Microbiological analysis and assessment of the Biotechnological potential of Lactobacillus sp. and Pediococcus sp. strains isolated from Togolese traditional Zea mays fermented food
Article Information
Banfitebiyi Gambogou1,3, Essodolom Taale2*, Kokou Anani3, Messanh Kangni-Dossou3, Damintoti Simplice Karou3, Yaovi Ameyapoh3
1Institut Togolais de Recherche Agronomique (ITRA), Direction des Laboratoires, Togo
2Laboratoire des Sciences Agronomiques et Biologiques Appliquées (LaSABA), Institut Supérieur des Métiers de l’Agriculture (ISMA), Université de Kara, Togo
3Laboratoire de Microbiologie et de Contrôle qualité des Denrées Alimentaire, Université de Lomé, Togo
*Corresponding author: Essodolom Taale, Laboratoire des Sciences Agronomiques et Biologiques Appliquées (LaSABA), Institut Supérieur des Métiers de l’Agriculture (ISMA), Université de Kara, Togo
Received: 27 July 2023; Accepted: 16 August 2023; Published: 16 October 2023
Citation: Banfitebiyi Gambogou, Essodolom Taale, Kokou Anani, Messanh Kangni-Dossou, Damintoti Simplice Karou, Yaovi Ameyapoh. Microbiological analysis and assessment of the Biotechnological potential of Lactobacillus sp. and Pediococcus sp. strains isolated from Togolese traditional Zea mays fermented food. Journal of Food Science and Nutrition Research. 6 (2023): 155-164.
View / Download Pdf Share at FacebookAbstract
A majority of traditional fermented products consumed in Africa are processed by natural fermentation of cereals, tuber, fruits, or fish, and are particularly used as weaning foods for infants and dietary staples for adults. The aim of this study is to evaluate the quality of “Akpan”, “Kom” and “Emakumè” by using standard microbiological methods. We used Biochemistry and molecular biology methods to identify presumptive lactic acid bacteria (LAB) strains and their biotechnological proprieties screened. The count of germs in CFU/g of Total Mesophilic Aerobic Microorganisms; Staphylococcus aureus, Total coliforms, fungi and LAB shows that the analysed samples are good for consumption (AFNOR, 1996). LAB strains were identified as Lactobacillus plantarum, Lactobacillus fermentum, Pediococcus pentosaceus and Pediococcus acidilactici. Lactobacillus fermentum shown better acidification and antioxidant activity. Pediococcus pentosaceus produced high lactic acid and showed better proteolytic activity. Pediococcus acidilactici produced high exopolysaccharide while the best amylolytic activity was observed by Lactobacillus plantarum and Lactobacillus fermentum. The mix culture of these strains into “Emakoumè”, “Akpan” and “Kom” guaranties better microbiological and nutritional quality of these feeds.
Keywords
Emakumé, Akpan, Kom, Lactobacillus, Pediococcus, Cereals, Dough
Emakum articles; Akpan articles; Kom articles; Lactobacillus articles; Pediococcus articles; Cereals articles; Dough articles
Article Details
Introduction
A majority of traditional fermented foods consumed in Africa are processed by natural fermentation of cereals, tuber, fruits, or fish, and are particularly used as weaning foods for infants and dietary staples for adults [1,2]. As reported by Blandino [3] and Soro-Yao [4] a wide range of cereal-based fermented foods exist in Africa and among them, we can find Ogi and Mawè in Benin, Kenkey in Ghana, Injera in Ethiopia, Poto-poto in Congo, Ogi and Kunu-zaaki in Nigeria, Uji and Togwa in Tanzania, Kisra in Sudan. In Togo, “Agbelima”, “Eblima”, and “Epoma” produced by spontaneous traditional fermentation of Manihot esculenta Crantz, Zea mays, and Sorghum bicolor respectively are widely consumed by the population from the south and other parts of the country [5-7]. African fermented food is obtained by natural uncontrolled fermentation mediated by a mixed cultures of lactic acid bacteria (LAB), acetic acid bacteria, Bacillus, Enterobacteriaceae and fungi (molds and yeasts) species [8-11]. According to Taale et al. [12], Taale et al. [13] and Chen [14]; lactic acid fermentation has many advantages: reduction of the risk of growth of pathogenic microorganisms by acidification of the medium and the production of bacteriocins, the degradation of some anti-nutritional factors (phytates, α-galactosides), the development of specific organoleptic properties by synthesis of organic acids and aroma. Even Kogno and his team [5-7] studied microbes colonizing “Akpan”, “Kom” and “Emakumè”, we still have few data on the types of microbes and their biotechnological applications. Togolese foods microbiology and biotechnology researchers need to have data on acidification, antioxidant, proteolytic, amylolytic, antimicrobial, and antifungal activity; exopolysaccharides, protease, and amylase production of involved microbes in order to propose cultures starters and /or with probiotic properties with a final objective to stabilize the quality of our local food and increase their attractiveness. This study aimed to evaluate the sanitary and nutritional aspect of local cereal-based fermented foods such as “Akpan”, “Kom” and “Emakumè”. Especially the work done was to (i) evaluate the microbiological quality of “Akpan”, “Kom” and “Emakumè” produced in maritime region of Togo by searching the density of Total coliforms, Total Mesophilic Aerobic Microorganisms, Staphylococcus aureus, Lactic Acid Bacteria (LAB), mold and yeasts; and (ii) characterize Lactic Acid Bacteria strains isolated for application as the starter cultures and probiotics.
Materials and Method
Sampling and microbiological analyses
We collected six samples of fermented dough (“Akpan”, “Kom”, and “Emakumè”) from street food sellers in Lomé, capital of Togo. They were put in Stomacher bags and were kept on ice box and quickly transported to the Laboratoire de Microbiologie et de Contrôle qualité des Denrées Alimentaire (LaMICODA, Université de Lomé). Research of total mesophilic aerobic microorganisms, total coliforms, S. aureus, yeasts, molds, and LAB (Table 1) were the microbiological analysis performed. Ten grams of fermented maize dough was homogenized with 90 mL of sterile peptone water solution (peptone 0.1% and NaCl 0.85%) (ISO 6887-1:2017). From this solution, decimal dilutions have been performed for seeding on suitable media depending on the germs. For enumeration of Total mesophilic aerobic microorganisms; 1mL of each decimal dilution chosen has been inoculated into Petri dish and 15 mL of the PCA medium have been added and mixed. After solidification of the medium, petri dish were incubated at 30°C for 72 hours (NF V 08-051:1999). Total coliforms and S. aureus were counted respectively on VRBL (violet Red Bile Lactose) and Baird Parker respectively. Enumeration of total yeasts and molds: 0.1 mL of each dilution was spread on the surface of Sabouraud dextrose agar with chloramphenicol 5% (SD+C) medium. Then, the petri dish were incubated at 25°C for five days (ISO 21527-2:2008).
|
Microorganisms |
Medium |
Incubation temperature (°C) |
Incubation time (h) |
Norms references |
|
Total Mesophilic Aerobic Microorganisms |
PCA |
30 |
72 |
NF V 08-051:1999 |
|
Total Coliform |
VRBL |
37 |
24 |
NF ISO 4832:1991 |
|
Staphylococcus aureus |
BP |
37 |
24 |
NF EN ISO 6888-1:1999 |
|
Yeasts and Molds |
SD+C |
30 |
72 |
ISO 21527-2:2008 |
|
Lactic Acid Bacteria (LAB) |
MRS |
30 |
72 |
ISO 15214:1998 |
BP: Baird Parker; PCA: Plate Count Agar; VRBL: Violet Red Bile Lactose; SD+C: Sabouraud Dextrose + Chloramphenicol; MRS: Man Rogosa Sharp
Table 1: Referenced methods used for microbial assessment
Isolation and phenotypical identification of lactic acid bacteria
The isolation of lactic acid bacteria (table 1) was carried out by enriching 10 g of each sample into 90 mL of modified-MRS (mMRS) broth (1% maltose, 1% lactose, and 10% yeast extract). Cultures were incubated at 30 °C for 3 days. Series of decimal dilutions of each resulting culture were inoculated onto MRS agar supplemented with 0.0025% of bromocresol green (MP Biomedicals) and 0.01% cycloheximide (MP Biomedicals) and incubated under anaerobic conditions at 30 °C for 48-72 h. Acid-producing bacteria characterized by a yellow zone around each colony were picked and purified on MRS agar. The identification of lactic acid bacteria isolates at the genus stage was carried out using a morphological and physiological study. Gram stain, presence of the spores, catalase, growth at different temperatures (10, 30, 40, 45 and 50°C), at different pHs (3.5, 6.5 and 9.6), NaCl (2, 3, 4, 6.2 and 10%), fermentation and production of gas (CO2) and acids from the carbohydrates were performed. Lactobacillus and Pediococcus strains presenting the best performances were selected for further studies.
Molecular identification of Lactobacillus and Pediococcus strains
For the identification of 16S rRNA sequences, bacterial genomic DNA was isolated using the phenol extraction method. The 16S rRNA genes were amplified using polymerase chain reaction (PCR) with the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R and (5′-TACGGTYACCTTGTTGTTACGACTT-3′) [15]. The conditions of amplification were : 94 °C for 3 min followed by 35 cycles at 94 °C for 45s, 50 °C for 1 min, and 72 °C for 2 min, with a final 5 min extension at 72 °C. After amplification, the PCR products were purified using Wizard® SV Gel and PCR Clean-Up System (Promega, USA) by dissolving the gel slice and processing PCR products purification (binding of DNA, washing and elution) [16]. The purified fragment were sequenced with the ABI prism™Bigdye™ Terminator Cycle sequencing ready reaction kit in an automated ABI 3730XL capillary DNAr (Applied Biosystems, Foster City, CA, USA). The sequences obtained were trimmed to eliminate the ambiguous bases and bases with poor quality. Sequence identification of the closest relative taxa of the strains was achieved using the BLAST analysis tool [17] in the GenBank DNA database (www.ncbi.nih.gov). Phylogenetic analysis of 16S rRNA gene sequences was conducted with MEGA-X software. The phylogenetic tree was constructed by using the neighbor-joining method. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 11 [18].
Selective LAB strains biotechnological application
This aim to study some properties of selected LAB strains for their use as starter culture and probiotic.
Acidification activity
A sterile maize flour extract (SMFE) was prepared: 200 g of flour was suspended in 1L distilled water and sterilized at 121°C/20 min. After precipitation, the flour was removed. The supernatant was used as liquid broth in subsequent experiments. Overnight LAB cultures on MRS broth, were centrifuged (5000g/5min) and washed three times with sterile distilled water. LAB cells (109 CFU/mL) were inoculated in 50 mL SMFE solution (1% v/v); and incubated at 30°C. The pH measurements were taken initially, after 24 h, 48 h, and 72 h of inoculation. Strains that rapidly and greatly decreased SMFE pH were analyzed for their ability to produce lactic and acetic acids, following 24 h, 48 h, and 72 h of fermentation. The configuration and quantity of lactic acid produced were determined by an enzymatic method involving lactate dehydrogenase (LDH, Boehringer Mannheim). The assays were carried out on 48-hour culture supernatants in MRS broth.
Screening for Exopolysaccharides production
The measurement of the amount of exopolysaccharides was done by the colorimetric test developed by Dubois [19]. 0.4ml supernatant of an overnight culture of LAB strain was mixed with 0.2 mL of phenol solution (5%) and 0.5 mL of concentrated sulphuric acid. After incubation in the dark for 30 min, the absorbance was measured at 490nm against a blank without LAB strain.
Screening for amylase production and amylolytic activity
The production of amylases is demonstrated by the hydrolysis test of wheat starch in an agar medium. The culture medium used is composed of 9 g/l of peptone casein, 9 g/l of yeast extract, 9 g/l of wheat starch and 14 g/l of agar. For the preparation of the medium, the wheat starch is placed in an aqueous solution and sterilized separately, it is added to the rest of the medium before being poured into the Petri dishes. A young colony is transplanted into two well separated points on the surface of the solidified medium. Petri dishes were incubated at 30°C for one week. At the end of the incubation period, the surface of the agars is sprayed with absolute ethanol. The appearance of a transparent halo around the colonies characterizes the production of amylases in the medium. The amylolytic activity of LAB was measured using the starch-iodine method as described by Bartkiene et al. [20]. To evaluate this activity, the SMFE was enriched with glucose 0.15% in order to enhance bacterial growth. The absorbance was measured at 670 nm using a spectrophotometer (Thermo Scientific Multiskan GO). One unit of α-amylase activity (1 AU) was defined as an amount of enzyme that catalyzes 1g soluble starch hydrolysis to dextrins in 10 min at 30 °C.
Proteolytic activity and protease production
The production of proteases is demonstrated by the test of the use of casein from skimmed milk as a source of carbon by microorganisms. The culture medium used consists of 9 g/l of peptone casein, 9g/l of yeast extract, 9g/l of skimmed milk and 14 g/l of agar. In the preparation of this medium, the skimmed milk is sterilized separately and then added to the other constituents before being poured into the dishes. A young 24-hour colony is transplanted into two very distinct points on the surface of the agars. The Petri dishes are then incubated at 30°C for one week. The production of proteases is evidenced by the appearance of a transparent halo around the colonies formed. Extracellular proteolytic activity of LAB strains inoculated for 72 h in SMFE was measured using Azocasein as substrate. Absorbance was measured at 440 nm against a blank containing only the Azocasein. Proteolytic activity was expressed as ΔDO440 × h−1 × mg−1 dry weight.
Antioxidant activity
The antioxidant activity was measured using DPPH according to the method described by Lin and Chang. LAB were harvested by centrifugation at 4400g/10min after culture of 18h incubation at 37 °C. The test was performed on the intact bacterial cells. Cells were washed three times with phosphate buffer solution (PBS 0.85% NaCl, 2.68 mM KCl, 10 mM Na2HPO4, and 1.76 mM KH2PO4, pH 7.4) and resuspended in the same buffer. The sample (0.8 mL) was reacted with 1 mL DPPH solution (0.2 mM in 0.5% ethanol). The control was prepared by using ultra-pure water. The tubes were incubated for 30 min in the dark. Absorbance was measured by spectrophotometer (Thermo Scientific Multiskan GO) at 517 nm in triplicate. The results were expressed as a percentage of the antiradical activity: Antioxidant activity (%) = (A517control−A517sample)/A517 control x 100
Antimicrobial activity
All isolate strains of lactic acid bacteria were grown in MRS broths, which showed optimal antagonistic effect under the anaerobic condition at 37°C for 48h. Cell-free supernatants were collected by centrifugation at 1000g/15 min and by aseptic filtration (0.45 μm) to appreciate antibacterial activity. E. coli ATCC25922, S. aureus ATCC25923, Aspergillus niger ATCC1015, Candida albicans ATCC14053, Listeria monocytogenes ATCC19115, and S. typhi ATCC14028 were used. Subculture was done in Muller Hinton broth (OXOID, UK) at 37°C. After adjusting the inoculum to a 0.5 McFarlane standard; a sterile cotton swab was dipped into the inoculum and rotated against the wall of the tube above the liquid to remove excess inoculum. The entire surface of Mueller Hinton Agar was swabbed three times, rotating the plates approximately 60° between streaks to ensure even distribution. The inoculated Petri dishes were allowed to stand for at least 3 minutes but not more than 15 minutes before punching the 5mm holes (four holes per petri dish) in the surface of the inoculated agar. Then a volume of 50 μl of cell-free supernatants was added to the wells. The Petri dishes were incubated for 18 to 24 hours at 37 °C within 15 minutes following the application of the cell-free supernatant extracts. The diameters of the zones of inhibition were measured after overnight incubation.
Results
The figure 1 shows the samples used in this study.
Emakumè: Fermented dough cooked maize; Kom: Fermented dough baked and packaged maize; Akpan: Fermented dough baked and packaged maize.
Microbial assessment
The count of germs in CFU/ml shows that the germs sought in our sample analyzed do not exceed the number of germs set as the criteria of satisfaction (m) or acceptability (M) of the quality (Table 2).
|
Sample |
TMAM |
TC |
S. aureus |
Mold |
Yeast |
LAB |
|
Fermented dough cooked Zea mays "Emakumè" |
2.05 |
< 1 |
< 1 |
< 1 |
1.82 |
5.45 |
|
Fermented dough baked and packaged Zea mays "Kom" |
2.37 |
< 1 |
< 1 |
< 1 |
2.33 |
5.21 |
|
Fermented dough baked and packaged Zea mays "Akpan" |
2.31 |
< 1 |
< 1 |
< 1 |
2.54 |
4.45 |
|
AFNOR criteria, 1996 (m – M) |
5-6 |
2-3 |
1-2 |
3-4 |
3-4 |
TC: Total coliforms; TMAM: Total Mesophilic Aerobic Microorganisms; Staph: Staphylococcus aureus, LAB: Lactic Acid Bacteria, m: number of microorganisms that show quality satisfaction of the product; M: number of microorganisms that shown acceptance quality of the product.
Table 2: Count of germs sought in log (UFC/ml)
Morphological identification and biochemical property
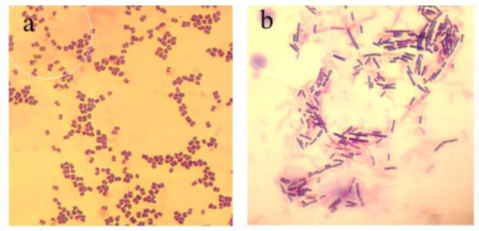
The morphological, biochemical and phylogenetic identification are represented respectively by the Gram stain test (Figure2), biochemical test (Table 3) and the dendrogram (Figure 3). Figure 2 shows that the Pediococcus are Gram-positive cocci grouped in a tetrad (figure 2a) and the Lactobacillus represented in figure2b shows that they are Gram-positive bacilli. Biochemical tests identified Lactobacillus fermentum, Lactobacillus plantarum, Pediococcus acidilactici and Pediococcus pentosaceus. Molecular identification was confirmed by phylogenetic analysis with the construction of the phylogenetic tree shown in figure 4.
VP: voges-proskauer; ADH: Arginine dihydrolase; Ma: Mannitol; CIT: citrate GL: Glucose; F: Fructose; G: Galactose; X: Xylose; A: Arabinose; R: Ribose; S: Sorbose; Me: Mélézitose; Sa: Saccharose; Man: Mannose; M: Maltose; D: Dextrose; L: Levulose; +: Positive test; -: Negative test.
Table 3: Biochemistry characteristics of Lactobacillus sp. and Pediococcus sp. isolated from Togolese fermented maize dough
Molecular identification of selective LAB strains
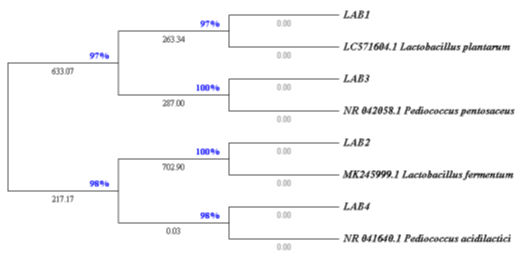
Phylogenetic and molecular evolutionary analyses of obtained sequences using MEGA version 11 showed that LAB1, LAB2, LAB3 and LAB4 are respectively close to Lactobacillus plantarum, Lb. fermentum, Pediococcus pentosaceus and Ped. acidilactici (figure 3).
The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree is shown (next to the branches). The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). This analysis involved 8 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1569 positions in the final dataset. Evolutionary analyses were conducted in MEGA11.
Total Biotechnologies proprieties
Acidification activities and organic acid production
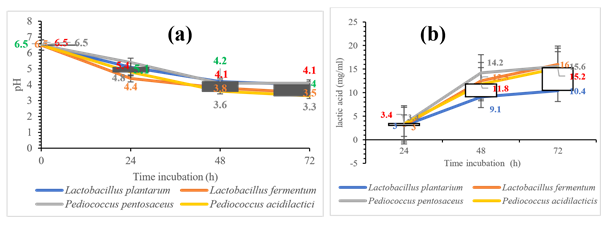
All selected LAB strains produce the same stereoisomer (+) of lactic acid. Only Lactobacillus fermentum produces (DL) lactate after 48 hours of fermentation (Table 4). Pediococcus pentosaceus produced the most lactic acid (14.20mg/ml) and lower pH (pH3.6) after 48h fermentation. Lactobacillus fermentum produced more lactic acid after 72 hours of fermentation (figure 4).
|
LAB strains |
Isomer |
Lactic acid (mg/ml/108 CFU) |
pH at 48h |
|
Lactobacillus plantarium |
L(+) |
9.1 |
4,2 |
|
Lactobacillus fermentum |
DL (+) |
12,5 |
3,8 |
|
Pediococcus pentosaceus |
L (+) |
14,20 |
3.6 |
|
Pediococcus acidilacticis |
L (+) |
11,80 |
4.1 |
Table 4: Profils de production de l’acide lactique par les bactéries étudiées
Exopolysaccharide production and proteolytic, amylolytic, and antioxidant activities
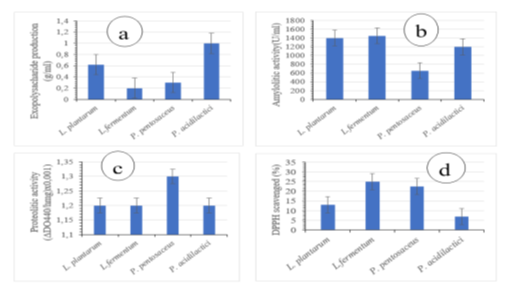
The ability of microorganisms to hydrolyze the main macromolecules (proteins, polysaccharides, lipids, etc.) is an interesting property in the selection of probiotic bacteria. Proteolytic, amylolytic, exopolysaccharide production, and antioxidant activity exhibited by Lb. fermentum, Lb. plantarum, Ped. acidilactici and Ped. acidilactici are shown in figure 5. Pediococcus acidilactici showed better production of exopolysaccharide (1g/ml) followed by Lactobacillus plantarum (0.6g/ml) (figure 5a). The amylolytic activity was greater with strains of Lactobacillus plantarum and Lactobacillus fermentum (1400 U/ml) followed by Pediococcus acidilactici (1200 U/ml) (Figure 5b). The Pediococcus pentosaceus strain is the strain that showed high proteolytic activity (1.3UI) (Figure 5c). Antioxidant activity was higher in Lactobacillus fermentum strains (25%) and in Pediococcus pentosaceus strains (22%) (Figure 5d).
|
Extract of Lb. plantarum |
Extract of Lb. fermentum |
Extract of Ped. acidilactici |
Extract of Ped. pentosaceus |
|
|
Escherichia coli ATCC25922 |
++ |
++ |
+++ |
+++ |
|
Salmonella typhi ATCC14028 |
++ |
++ |
++ |
++ |
|
Listeria monocytogenes ATCC19115 |
- |
+ |
++ |
+++ |
|
Staphylococcus aureus ATCC25923 |
+ |
+++ |
+++ |
+++ |
|
Aspergillus niger ATCC1015 |
++ |
+ |
++ |
++ |
|
Candida albicans ATCC14053 |
++ |
- |
++ |
++ |
+++: diameter of the inhibition zone (Ø) ≥ 15mm; ++: diameter of the inhibition zone (Ø) between 10 and 15 mm; +: diameter of the inhibition zone (Ø) between 5 and 10 mm; -: diameter of the inhibition zone (Ø) ≤ 5; Lb. : Lactobacillus; Ped.: Pediococcus
Table 5: Antimicrobial activity of bacteriocins extracts of Lactobacillus sp. and Pediococcus sp. strains
Discussion
The traditional fermentation of cereal products widely practiced in Africa usually involves the spontaneous development of different LAB. The final microbiological status of the product is influenced by the raw material and the process method [10]. The usual chance inoculation in the fermentation of African fermented foods largely contributes to the short shelf life of the products [21,22]. To overcome the perennial problem of short shelf-life often associated with African fermented foods and increase micronutrients, the use of starters with an ability to produce micronutrients and antimicrobials (including bacteriocins as potential biopreservatives) against food-borne pathogens should be encouraged in the preparation of our indigenous fermented foods. The quality of the food studied is of satisfactory quality and complies with the acceptability criteria, however, the presence of total coliforms (Table 2) which are indicators of fecal contamination may be due to labor or equipment during processing product packaging [23]. In addition to the presence of TMAM, the presence of Yeast and especially the majority presence of LAB would be at the origin of the fermentation, texture, taste, and even the nutritional characteristics of the products. In several studies, the authors have shown that yeasts and LAB are used both to activate fermentation, act on particular aromas, and also to enhance certain taste characteristics. In the case of co-fermentation, different starters may allowed a reduction in the duration of lactic fermentation (9 hours instead of 12-16 hours) and of lactic + alcoholic fermentation (12 hours instead of 21-48 hours). Thus, to determine the technological interest of the LAB in “Akpan”, “Kom” and “Emakumè”, our study firstly based on morphological (Figure 2) and biochemical (Table 3) properties identified two groups of LAB: Pediococcus (Ped. pentosaceus and Ped. acidilactici) and Lactobacillus (Lb. plantarium and Lb. fermentum). Pediococcus sp. are Gram-positive cocci grouped into tetrads whereas Lactobacillus sp. are Gram-positive bacilli grouped by two or alone. Secondly, the identity of the four bacterial strains is confirmed by 16S rRNA phylogenetic analysis (Figure 3). Previous study done by Kogno et al.[5-7] showed that Pediococcus sp. and Lactobacillus sp. strains predominate in fermented cereal products in Togo. The evaluation of the technological properties, of the selected LABs for better production of nutrient-rich and long shelf-life foods revealed that the four strains have the ability to ferment rapidly (under 24 hours) cornflour and produce lactic acid, as is the case with Lactobacillus plantarum (ΔpH= 1.4, ie 1.02 mg. Ml-1 of lactic acid for 108 CFU about 48 hoursat pH4.2). Lb. fermentum is a heterofermentative (producing D and L isomer lactic acid) LAB with important characteristics such as low time use of fermentation (ΔpH= 2.1, i.e. 2.17 mg/ml of lactic acid in 48h at pH 3.8), high acid lactic producer in 72h (16 mg/ml). According to Holzapfel et al. [24], Pediococcus sp. is a type of LAB, of which the most prominent characteristics are acid tolerance, and possessing a strictly fermentative (homofermentative), and optionally anaerobic metabolism, with lactic acid as the major metabolic end-product. Our study confirmed that Ped. acidilactici and Ped. pentosaceus are a homofermentative bacteria, with acid tolerance, and optional anaerobic metabolism with respectively ΔpH= 1.7 and 1.1 in 24h (Table 4 and Figure 4). As previously described by Papalexandratou et al. [25] Lactobacillus sp. and Pediococcus sp. are broad ecological distribution, including fermented food, plants, animal/human gut, wine, etc. This broad ecological distribution reflects metabolic flexibility that has fuelled the widespread application of the species in the food and health industry. For example, strains of Ped. acidilactici are employed as acid-producing starter cultures in milk fermentation, as adjunct cultures to accelerate or intensify flavour development in bacterial ripened cheeses, and as probiotics to enhance human or animal health [26-28]. Another example is the use of Lb. plantarum in the fermentation of dairy products such as cheese, kefir, sauerkraut, fermented meat products, fermented vegetables, and beverages [29-32]. Amylolytic, proteolytic activities, the production of exopolysaccharides (EPS) and antioxidant activities are other technological properties searched amount our four LAB because amylolytic and proteolytic activities have the respective interest of degrading starch into simple sugars and proteins into amino acids. Antioxidant activity and EPS will neutralize or reduce the effects of free radicals in the body and texture of food products respectively. Thus, they are frequently used by the food industry to improve the textural and organoleptic properties of fermented milk and different types of cheese [33-35]. They can also be used to product low-fat dairy products allowing the design of healthy products with acceptable sensory characteristics [36,37]. The results obtained shows that the four LAB strains produce EPS: 1g/ml of EPS for Ped. acidilactici; 0.6g/ml for Lb. planetarium; 0.3g/ml for Ped. pentosaceus and 0.2g/ml for Lb. fermentum. Pediococcus. pentosaceus, Ped. acidilactici, Lb. plantarium and Lb. fermentum possess amylolytic (620U/ml, 1200U/ml, 1400U/ml and 1400U/ml respectively); proteolytic (1.3U, 1.2U, 1.2U and 1.2U respectively) and antioxidant (13%, 7%, 23% and 25% respectively) properties. Similar studies shown that some strains of Ped. acidilactici are able to improve antioxidant status and promote nutrient digestibility [38,39]. Moreover, it had been reported that Ped. pentosaceus had probiotic functions including anti-inflammation, anti-cancer, antioxidant, detoxification, and cholesterol-lowering [40-43]. Bacteriocins produced by Ped. acidilactici and Ped. pentosaceus inhibit the growth of all reference strains used in this study (Table 5); while those of Lb. plantarium and Lb. fermentum inhibit the growth Escherichia coli ATCC 25922, Salmonella typhi ATCC 14028, Aspergillus niger ATCC 1015 and Candida albicans ATCC 14053. Several studies related that the health claims of Lb. plantarum permitted the development of different probiotic formulations and its antibacterial properties are used for food safety as in the biopreservation technology [38,44-46]. Bacteriocins produced by Pediococcus acidilactici in this study could be assimilate to pediocin because of his ability to produce antimicrobial agents [47], such as pediocin [48,49]. Indeed, pediocin is used for the preservation and safety assurance of food and beverage during storage by inhibiting pathogenic and spoilage bacteria [48], such as Listeria monocytogenes [50-52]. Ha, Rossi and Lathrop [44,50] demonstrated that the strains of Pediococcus acidilactici. prevent the colonization of pathogens such as Shigella spp., Salmonella spp., Clostridium difficile, and Escherichia coli in the small intestine. To make better use of LAB in the food industry, it is known that the diversity of strains may contribute to dissimilar fermentation products with various probiotic functions. As confirmed by experiments, strains not only could be applied as biopreservatives for foods, crops, and livestock but also could be used to accelerate beneficial physical activities. Adding freshness and flavour to food, LAB strains can be used in the field of food research to promote the development of food engineering from chemical treatment to biological treatment.
Conclusion
The “Emakoumè”, “Akpan” and “Kom” quality evaluated showed the microbiological quality of these feeds. For biotechnological proprieties assess, all strains studied have some interesting properties such as fermentation of dough, amylolytic, proteolytic and antioxidant activities, lactic acid and EPS production, and antimicrobial activities. The productivities obtained in the cultures in flasks for all the bacteria, suggest possibilities for optimization through controlled cultures in fermenters. The production of L (+) lactic acid by the strains studied is in favor of their use as probiotic bacteria. The amount of lactic acid and EPS produced suggest that production can be optimized, particularly with adequate culture conditions. The acidity tolerance, as well as the production of amylases and proteases in its bacterial isolates, are also encouraging for their use as probiotic bacteria for certain types of applications. However, we are continuing research for an application of its properties in fermented foods such as starters.
References
- Vieira-Dalodé G, Jespersen L, Hounhouigan J, et al. Lactic acid bacteria and yeasts associated with gowé production from sorghum in Bénin. Journal of Applied Microbiology 103 (2007): 342-349.
- Achi OK, Ukwuru M. Cereal-based fermented foods of Africa as functional foods. International Journal of Microbiology and Application 2 (2015): 71-83.
- Blandino A, Al-Aseeri ME, Pandiella SS, et al. Cereal-based fermented foods and beverages. Food research international 36 (2003): 527-543.
- Soro-Yao AA, Brou K, Amani G, et al. The use of lactic acid bacteria starter cultures during the processing of fermented cereal-based foods in West Africa: A Review. Tropical life sciences research 25 (2014): 81-100.
- Kogno E, Anani K, Djeri B, et al. Microbiological profile of agbelima, eblima and epoma, three traditional fermented products in Togo. International Journal of Applied Biology and Pharmaceutical Technology 8 (2017): 31-38.
- Kogno E, Djeri B, Anani K, et al. Screening of fermentative microorganisms with amylolytic potentials involved in the fermentation of traditional fermented products in Togo. Int. J. Curr. Microbiol. App. Sci 6 (2017): 1067-1077.
- Kogno E, Soncy K, Taale E, et al. Molecular characterization of lactic acid bacteria involved in Togolese traditional fermented cereal foods. International Journal of Recent Advances in Multidisciplinary Research 4 (2017): 2308-2312.
- Coulin P, Farah Z, Assanvo J, et al. Characterisation of the microflora of attiéké, a fermented cassava product, during traditional small-scale preparation. International journal of food microbiology 106 (2006): 131-136.
- Ouattara HG, Reverchon S, Niamke SL, et al. Molecular identification and pectate lyase production by Bacillus strains involved in cocoa fermentation. Food Microbiology 28 (2011): 1-8.
- Alfred KK, Jean Paul BKM, Théodore DND, et al. Microbiological and chemical hazards of commercial attieke (a fermented cassava product) produced in the south of Côte d’Ivoire. Food Quality and Safety 3 (2019): 187-190.
- Flibert G, Siourime SN, Hamidou C, et al. Lactic acid bacteria and yeasts associated with cassava fermentation to attieke in burkina faso and their technological properties. American Journal of Food Science and Technology 9 (2021): 173-184.
- Taale E, Savadogo A, Zongo C, et al. Bioactive molecules from bacteria strains: case of bacteriocins producing bacteria isolated from foods. Current Research in Microbiology and Biotechnology 1 (2013): 80-88.
- Taale E, Djery B, Sina H, et al. Indigenous food and food products of West Africa: Employed microorganisms and their antimicrobial and antifungal activities non ribosomal peptide synthetase view project fight against diabetes in benin view project. In: Verma DK, Patel AR, Srivastav PP, Mohapatra B, Niamah AK. (Eds.), Microbiology for food and health: Technological development and advances. Apple Academic Press, Inc., Canada (2020): 149-194.
- Chen P. Lactic acid bacteria in fermented food. Advances in Probiotics. Elsevier (2021): 397-416.
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt, E, Goodfellow, M.(eds.), Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Son, New York (1991): 115-136.
- Taale E, Sina H, Zongo C, et al. Searching for bacteriocin pln loci from lactobacillus spp. Isolated from fermented food in burkina faso by molecular methods. Article in International Journal of Applied Biology and Pharmaceutical Technology 7 (2016): 86-94.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30 (2013): 2725-2729.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution 35 (2018): 15-47.
- Dubois M, Gilles KA, Hamilton JK, et al. Colorimetric Method for Determination of Sugars and Related Substances. Analytical Chemistry 28 (1956): 350-356.
- Bartkiene E, Schleining G, Rekstyte T, et al. Influence of the addition of lupin sourdough with different lactobacilli on dough properties and bread quality. International journal of food science & technology 48 (2013): 2613-2620.
- Houngbédji M, Johansen P, Padonou SW, et al. Occurrence of lactic acid bacteria and yeasts at species and strain level during spontaneous fermentation of mawè, a cereal dough produced in West Africa. Food Microbiology 76 (2018): 267-278.
- Obafemi YD, Oranusi SU, Ajanaku KO, et al. African fermented foods: overview, emerging benefits, and novel approaches to microbiome profiling. Npj Science of Food 2022 6 (2022): 1-9.
- Anyogu A, Olukorede A, Anumudu C, et al. Microorganisms and food safety risks associated with indigenous fermented foods from Africa. Food Control 129 (2021): 108227.
- Holzapfel WH, Franz C, Ludwig W, et al The genera pediococcus and tetragenococcus. Prokaryotes 4 (2006): 229-266.
- Papalexandratou Z, Camu N, Falony G, et al. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiology 28 (2011): 964-973.
- Ruiz-Moyano S, Martín A, Benito MJ, et al. Application of Lactobacillus fermentum HL57 and Pediococcus acidilactici SP979 as potential probiotics in the manufacture of traditional Iberian dry-fermented sausages. Food Microbiology 28 (2011): 839-847.
- Abbasiliasi S, Tan JS, Bashokouh F, et al. In vitro assessment of Pediococcus acidilactici Kp10 for its potential use in the food industry. BMC microbiology 17 (2017): 1-11.
- Pakroo S, Tarrah A, Bettin J, et al. Genomic and phenotypic evaluation of potential probiotic pediococcus strains with hypocholesterolemic effect isolated from traditional fermented food. Probiotics and Antimicrobial Proteins 14 (2022): 1042-1053.
- Liu YW, Liong MT, Tsai YC. New perspectives of Lactobacillus plantarum as a probiotic: The gut-heart-brain axis. Journal of Microbiology 56 (2018): 601-613.
- Cao P, Wu L, Wu Z, et al. Effects of oligosaccharides on the fermentation properties of Lactobacillus plantarum. Journal of Dairy Science 102 (2019): 2863-2872.
- Bartkiene E, Zokaityte E, Lele V, et al Combination of extrusion and fermentation with Lactobacillus plantarum and L. uvarum strains for improving the safety characteristics of wheat bran. Toxins 13 (2021): 163.
- Yuan S, Yang F, Yu H, et al. Biodegradation of the organophosphate dimethoate by Lactobacillus plantarum during milk fermentation. Food Chemistry 360 (2021): 130042.
- Zannini E, Waters DM, Coffey A, et al. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Applied Microbiology and Biotechnology 100 (2016): 1121-1135.
- Adesulu-Dahunsi AT, Jeyaram K, Sanni AI. Probiotic and technological properties of exopolysaccharide producing lactic acid bacteria isolated from cereal-based nigerian fermented food products. Food Control 92 (2018): 225-231.
- Ale EC, Rojas MF, Reinheimer JA, et al. Lactobacillus fermentum: Could EPS production ability be responsible for functional properties? Food microbiology 90 (2020): 103465.
- Kanamarlapudi SLR, Muddada S. Characterization of exopolysaccharide produced by Streptococcus thermophilus CC30. BioMed research international 12 (2017): 4201089.
- Domingos-Lopes MFP, Lamosa P, Stanton C, et al. Isolation and characterization of an exopolysaccharide-producing Leuconostoc citreum strain from artisanal cheese. Letters in applied microbiology 67 (2018): 570-578.
- Ali AMM, Gullo M, Rai AK, Bavisetty SCB. Bioconservation of iron and enhancement of antioxidant and antibacterial properties of chicken gizzard protein hydrolysate fermented by Pediococcus acidilactici ATTC 8042. Journal of the Science of Food and Agriculture 101 (2021): 2718-2726.
- Li H, Huang J, Wang Y, et al. Study on the nutritional characteristics and antioxidant activity of dealcoholized sequentially fermented apple juice with Saccharomyces cerevisiae and Lactobacillus plantarum fermentation. Food Chemistry 363 (2021): 130351.
- Ilavenil S, Vijayakumar M, Kim DH, et al. Assessment of probiotic, antifungal and cholesterol lowering properties of Pediococcus pentosaceus KCC-23 isolated from Italian ryegrass. Journal of the Science of Food and Agriculture 96 (2016): 593-601.
- Ayyash M, Abu-Jdayil B, Olaimat A, et al. Physicochemical, bioactive and rheological properties of an exopolysaccharide produced by a probiotic Pediococcus pentosaceus M41. Carbohydrate polymers 229 (2020): 115462.
- Jiang S, Cai L, Lv L, et al. Pediococcus pentosaceus, a future additive or probiotic candidate. Microbial cell factories 20 (2021): 1-14.
- Qi Y, Huang L, Zeng Y, et al. Pediococcus pentosaceus: Screening and Application as Probiotics in Food Processing. Frontiers in Microbiology 12 (2021).
- Ha EM. Escherichia coli-derived uracil increases the antibacterial activity and growth rate of Lactobacillus plantarum (2016).
- Wang L, Zhang H, Rehman MU, et al. Antibacterial activity of Lactobacillus plantarum isolated from Tibetan yaks. Microbial Pathogenesis 115 (2018): 293-298.
- Mao Y, Zhang X, Xu Z. Identification of antibacterial substances of Lactobacillus plantarum DY-6 for bacteriostatic action. Food Science & Nutrition 8 (2020): 2854-2863.
- Fugaban JII, Vazquez Bucheli JE, Park YJ, et al. Antimicrobial properties of Pediococcus acidilactici and Pediococcus pentosaceus isolated from silage. Journal of Applied Microbiology 132 (2022): 311-330.
- Reda FM. Antibacterial and anti-adhesive efficiency of Pediococcus acidilactici against foodborne biofilm producer Bacillus cereus attached on different food processing surfaces. Food science and biotechnology 28 (2019): 841-850.
- Lee DH, Kim BS, Kang SS. 2020. Bacteriocin of Pediococcus acidilactici HW01 inhibits biofilm formation and virulence factor production by Pseudomonas aeruginosa. Probiotics and antimicrobial proteins 12 (2020): 73-81.
- Rossi F, Lathrop A. Effects of Lactobacillus plantarum, Pediococcus acidilactici, and Pediococcus pentosaceus on the Growth of Listeria monocytogenes and Salmonella on Alfalfa Sprouts. Journal of Food Protection 82 (2019): 522-527.
- De Azevedo POS, Mendonça CMN, Seibert L, et al. Bacteriocin-like inhibitory substance of Pediococcus pentosaceus as a biopreservative for Listeria sp. control in ready-to-eat pork ham. Brazilian Journal of Microbiology 51 (2020): 949-956.
- Ramos B, Brandão TRS, Teixeira P, et al. Biopreservation approaches to reduce Listeria monocytogenes in fresh vegetables. Food Microbiology 85 (2020): 103282.
- Lin MY, Chang FJ. Antioxidative effect of intestinal bacteria bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Digestive Diseases and Sciences 45 (2000): 1617-1622.