Minimal Detectable Amounts of Exogenous DNA after Systemic Cell Engraftment
Article Information
Daniel Rodrigo Marinowic1,2, Daniele Vieira Da Silva1,2, Alessandra Deise Sebben1,2, Denise Cantarelli Machado1,2, Jaderson Costa Da Costa1*
1Instituto do Cérebro do Rio Grande do Sul (InsCer), Av. Ipiranga, 6690, building 63, Jardim Botânico, Porto Alegre Rio Grande do Sul - 90610.000, Brazil
2Instituto de Pesquisas Biomédicas da Pontifícia Universidade Católica do Rio Grande do Sul (PUCRS), Av. Ipiranga, 6690, building 60, second floor, Jardim Botânico, Porto Alegre Rio Grande do Sul - 90610.000, Brazil
*Corresponding Author: Jaderson Costa Da Costa, Instituto do Cérebro do Rio Grande do Sul (InsCer). Av. Ipiranga, 6690, building 63, Jardim Botânico, Porto Alegre Rio Grande do Sul - 90610.000, Brazil
Received: 29 July 2016; Accepted: 6 August 2016; Published: 10 August 2016
View / Download Pdf Share at FacebookAbstract
In cellular therapy models, the establishment of accurate methods to track the homing, cell fate and survival are fundamental to ensure safety and efficacy. The PCR is used to detect donor cells DNA in xenotransplant models. Five different concentrations of EGFP+ DNA from C57/BL6 mouse muscle tissue: 1,000 pg/μL, 500 pg/μL, 100 pg/μL, 10 pg/μL e 1 pg/μL were used to contaminate 10 ng/μL of DNA Wistar rat. Polymerase chain reaction was performed using 2 μL of each sample. EGFP+DNA were detected in samples containing 2,000 pg, 1,000 pg, 200 pg and 20 pg. Samples with 2 pg EGFP+DNA did not show signal. Here we demonstrate an alternative to proving the presence or confirm the absence, of exogenous DNA in organs and tissues. The concentration curve for cell migration detection by real time PCR will allow the use in systemically transplanted cells in animal models subjected to cell therapy.
Keywords
Cell migration; Molecular detection; Cell therapy
Article Details
1. Introduction
In recent decades, stem cell transplantation have been experimentally investigated as a therapeutic alternative to treat several diseases [1-4 ]. Despite its potential benefits, the clinical application remains limited due to lack of an effective route of delivery as well as the definition of the best cell linage. The search for accurate methods to track the homing, cell fate and survival is fundamental to ensure safety and efficacy [5 ]. Many techniques such as histological methods, real-time quantitative polymerase chain reaction, radiolabeling, bioluminescence, SPECT, PET and magnetic resonance imaging (MRI) are used to investigate the route of cell migration (REFs). All these procedures have limitations: the likelihood to produce artifacts (false-positives or negatives); variability regarding the quantitative results; hardship to track cell fate, and prejudicial effects relate to radioactivity [5 ].
Several studies have used the green fluorescent protein (GFP+) as a reporter to allow one to detect the transplanted cells, as well as to identify the migration route and survival in the host organism. GFP is one of the most widely used fluorescent probes for this purpose. The detection becomes more efficient when the xenotransplantation is realized and emerges as an alternative to avoid non-specific signals ensuring precise cell detection by immunofluorescent techniques or real-time polymerase chain reaction (QT-PCR). The PCR is used to detect DNA from donor cells from xenotransplants models and several experimental studies showing cell quantification and survival after transplantation have been published. One of the main advantages of this technique is that the limitations of cell labeling are avoided, and cell migration can be assessed, due to the presence of the stable genetic marker in the donor cell genome [6, 7 ]. The present study shows a new strategy to determine the minimum of DNA that can be detect by PCR, in other words, how to determine the cut off of your own experimental settings. We present detectable levels of EGFP+DNA contaminants in DNA from Wistar rats as a reference to the minimal amounts of exogenous DNA that ought to be present to allow the discrimination, mainly in cell therapy studies.
2. Methods
2.1 DNA Extraction
Gastrocnemius muscle from Wistar rats and C57/BL6 mouse were used as a source of DNA. DNA was extracted using TRIzol (Life Technologies) according to manufacturer instructions. Briefly, the tissue samples were minced in a small grail containing 300 µL of PBS followed by the addition of 600 µL of TRIzol (Life Technologies) and 120 µL of chloroform (Merk). The samples were homogenized using a vortex (Genie) and centrifuged at 12,000 x g for
10 minutes. The upper aqueous phase was transferred to a new tube, 0.7 x the volume of isopropyl alcohol (Merk) was added, and the samples were stored at -20°C for 12 hours. The sample was thawed and centrifuged at 12,000 x g for 10 minutes and the supernatant was discarded. The precipitate was resuspended with 40 µL of ultrapure DNAse and RNAse free water (Life Technologies). The DNA was quantified using fluorometer Qubit 2.0 (Life Technologies).
2.2 DNA Contamination Curve
Five different concentrations of EGFP+DNA from C57/BL6 mouse muscle tissue: 1,000 pg/µL, 500 pg/µL, 100 pg/µL, 10 pg/µL e 1 pg/µL were used to contaminate 10 ng/µL of Wistar rat DNA. (Table 1).
| Sample | Rat DNA | Mouse EGFP+ DNA | PCR * |
|---|---|---|---|
| 1 | 10 ng/µL | 1,000 pg/µL | 2,000 pg EGFP+DNA |
| 2 | 10 ng/µL | 500 pg/µL | 1,000 pg EGFP+DNA |
| 3 | 10 ng/µL | 100 pg/µL | 200 pg EGFP+DNA |
| 4 | 10 ng/µL | 10 pg/µL | 20 pg EGFP+DNA |
| 5 | 10 ng/µL | 1 pg/µL | 2 pg EGFP+DNA |
Table 1: EGFP+DNA concentration used to contaminate Wistar DNA to perform PCR. After mixing the amounts showing in the column rat DNA with mouse EGFP+DNA, 2 µL of each aliquot was taken to perform the PCR. Therefore, the column PCR* shows the real amount of amplified DNA.
3. PCR
Polymerase chain reaction was performed using the thermocycler StepOne Plus (Life Technologies) with Sybr Select Master Mix (Life Technologies) using 2 µL of each sample (Table 1) in duplicated. Specific primers complementary to EGFP (forward 5’ ? ATGTTGTGGCGGATCTTGA ? 3’/ reverse: 5’ ? GGGCACAAGCTGGAGTACA ? 3’) and to ?-actin gene (foward 5’ ? TAGCCATCCAGGCTGTGTTG ? 3’/reverse: 5’ ?GGAGCGCGTAACCCTCATAG ? 3’) obtained by sequence alignment using the Primer Blast database, in the NCBI website (http://www.ncbi.nlm.nih.gov/blast) were used.
The PCR conditions were as follows: 95 °C for 5 minutes, 40 cycles of 95 °C for 45 seconds, 60 °C for 30 seconds, and 72 °C for 30 seconds. The results were analyzed using the software specific for the StepOne Plus.
4. Electrophoreses
The products generated by PCR reaction were separated by electrophoresis in a 2 % agarose gel containing ethidium bromide in TAE buffer for 45 minutes at 100 V. The DNA bands were visualized by ultraviolet transillumination (Gel Doc EZ Imager) and the images were captured using the software Image Lab 4.1.
5. Results
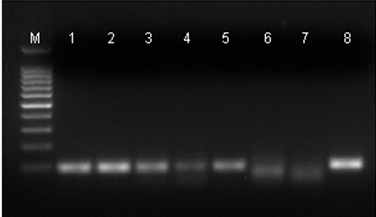
The dilution curve of EGFP+DNA used to contaminate the rat DNA have allowed to show the minimum amount of DNA needed to be detected by PCR. The 114 bp fragment generated by PCR corresponds to the ?-actin gene and 100 bp to the EGFP gene (Figure 1).
Figure 1: PCR Products after electrophoresis on a 2% agarose gel. (M): Molecular weight marker 100 bp (100 bp ladder); (1): positive control to EGFP+DNA gene; (2): rat DNA with 2,000 pg of EGFP+DNA; (3): rat DNA with 1,000 pg of EGFP+DNA; (4) rat DNA with 200 pg of EGFP+DNA; (5): rat DNA with 20 pg of EGFP+DNA; (6): rat DNA with 2 pg of EGFP+DNA; (7) negative control (rat DNA without EGFP+DNA); (8) positive control to ?-actin gene. The 114 bp products correspond to the endogenous control (?-actin gene), and the 100 bp products correspond to the EGFP+DNA.
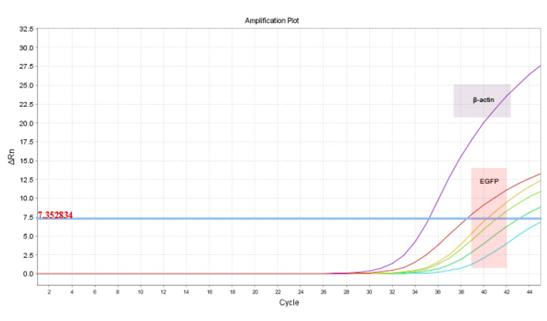
Analyzing the Ct curve, the threshold cut off was determined based on the plateau obtained by amplification of rat
DNA with EGFP primers, this is, negative and/or undetectable. EGFP+DNA were detected in samples containing 2,000 pg, 1,000 pg, 200 pg and 20 pg. Samples with 2 pg EGFP+DNA did not show signal strength over the threshold which means that the signal corresponds only to the noise inherent to the technique (Figure 2).
Figure 2: Amplification plot of qPCR: The endogenous controls (?-actin) are detectable for every tested samples (purple highlighted). Different EGFP+DNA concentration (2,000 pg, 1,000 pg, 200 pg and 20 pg) were detectable. Sample contaminated with 2 pg of EGFP+DNA was below the threshold and considerate undetectable (pink highlighted).
6. Discussion
Usually, in studies with cell therapy, the detection of DNA from infused cells is not easily to distinguish. Here we demonstrate an alternative to prove the presence, or confirm the absence, of exogenous DNA in organs and tissues from experimental animal models. Moreover, to detect cell migration will not be necessary any genetic modification or previous cell labeling before transplantation which will speed and facilitate the investigations using stem cell therapy [5 ].
Experiments using xenotransplants, specific DNA sequences from donor organism can be detected by real time PCR in the receptor tissue using a specific sequence of a gene present only in the donor genome. Other strategy to detect DNA is sex-mismatched transplantation where one have to consider the gender of donor and receptor. The use of males as cells donors are preferable since it became easy to detect the DNA sequences corresponding to some region of the Y chromosome [5, 8, 9 ].
Some studies were able to detect donor genome, however other were inconclusive. Cell migration assay using PCR confirmed the presence of grafted human cells in a hypoxic ischemia model in newborn rats. Different concentrations of umbilical cord human cells (106, 107 e 108 cells) were transplanted into the left jugular vein, and seven days after cells were detected in the ipsilateral, and contralateral brain hemisphere to the lesion [10 ]. Venturin and colleagues in 2011, showed a cell migration assay in a model of epilepsy induced with lithium/pilocarpine in rats. Bone marrow mononuclear cells from EGFP+ mouse were injected into the tail vein (107 cells). Twenty four hours, two, and four weeks the investigators were able to detect EGFP+ cells in the animal brains. However eight weeks later the donors DNA were not detectable [11 ].
Male dead cardiocytes injected into female hearts were detected by conventional PCR up to one hour after transplantation, after 24 hours only a faint signal was detectable, and seven days later it disappear. To analyze cell migration, the authors injected live cardiocytes into the right ventricle and atria, and investigate the presence of these cells into the left region. They did not found any signal meaning that there no cell migration [7 ].
There are some limitations to cell migration detection by PCR because its positivity does not mean the presence of live cells, neither transdifferentiated nor a site specific location [5 ]. The failure to detect cell migration by the polymerase is attributed to the absence of transplanted cells at the site investigated. Qualitative PCR assays show the presence or absence of exogenous genetic material into DNA extraction from a target organ. The amounts of material detectable by PCR in cell migration assays are not yet established. The lack of a standard is also detrimental. Indeed, a negative result by PCR cannot be related to the absence of cells into tissue site, but maybe with scarce DNA amounts to allow polymerase amplification and discrimination between endogenous and exogenous genetic material.
We present detectable levels of EGFP+DNA contaminants in DNA from Wistar rats as a reference to the minimal amounts of exogenous DNA that ought to be present to allow the discrimination, mainly in cell therapy studies. Here we show that DNA concentrations below 20 pg into the target organ are undetectable by real time PCR assay and even after separation electrophoresis in agarose gels. The concentration curve for cell migration detection by real time PCR and subsequent electrophoresis will allow the use of molecular parameters to prove the presence or absence of genetic material from transplanted cells systemically or at the injury site in animal models subjected to cell therapy.
Acknowledgement
The authors wish to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support.
Conflict of Interest
The authors declare no conflicts of interest.
References
- Costa-Ferro, Zaquer SM, et al. Transplantation of bone marrow mononuclear cells decreases seizure incidence, mitigates neuronal loss and modulates pro-inflammatory cytokine production in epileptic rats. Neurobiol Dis 46 (2012): 302-313.
- Liu, Shan, et al. Strategies to Optimize Adult Stem Cell Therapy for Tissue Regeneration. Int J Mol Sci 17 (2016): 982.
- Rodríguez-Frutos, Berta, et al. Stem Cell Therapy and Administration Routes After Stroke. Transl Stroke Res 1 (2016): 1-10.
- Zanirati, Gabriele, et al. Transplantation of Bone Marrow Mononuclear Cells Modulates Hippocampal Expression of Growth Factors in Chronically Epileptic Animals. CNS Neurosci Ther 21 (2015): 463-471.
- Terrovitis, John V., Rachel Ruckdeschel Smith, and Eduardo Marbán. Assessment and optimization of cell engraftment after transplantation into the heart. Circ Res 106 (2010): 479-494.
- Robey, Thomas E., et al. Systems approaches to preventing transplanted cell death in cardiac repair. J Mol Cell Cardiol 45 (2008): 567-581.
- Müller-Ehmsen, Jochen, et al. Survival and development of neonatal rat cardiomyocytes transplanted into adult myocardium. J Mol Cell Cardiol 34 (2002): 107-116.
- Suzuki, Ken, et al. Role of interleukin-1? in acute inflammation and graft death after cell transplantation to the heart. Circulation 110 (2004): II-219.
- Johnston, Peter V., et al. Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation 120.12 (2009): 1075-1083.
- De Paula, S., et al. The dose-response effect of acute intravenous transplantation of human umbilical cord blood cells on brain damage and spatial memory deficits in neonatal hypoxia-ischemia. Neurosci 210 (2012): 431-441.
- Venturin, Gianina Teribele, et al. Bone marrow mononuclear cells reduce seizure frequency and improve cognitive outcome in chronic epileptic rats. Life Sci 89 (2011): 229-234.