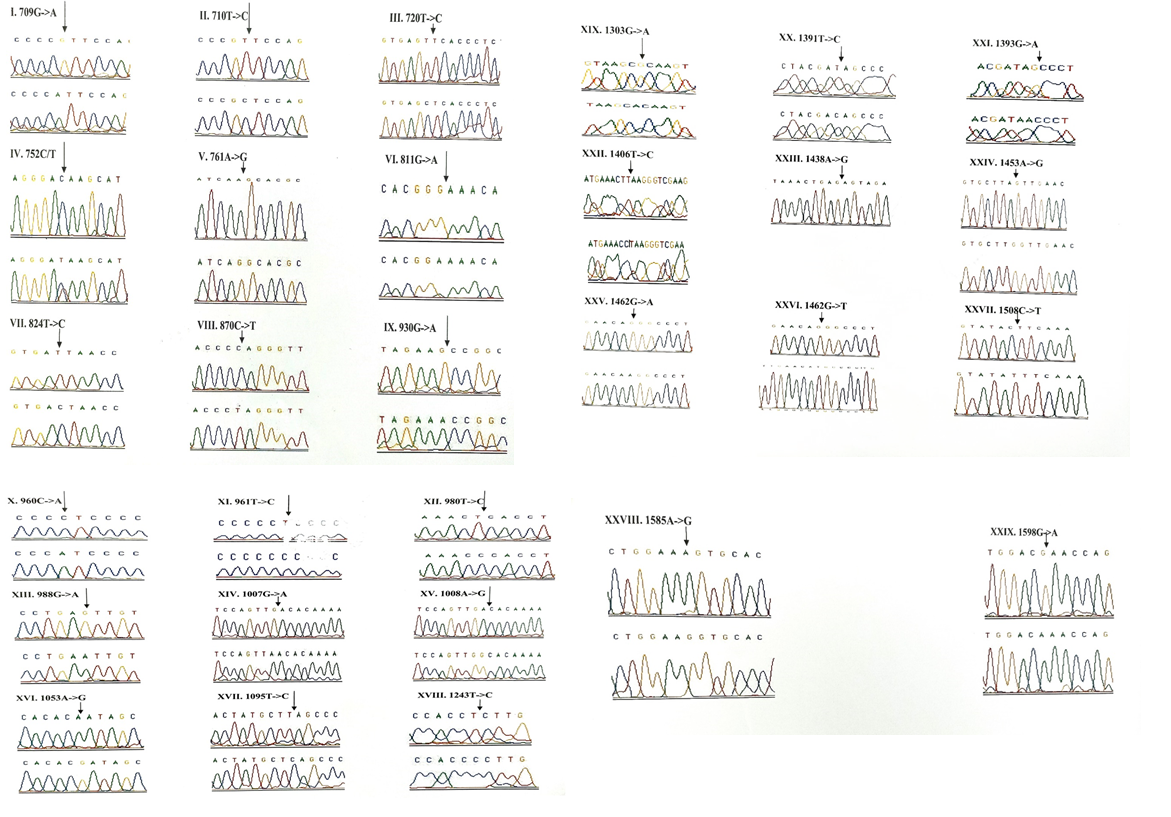
Out of 29 variants, three deafness associated homoplasmic variants namely, G709A, T961C and T1095C were observed. The homoplasmic G to A transition at the nucleotide position 709 was observed in 33 patients and 4 controls. Poor degree of conservation of the nucleotide was observed across the mammals. Multivariate analysis between the patients and controls revealed that the G709A mutation was significantly associated with deafness (odds ratio [OR], 0.1956; 95% confidence interval CI, 0.0932-0.4109; P=0.00001 (20.07014)). The homoplasmic T to C transition at 961 nucleotide position was observed in 2 patients and was absent in controls. The variant was found to be localized in a poorly conserved nucleotide position of the mammals. The T1095C mutation was observed in homozygous condition in 5 patients and 1 control. The mutant allele accounted for 2.85% in the patients and 1% in the controls. This mutation was localized at sites which are highly conserved in human, mouse, bovine and Xaenopuslaevis. However, the Odds ratio and the P-value ([OR] 0.3736; CI=0.0912-1.5366; P=0.31518 (1.00886)) of the mutant allele between patients and controls indicated that the presence of mutation in patients with deafness was not significant. A to G homozygous transition at the nucleotide position 1438 was considered a poly-morphism and was present in all children with congenital deafness and control subjects. The chromatograms of all the 12S rRNA variants are presented in Figure-1.
3.1.2 Frequency of 16S rRNA (MTRNR2) gene mutations: 16 variants were observed in the 16S rRNA gene of the children with congenital nonsyn-dromic hearing impairment (Table 3). Of these, a homoplasmic variant, A1978C was found to be novel and was observed in one patient with an allele frequency of 0.57%. It was not reported in the control group. Multivariate analysis of all the 16S rRNA variants predicted that only G1888A variant (odds ratio [OR], 0.318; 95% confidence interval CI, 0.1753-0.5774; P=0.00011 (14.99836)) is significa-ntly associated with deafness. The chromatograms of all the 16S rRNA variants are presented in Figure-2.
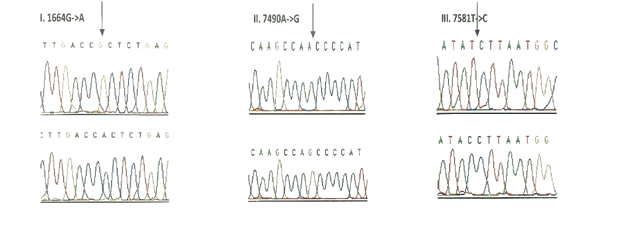
3.1.3 Frequency of mutations in the tRNA genes: Seven tRNA genes (tRNAVal, tRNALeu(UUR), tRNAIle, tRNAGln, tRNAMet, tRNASer(UCN) and tRNAAsp) were screened in all the 175 children with congenital deafness and controls. However, mutations were detected in only three genes encoding tRNA namely, tRNA Valine, tRNA Serine (UCN) and tRNA Asp (Table 4). Homozygous G1664A variant in tRNA Valine was observed in 8 probands with an allele frequency of 4.57% followed by T7581C variant in tRNA Asp gene in 6 probands (3.42%) and A7490G homozygous variant in tRNA Ser (UCN) gene in 3 probands (1.71%). Only T7581C variant (1.08%) was observed in controls. The chromatograms of all the tRNA variants are presented in Figure-3. None of the three variants were predicted to disrupt the Watson-Crick base pairing in the secondary structure of tRNA sequences.
3.1.4 Frequency of mutations in the protein coding genes (ND1, ND2, COI & COII): A total of 58 variants were observed in the protein coding genes of which 43 were synonymous and 15 were missense mutations resulting in a change in the amino acid. Out of the 58 variants, 19 were not reported earlier and were novel (Table 5). The chromatograms of the novel and missense mutations are shown in Figures 4 and 5.
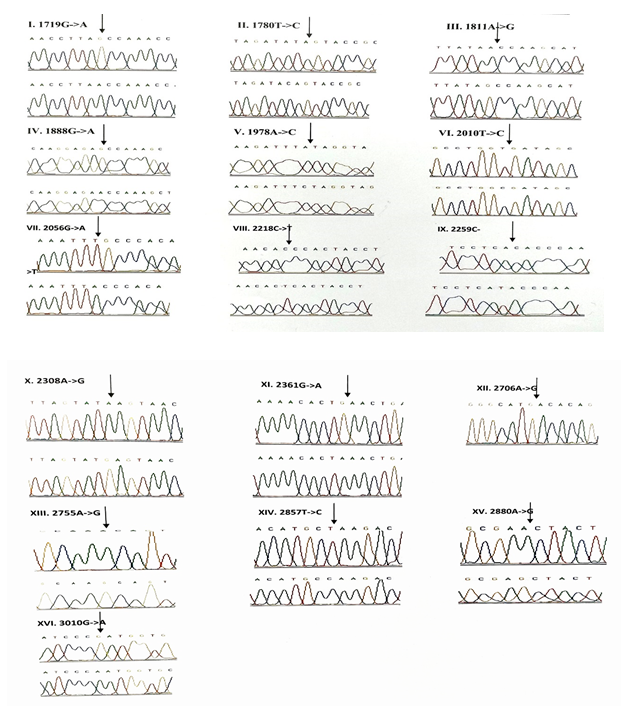
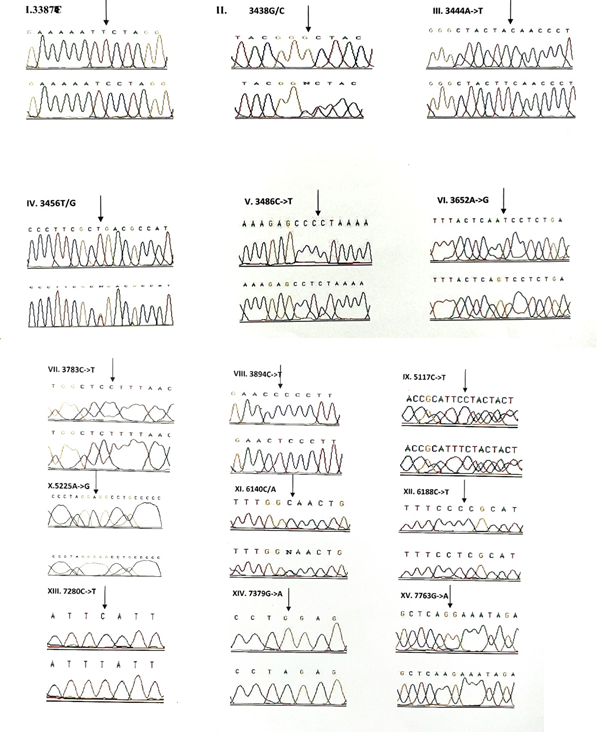
3.1.5 NADH dehydrogenase I gene (ND1): 22 variants were found in ND1 region of the mitochondria of which 8 were novel and 5 were missense. Out of the 8 novel mutations, 6 (T3387C, G3438C, A3444T, C3486T, C3783T, C3894T) were homozygous synonymous, one was homozygous missense (A3652G) and one was heterozygous silent (3456T/G) variants. The homozygous missense variant, A3652G resulted in the replacement of Isoleucine by Valine. It was found in 2 patients (1.14%) and absent in controls. The other missense variants observed in ND1 region were T3335C, A3505G, C3571T and A4225G. A3505G missense variant was observed in majority of the probands (6/3.42%) resulting in a replacement of Threonine by Alanine followed by A4225G variant in 4 probands (2.28%) resulting in replacement of Methionine by Valine. The T3335C variant resulting in replacement of Isoleucine by Threonine and C3571T variant causing the replacement of Leucine with Pheny-lalanine were found in 1 patient each (0.57%). Multivariate analysis of the ND1 variants in the patients and controls predicted two silent homozygous mutations namely C3486T (odds ratio [OR], 2.8; 95% confidence interval CI, 1.2422-6.3075; P=0.02151 (5.28465)) and C3894T (odds ratio [OR], 0.2027; 95% CI, 0.05180-0.7966; P=0.01898 (5.50312)) are found to be significantly associated with deafness.
3.1.6 NADH dehydrogenase 2 gene (ND2): 6 mutations were observed in ND2 region of the mitochondrial genome out of which 2 (C5117T and A5225G) were found to be novel. Novel 5117C to T transition was observed in one proband and 5225G was observed in 3 probands. Two variants namely, A5301G and G5460A were homozygous missense and the remaining 4 variants, A4259T, G4580A, C5117T, A5225G were silent homozygous. 16 probands with deafness were shown having G4580A variant with an allele frequency of 9.14%. 4 (2.28%) probands had A4529T transversion. Both the variants were also reported in one control each. The odds ratio [OR] for the G4580A variant was found to be significant with P-value less than 0.05 (OR, 1.092; 95% CI, 0.0286-0.4175; P=0.00029 (13.12824)) suggesting that the variant is associated with congen-
congenital hearing impairment.
3.1.7 Cytochrome Oxidase I (COI) gene: A total of 14 variants were observed in the COI region of the genome of which four were novel (6140C/A, C6188T, C7280T, G7379A) and 2 were missense (T7258C and G7269A). The novel variants, namely C6188T, C7280T and G7379A were homozygous silent transitions. Heterozygous 6140C/A and homozygous C6188T were observed in one patient each. While C7280T variant was observed in 7 (4%) probands and one control, the G7379A variant was observed in 6 (3.42%) probands and was absent in controls. The two missense variants namely, T7258C resulting in a replacement of Isoleucine with Threonine and G7269A causing the replacement of Valine by Isoleucine were observed in 3 patients (1.71%) and were absent in controls. The remaining 8 homozygous synonymous variations include: C7028T, G7211A, T7268G, A7271G, A7298G, A7307G, A7325G and G7337A.
3.1.8 Cytochrome Oxidase II (COII) gene: 16 variants were seen after screening 175 patients in the COII region. Out of 16 variants, 6 were found to be missense changing the amino acid in the protein sequence and 5 of them were novel and not reported elsewhere. Out of the 5 novel mutations namely, G7763A, G7830A, T7961C, A7963G, 8115G/A, three of them (G7763A, G7830A and 8115G/A) were missense causing a change in the amino acid sequence. The heterozygous variant 8115G/A was found in majority of the probands (30/17.14%) with an allele frequency of 8.57% resulting in an amino acid replacement of Glycine with Glutamic acid. None of the controls showed this variant. The G7830A homozygous variant resulting in a replacement of Arginine by Histidine was observed in 2 probands and one control. The G7763A resulting in the replacement of Glutamic acid to Lysine was observed in one proband and was not found in controls. The remaining three missense mutations that were observed in the probands include: T7679C, G7775A and G7859A. The homozygous T7679C variant resulted in the replacement of Phenylalanine by Leucine in 2 probands (1.14%). The G7775 homoplasmic variant resulting in the replacement of Valine by Isoleucine and G7859A variant resulting in the replacement of Aspartic acid by Asparagine were observed in 2 probands and 2 controls. The known synonymous variations observed in the present study include T7684C, T7711C, G7762A, T7759C, A7843G, T7870C, C7948T and T8023C. Multivariate analysis predicted that the silent variants namely, A7843G (odds ratio [OR], 0.2294; 95% confidence interval CI, 0.0582-0.9075; P=0.03398 (4.49551)) and T7961C (odds ratio [OR], 0.3596; 95% confidence interval CI, 0.1508-0.8586; P=0.03201 (4.59803)) were significantly associated with congenital hearing impairment.
3.2 Novel mutations identified in the mtDNA of children with congenital nonsyndromic hearing impairment
Out of the 26 novel variations, majority (8) of them were observed in the protein coding, ND1 gene followed by ribosomal 12S rRNA (6), COII (5) and COI (4) genes. Protein coding, ND2 gene showed 2 novel variants and ribosomal 16S rRNA gene showed one novel variant. Three variants, each belonging to ND1 (3456T/G), COI (6140C/A) and COII (8115G/A) genes were found to be heteroplasmic. Out of 27, 8 variants were observed to be transversions and 11 as transitions. Out of 19 novel variants of protein coding genes, 3 missense (A3652G, G7830A, 8115G/A) mutations and 16 silent mutations were observed. Seven (G1462T, C3486T, C3894T, C7280T, G7830A, T7961C, A7963G) out of 26 novel variants were observed in controls also.
3.3 Point mutations observed in the mtDNA of protein coding genes and their degree of conservation
The point mutations observed in the present study and their degree of conservation across the mammalian species are given in Table 7.A total of 15 missense mutations were identified of which 3 (A3652G, G7830A, 8115G/A) were novel and not reported earlier. The highly conserved amino acid sequences across the mammalian species were altered by 6 (C3571T, A3652G, T7679C, G7763A, G7830A, 8115G/A) missense mutations, thereby satisfying one of the canonical criteria for pathogenesis. The A to G homozygous missense transition at nucleotide position 3652 in the ND1 gene causes the replacement of Isoleucine by Valine and was observed in 2 patients. G7830A transition in COII region causes Arginine to be replaced by Histidine and was reported in 2 patients and 1 control. The heterozygous novel missense variant in COII gene, 8115G/A accounted for 17.14% of the patients and was shown to replace Glycine with Glutamic acid. This variant was not observed in any of the control cases. Out of 15, 2 missense mutations were found to be associated with two mitochondrial diseases. The ND2 gene mutation G5460A associated with Alzheimer’s and Parkinson’s disease was observed in 3 (1.71%) out of 175 probands and was shown to be poorly conserved. The variant was not detected in the control group. The missense variant, G7859A of COII gene associated with progressive encephalomyopathy, was observed in 2 patients (1.14%) and controls (2.17%) each. It was observed to be poorly conserved among the mammalian species.
3.4 In silico analysis
In silico analysis of the mutations revealed the alteration of the predicted secondary structure in the protein coding subunits due to 15 missense mutations (Table 8). The GOR4 expasy tool revealed the change in the secondary structure of the protein of all the missense variants. The polyphen bioinformatics tool based on the PSIC score difference predicted 3 missense variants (T3335C, G7830A, 8115G/A) to be pathogenic or possibly damaging due to alteration in the tertiary structure by improper substitution in the transmembrane region. However, the PMut bioinformatics tool based on the protein NN output predicted 6 missense variants (T3335C, T7679C, G7763A, G7830A, G7859A, 8115G/A) out of 15 to be pathological. The remaining variants were shown to cause neutral effect. The SIFT tool based on the median sequence conservation predicted 12 missense variants to affect protein function. Three missense variants (A5301G, T7679C, G7775A) were found to be tolerated or non-pathogenic.
Table 1: Primers used for amplifying 13 mitochondrial genes (2 Ribosomal genes, 7 tRNA genes, 4 Protein coding genes) in 175 children with nonsyndromic congenital deafness.
Note: All the Variants, except 752C/T were observed only in homozygous condition in hearing impairment and control groups. Cto T transition was observed only in heterozygous condition in the patients.
Table 2: Allele frequency and novelty of the mitochondrial 12S rRNA gene variants observed in children with congenital non-syndromic hearing impairment.
Note: All the Variants were observed only in homozygous condition in hearing impairment and control groups.
Table 3: Allele frequency and novelty of the mitochondrial 16S rRNA gene variants observed in children with congenital nonsyndromic hearing impairment.
Note: All the Variants were observed only in homozygous condition in hearing impairment and control groups.
Table 4: Allele frequency and novelty of the mitochondrial tRNA gene variants observed in children with congenital nonsyndromic hearing impairment.
Note: All the variants, except 3438G/C, 3456T/G, 6140C/A and 8115G/A were observed only in homozygous condition in hearing impairment and control groups. 3438G/C, 3456T/G, 6140C/A and 8115G/A variants were observed only in heterozygous condition in the patients.
Table 5: Allele frequency and novelty of the mitochondrial variants observed in protein coding genes (ND1, ND2, COI, COII) Analyzed in patient and control groups.
Table 6: Novel mutation observed in the mitochondrial DNA of children with congenital hearing impairment.
Degree of conservation: Conservation of amino acid for the protein coding genes in Homosapiens (H), Bos Taurus (B), Canis familiaris (C), Gorilla gorilla (G), Mus musculus (M), Pan troglodytes (P), Rattus norvegicus(R)
Table 7: Missense mutation observed in the mitochondrial genes and their degree of conservation across various mammalian species.
Table 8: Alterations of the secondary structure and functional effect predictability due to missense mutation observed in the protein coding genes in children with congenital nonsyndromic hearing impairment.
4. Discussion
Mitochondrial hearing loss constitutes less than 1% of all hereditary hearing loss cases. A mitochondrial genome is 16,569-bp comprising of 22 tRNA and 2 rRNA, that codes for 13 proteins. Only maternal DNA is inherited to the child not paternal, which makes all the children at risk of having hearing impairment when the mitochondrion of the mother has a disease-causing mutation. Mitochondrial hearing loss might be nonsyndromic or syndromic. Most of the of deaf children are born to normal hearing parents. Sensorineural hearing loss (SNHL) might be the main and only presenting feature of a mtDNA defect.Nonsyndromic deafness has been appeared to occur in most of the cases after exposure to aminoglycosides. In fact, it has been anticipated that these antibiotics target the 12S rRNA, the evolutionary equivalent to the 16S rRNA in bacteria, causing defects in mitochondrialtranslation [13].
Mutations in two mitochondrial genes have been evidently associated with nonsyndromic deafness. One is ribosomal RNA 12S gene (MT-RNR1) with m.1494C>T and m.1555A>G mutations [14, 15] was shown to cause increased susceptibility to aminogly-coside antibiotic induced hearing loss and nonsyn-dromic sensorineural hearing loss [16]. The other gene is transfer RNA Serine (UCN) gene (MT-TS1), with mutations m.7511T>C, m.7445A>G, and m.7472_7473insC [17-19] which causes a reduction of tRNA level [20] resulting in nonsyndromic deafness. In the present study a total of 29 variants were observed in 12S rRNA gene. Out of these 29 variants, 6 variants (T720C, G811A, C960A, A1053G, A1453G and G1462T) were found to be novel and were not reported elsewhere. None of the novel mutations, except the G to T transition at the nucleotide position 1462 was observed in the control group. Three deafness associated homoplasmic variants namely, G709A, T961C and T1095C were also observed among the 29 variants,while, 16 variants were observed in the 16S rRNA gene. Of these, a homoplasmic variant, A1978C was found to be novel and was observed in one patient, and not reported in the control group.
Mitochondrial DNA mutations in tRNA genes may cause tRNA modification and affect tRNA metabolism, therefore impairing the protein synthesis and reducing the ATP synthesis, that are considered to be the key pathogenic factors [21-23]. Kai et al. screened 300 deaf infants and 200 healthy subjects for mt-tRNA gene mutations and analyzed the mtDNA copy number and reactive oxygen species (ROS) levels in patients carrying the mt-tRNA mutations. Subsequently identified three mt-tRNA mutations; tRNALeu (UUR) A3243G; tRNAAla T5655C and tRNAGlu A14692G in patients. A3243G mutation formed a novel base-pairing (13G-23A) in the D-stem of tRNALeu (UUR). The T5655C mutation was present at the very conserved acceptor arm of tRNAAla. While the A14692G mutation was located at 55 positions in the TWC loop of tRNAGlu. Lower level of mtDNA copy number and increased ROS levels were observed in patients with A3243G, T5655C and A14692G mutations [24]. In the current study, although 7 tRNA genes (tRNAVal, tRNALeu (UUR), tRNAIle, tRNAGln, tRNAMet, tRNASer(UCN) and tRNAAsp) were screened in all the 175 children with congenital deafness and controls, mutations were detected in only three genes encoding tRNA namely, tRNA Valine (G1664A), tRNA Serine (UCN) (A7490G) and tRNA Asp (T7581C). None of the variants exce-
pt T7581C (1.08%) was observed in controls.
The search for novel candidate mtDNA mutations responsible for nonsyndromic deafness made Leve-que et al, to sequence the whole mitochondrial DNA of 29 families using a microarray resequencing chip which showed a clear maternal pattern of inheritance (three to five generations) and did not carry any known mutations [25]. The results of the study lead to the identification of four other genes, subunit ND1 of complex I of the respiratory chain (MT-ND1); the tRNA for Isoleucine (MT-TI); subunit COII of complex IV (MT-CO2); and the tRNA of Serine 2 (AGU/C) (MT-TS2) that might be involved in the pathology of nonsyndromic deafness [25].
Nicol et al has characterized five candidate mutat-ions localized in subunit ND1 of complex I of the respiratory chain (m.3388C>A [p.MT-ND1:Leu28 Met]), the tRNA for Isoleucine (m.4295A>G), subunit COII of complex IV (m.8078G>A [p.MT-CO2: Val165Ile]), the tRNA of Serine 2 (AGU/C) (m.12236G>A), and Cytochrome B, subunit of complex III (m.15077G>A [p.MT-CYB: Glu111 Lys]), constructed cybrid cell lines for each of the mtDNA mutationsstudied and performed functional studies to assess the probable significances of these mutations on mitochondrial bioenergetics. Out of the five mutations studied, two showed a significant defect in mitochondrial function (m.3388C>A [p.MT-ND1:Leu28Met] and m.4295A>G); two presented mild defects (m.8078G>A [p.MT-CO2: Val165Ile] and m.12236G>A), and one had no effect (m.15077G>A [p.MT-CYB: Glu111Lys]). Nicol et al studiedfive patientswith mitochondrial deafness carrying m.1555A>G mutation in 12S rRNA, and never been treated with aminoglycosides. Interest ingly, the results showed that none of them having mutations in mitochondrial 12S ribosomal RNA. Their mutations affected OXPHOS complexes and their function, signifying that the origin of nonsyn dromic hearing loss with maternal inheritance might be certainly a direct dysfunction of mitochondrial bioenergetics. This study demonstrated that various mitochondrial genes including protein-coding genes might be responsible for nonsyndromic deafness and exposure to aminoglycosides is not essential to develop the disease [26].
In our study, 58 variants were observed in the protein coding genes of which 43 were synonymous and 15 were missense mutations resulting in a change in the amino acid. Out of the 58 variants, 19 were not reported earlier and were novel. 22 variants were found in the NADH dehydrogenase I gene (ND1) region of the mitochondria of which 8 were novel and 5 were missense. Out of the 8 novel mutations, 6 (T3387C, G3438C, A3444T, C3486T, C3783T, C3894T) were homozygous synonymous, one was homozygous missense (A3652G) and one was heterozygous silent (3456T/G) variants. While, 6 mutations were observed in NADH dehydrogenase 2 (ND2) region of the mitochondrial genome out of which 2 (C5117T and A5225G) were found to be novel. A total of 14 variants were observed in the Cytochrome Oxidase I (COI) region of the genome of which four were novel (6140C/A, C6188T, C7280T, G7379A) and 2 were missense (T7258C and G7269A). The novel variants, namely C6188T, C7280T and G7379A were homozygous silent transitions,whereas 16 variants were identified in the Cytochrome Oxidase II (COII) region. Out of 16 variants, 6 were found to be missense changing the amino acid in the protein sequence and 5 of them were novel and not reported elsewhere. Out of the 5 novel mutations namely, G7763A, G7830A, T7961C, A7963G, 8115G/A, three of them (G7763A, G7830A and 8115G/A) were missense causing a change in the amino acid sequence. Valeria et al identified novel sequence variants including C712A and the heteroplasmic G786A in MT-RNR1, A3213G in MT-RNR2, C7534T in the D-loop of TRND (tRNAAsp) and in the MT-CO2 gene, A7746G in idiopathic SNHL patients [27].
Interestingly, our study has identified 26 novel variants in 13 mitochondrial genes of children with nonsyndromic congenital hearing impairment. Out of the 26 novel variations, majority (8) of them were observed in the protein coding ND1 gene followed by ribosomal 12S rRNA (6), COII (5) and COI (4) genes. Protein coding, ND2 gene showed 2 novel variants and ribosomal 16S rRNA gene showed one novel variant. Three variants, each belonging to ND1 (3456T/G), COI (6140C/A) and COII (8115G/A) genes were found to be heteroplasmic. Out of 26, 8 variants were observed to be transversions and 11 as transitions. Out of 19 novel variants of protein coding genes, 3 missense (A3652G, G7830A, 8115G/A) mutations and 16 silent mutations were observed. Seven (G1462T, C3486T, C3894T, C7280T, G7830A, T7961C, A7963G) out of 26 novel variants were observed in controls also. The present study has made an important insight on the role of mitochondrial genome in the pathology of congenital nonsyndromic hearing impairment.
5. Conclusion
Though maternally inherited hearing loss account for nearly 1% of cases, nonetheless correct diagnosis is essential for affected individuals and their family members. Due to the absence of the extensively used new-born screening, the age at diagnosis is typically late. Still, genetic diagnosis, carrier detection, and reproductive risk counselling may be provided only for a limited number of affected individuals, as mutation screening is offered for only a limited number of genes. However, identification of additional genes will improve early diagnosis and may also offer future prospects for a rational therapy.
Acknowledgements
The authors would like to thank Dr. K. Thangaraj, Director, Centre for DNA Fingerprinting and Diagnostics and former Senior Scientist and Group Leader Centre for Cellular Molecular Biology (CCMB) for encouragement, support, advice and for providing facilities to carry out study at CCMB and Dr. M.V. Vishnuvardhan Reddy ENT specialist for the diagnosis of cases. Authors also like to thank shri. Mahendra Ranka, chairman, Bhagwan Mahavir Memorial Trustand Mrs. Sunita Kumar, Chairman, Managing Director, MAA Research Foundation for their support. Dr. L. Hema Bindu would like to thank Indian Council of Medical Research (ICMR) for providing her Senior Research Fellowship (SRF).
Conflicts of Interest
None to declare.
References
- Neumann K, ChadhaS, TavartkiladzeG, et al. Newborn and infant hearing screening facing globally growing numbers of people suffering from disabling hearing loss. Int. J. Neonatal Screen 5 (2019): 7.
- JamesM, Kumar P, Ninan P J. A study on prevalence and risk factors of hearing Impair-ment among newborns. Int. J. Contemp. Pediatr 5 (2018): 304-309.
- Olusanya B O, Neumann KJ, Saunders J. E.The global burden of disabling hearing impairment: a call to action. Bull. World Health Organ 92 (2014): 367-373.
- Sheffield A M, Smith R J. The Epidemiology of Deafness. Cold Spring Harb. Perspect. Med (2018).
- Zhou Y, LiC, LiM, et al. Mutation analysis of common deafness genes among 1,201 patients with non-syndromic hearing loss in Shanxi Province. Mol. Genet. Genomic Med 7 (2019): e537.
- Friedman TB, Griffith AJ. Human nonsyn-dromic sensorineural deafness, Annu. Rev. Genomics Hum. Genet 4 (2003): 341-402.
- Nance W.E. The genetics of deafness, Ment. Retard. Dev. Disabil. Res. Rev 9 (2003): 109-119.
- Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed, Hum. Mutat 13 (1999): 261-270.
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medi-cine. Annu Rev Genet 39 (2005): 359-407.
- Chinnery PF, Elliott C, Green GR, et al. The spectrum of hearing loss due to mitochondrial DNA defects. Brain 123 (2000): 82-92.
- Mark JR, Scott LT, Vincent OT, et al. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Research 26 (1998): 967-973.
- Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23 (1999): 147.
- Guan MX. Molecular pathogenetic mechan-ism of maternally inherited deafness. Ann NY Acad Sci 1011 (2004): 259-271.
- Prezant TR, Agapian JV, Bohlman MC, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 4 (1993): 289-294.
- Zhao H, Li R, Wang Q, et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet 74 (2004): 139-152.
- Usami S, Abe S, Shinkawa H, et al. Sensorin-eural hearing loss caused by mitochondrial DNA mutations: special reference to the A1555G mutation. J CommunDisord 31 (1998): 423-434.
- Sue CM, Tanji K, Hadjigeorgiou G, et al. Maternally inherited hearing loss in large kindred with a novel T7511Cmutation in the mitochondrial DNA tRNA (Ser (UCN)) gene. Neurology 52 (1999): 1905-1908.
- ToompuuM, Levinger LL, Nadal A, et al. The 7472insC mtDNA mutation impairs 5’ and 3’ processing of tRNA (Ser (UCN)). BiochemBioph ResCo 322 (2004): 803-813.
- Vernham GA, Reid FM, Rundle PA, et al. Bilateral sensorineural hearingloss in mem-bers of a maternal lineage with mitochondrial point mutation. ClinOtolaryngol Allied Sci 19 (1994): 314-319.
- KokotasH, Petersen MB, Willems PJ. Mitochondrial deafness. Clin Genet 71 (2007): 379-391.
- McKenzie M, Liolitsa D, Hanna MG. Mitochondrial disease: mutations and mecha-nisms. Neurochem Res 29 (2004): 589-600.
- Li X, Zhang LS, Fischel-Ghodsian N, et al. Biochemical characterization of the deafness-associated mitochondrialt RNA Ser(UCN) A7445G mutation in osteosarcoma cell cybrids.BiochemBiophys Res Commun 328 (2005): 491-498.
- Tang X, Li R, Zheng J, et al. Maternally inherited hearing loss is associated with the novel mitochondrialtRNA Ser (UCN) 7505T>C mutation in a Han Chinese family. Mol Genet Metab 100 (2010): 57-64.
- Kai Tang, Ziying Gao, Chunling Han, et al. Screening of mitochondrial tRNA mutations in 300 infants with hearing loss, Mito-chondrial DNA Part A (2018).
- Leveque M, Marlin S, Jonard L, et al. Wholemitochondrial genome screening in maternally inherited non-syndromic hearing impairment using a microarray resequencing mitochondrial DNA chip. Eur J HumGenet 15 (2007): 1145-1155.
- Nicol ´as Guti ´ errezCort ´ es, Claire Pertui-set, Elodie Dumon, et al. Novel Mitochon-drial DNA Mutations Responsible for Maternally Inherited Nonsyndromic Hearing Loss. Human Mutation 33 (2012): 681-689.
- Valeria Guaran, Laura Astolfi, Alessandro Castiglione, et al. Association between idiopathic hearing loss and mitochondrial DNA mutations: A study on 169 hearing-impaired subjects. International Journal of Molecular Medicine 32 (2013): 785-794.