Meningeal Melanocytoma - Focus on Molecular Aspects with 3 New Molecular Alterations: A Literature Review and Report of Two Cases
Article Information
Axel de BERNARDI1, Maureen BERNADACH1,3, Judith PASSILDAS-JAHANMOHAN1-4*, Julian BIAU1,2,6, Jean-Louis KEMENY7, Xavier DURANDO1-4
1Centre Jean Perrin, 58 rue Montalembert F-63011 Clermont-Ferrand, France
2Université Clermont Auvergne, Centre Jean Perrin, INSERM, U1240 Imagerie Moléculaire et Stratégies 3Théranostiques, 58 rue Montalembert F-63000 Clermont-Ferrand, France
4Division de Recherche Clinique, Délégation Recherche Clinique et Innovation, Centre Jean Perrin, 58 rue Montalembert F-63011 Clermont-Ferrand, France
5Centre d'Investigation Clinique, UMR501, 63011, Clermont-Ferrand, France
6Département de Radiothérapie, Centre Jean PERRIN, 63011, Clermont-Ferrand, France
7CHU de Clermont-Ferrand, Service d’Anatomie et de cytologie pathologique, 58 rue Montalembert 63000 Clermont-Ferrand, France
*Corresponding Author: Dr. Judith PASSILDAS-JAHANMOHAN, CLCC Centre Jean Perrin, Division de Recherche Clinique 58, rue Montalembert, BP 392, 63011 Clermont-Ferrand Cedex 1, France
Received: 22 April 2020; Accepted: 04 May 2020; Published: 03 July 2020
Citation: Axel de BERNARDI, Maureen BERNADACH, Judith PASSILDAS-JAHANMOHAN, Julian BIAU, Jean-Louis KEMENY, Xavier DURANDO. Meningeal Melanocytoma - Focus on Molecular Aspects with 3 New Molecular Alterations: A Literature Review and Report of Two Cases. Archives of Clinical and Medical Case Reports 4 (2020): 596-611.
View / Download Pdf Share at FacebookAbstract
Background: Meningeal melanocytoma (MM) is a locally aggressive, low-grade primary melanocytic tumor of the central nervous system (PMN-CNS). According to the literature, GNAQ and GNA11 mutations are relatively frequent. In contrast, BRAF and NRAS mutations are very rare. Other series have reported BAP1 mutation, monosomy 3 and gain of chromosome 6. In this paper, we discussed two cases of MM for whom a FoundationOne CDxTM assay was performed.
Results: Case report 1: A 48 year old man was diagnosed with intermediate-grade MM based on histological assessment, which is consistent with the clinical evolution. The FoundationOne CDxTM assay detected a SF3B1 mutation, usually found in 15% of uveal melanomas. This mutation is correlated with improved progression-free survival in uveal melanomas. Case report 2: A 39 year old women was diagnosed with MM. Her evolution wasn’t expected based on histology only; her history didn’t match with rather benign histological features microscopically. The FoundationOne CDxTM assay provided two interesting elements: first, the tumour was positive for CDKN2A p16INK4a M52fs*1 and p14ARF H66fs*106 mutations, most commonly found in cutaneous melanomas. Secondly, a GNA11 Q209L mutation, known to be a marker of aggressiveness in uveal melanoma, was detected.
Conclusion: The identification of molecular alterations in PMN-CNS, in addition to immunohistochemical analysis could provide a better understanding of the biological and clinical behaviour of these tumours, a better identification of melanocytic melanomas with poor prognosis and a high risk of relapse or metastasis, and the development of novel therapeutic options for these patients.
Keywords
Neuro-oncology; Meningeal melanocytoma; Genetic mutation; Case report
Neuro-oncology articles, Meningeal melanocytoma articles, Genetic mutation articles, Case report articles
Neuro-oncology articles Neuro-oncology Research articles Neuro-oncology review articles Neuro-oncology PubMed articles Neuro-oncology PubMed Central articles Neuro-oncology 2023 articles Neuro-oncology 2024 articles Neuro-oncology Scopus articles Neuro-oncology impact factor journals Neuro-oncology Scopus journals Neuro-oncology PubMed journals Neuro-oncology medical journals Neuro-oncology free journals Neuro-oncology best journals Neuro-oncology top journals Neuro-oncology free medical journals Neuro-oncology famous journals Neuro-oncology Google Scholar indexed journals oncology articles oncology Research articles oncology review articles oncology PubMed articles oncology PubMed Central articles oncology 2023 articles oncology 2024 articles oncology Scopus articles oncology impact factor journals oncology Scopus journals oncology PubMed journals oncology medical journals oncology free journals oncology best journals oncology top journals oncology free medical journals oncology famous journals oncology Google Scholar indexed journals Meningeal melanocytoma articles Meningeal melanocytoma Research articles Meningeal melanocytoma review articles Meningeal melanocytoma PubMed articles Meningeal melanocytoma PubMed Central articles Meningeal melanocytoma 2023 articles Meningeal melanocytoma 2024 articles Meningeal melanocytoma Scopus articles Meningeal melanocytoma impact factor journals Meningeal melanocytoma Scopus journals Meningeal melanocytoma PubMed journals Meningeal melanocytoma medical journals Meningeal melanocytoma free journals Meningeal melanocytoma best journals Meningeal melanocytoma top journals Meningeal melanocytoma free medical journals Meningeal melanocytoma famous journals Meningeal melanocytoma Google Scholar indexed journals melanocytoma articles melanocytoma Research articles melanocytoma review articles melanocytoma PubMed articles melanocytoma PubMed Central articles melanocytoma 2023 articles melanocytoma 2024 articles melanocytoma Scopus articles melanocytoma impact factor journals melanocytoma Scopus journals melanocytoma PubMed journals melanocytoma medical journals melanocytoma free journals melanocytoma best journals melanocytoma top journals melanocytoma free medical journals melanocytoma famous journals melanocytoma Google Scholar indexed journals Genetic mutation articles Genetic mutation Research articles Genetic mutation review articles Genetic mutation PubMed articles Genetic mutation PubMed Central articles Genetic mutation 2023 articles Genetic mutation 2024 articles Genetic mutation Scopus articles Genetic mutation impact factor journals Genetic mutation Scopus journals Genetic mutation PubMed journals Genetic mutation medical journals Genetic mutation free journals Genetic mutation best journals Genetic mutation top journals Genetic mutation free medical journals Genetic mutation famous journals Genetic mutation Google Scholar indexed journals treatment articles treatment Research articles treatment review articles treatment PubMed articles treatment PubMed Central articles treatment 2023 articles treatment 2024 articles treatment Scopus articles treatment impact factor journals treatment Scopus journals treatment PubMed journals treatment medical journals treatment free journals treatment best journals treatment top journals treatment free medical journals treatment famous journals treatment Google Scholar indexed journals CT articles CT Research articles CT review articles CT PubMed articles CT PubMed Central articles CT 2023 articles CT 2024 articles CT Scopus articles CT impact factor journals CT Scopus journals CT PubMed journals CT medical journals CT free journals CT best journals CT top journals CT free medical journals CT famous journals CT Google Scholar indexed journals Case report articles Case report Research articles Case report review articles Case report PubMed articles Case report PubMed Central articles Case report 2023 articles Case report 2024 articles Case report Scopus articles Case report impact factor journals Case report Scopus journals Case report PubMed journals Case report medical journals Case report free journals Case report best journals Case report top journals Case report free medical journals Case report famous journals Case report Google Scholar indexed journals kidney articles kidney Research articles kidney review articles kidney PubMed articles kidney PubMed Central articles kidney 2023 articles kidney 2024 articles kidney Scopus articles kidney impact factor journals kidney Scopus journals kidney PubMed journals kidney medical journals kidney free journals kidney best journals kidney top journals kidney free medical journals kidney famous journals kidney Google Scholar indexed journals Sequential therapy articles Sequential therapy Research articles Sequential therapy review articles Sequential therapy PubMed articles Sequential therapy PubMed Central articles Sequential therapy 2023 articles Sequential therapy 2024 articles Sequential therapy Scopus articles Sequential therapy impact factor journals Sequential therapy Scopus journals Sequential therapy PubMed journals Sequential therapy medical journals Sequential therapy free journals Sequential therapy best journals Sequential therapy top journals Sequential therapy free medical journals Sequential therapy famous journals Sequential therapy Google Scholar indexed journals
Article Details
Abbreviations:
CNS- central nervous system; MM: Meningeal melanocytoma; PMN-CNS- primary melanocytic tumor of the central nervous system; MRI- magnetic resonance imaging; UM- uveal melanoma; IHC- immunohistochemistry; Gry- Grays; NGS- next generation sequencing; TMB- tumor mutation burden; STIR- short inversion recovery; MMR- mismatch repair; MSI- microsatellite instability; pRB- retinoblastoma protein; FA- fanconi anemia; HRRD- homologous recombination repair deficiency
1. Introduction
According to the WHO 2016 classification, primary melanocytic tumors of the central nervous system (PMN-CNS) include meningeal melanocytoma (MM), meningeal melanoma, meningeal melanocytosis and meningeal melanomatosis. MM is a locally aggressive, low-grade PMN-CNS, arising from leptomeningeal melanocytes. These melanocytes derive from multipotent cells migration from the neural crest during embryogenesis [1] and are physiologically located at the highest concentrations in the upper cervical spinal cord and the ventrolateral surface of the medulla oblongata [2]. MM is a very rare disease (1 per 10 million per year), more frequent in females (1,5 : 1 ratio), with a peak incidence in the fifth decade and predilection for the spinal cord and posterior fossa [3-5]. Histologically, melanocytomas correspond to a very limited nodular mass of black or dark brown colour. Microscopic examination shows oval or fusiform cells, with highly pigmented cytoplasm, little cytonuclear atypia, tight nests, vasocentric fascicles and sheet-like arrangements [6] and low proliferative activity (zero to one mitosis per 10 high-power fields (HPFs) and Ki-67 <1-2%) [4]. Intermediate-grade melanocytic tumour shares with malignant melanoma common features of aggressiveness such as increased mitotic activity (1-3 mitoses per 10 HPFs and Ki-67 ranging from 1% to 4%) or microscopic invasion of central nervous system (CNS) structures, but lack cellular atypia found in high-grade lesions [7, 8].
Magnetic resonance imaging (MRI) is particularly interesting for diagnosis and extension assessment, due to the paramagnetic behaviour of the melanin contained in this tumour. It most often corresponds to a solitary extra cranial mass attached to the underlying dura, with a heterogeneous T1 hypersignal, less intense than fat, T2 hyposignal and a moderate increase after injection [9]. MM is often first referred to as meningioma on imaging reports despite different radiological features. The treatment of MM is based on complete surgical resection. Patients with complete surgical excision have good prognosis. In case of incomplete resection or recurrence, adjuvant radiation therapy is recommended and the risk of relapse rises up to 50% one year after surgery [10]. To date, there are no prognostic factors to identify tumours at risk of relapse. Genetic screening could help clinicians to better identify high-grade tumours with a high risk of recurrence. PMN-CNS genetic background remains limited, due to the rarity of these tumors.
According to the literature data, GNAQ and GNA11 mutations are relatively frequent. In contrast, BRAF and NRAS mutations are very rare[9]. Other series have reported BAP1 mutation [11], monosomy 3 and gain of chromosome 6 [12]. Early data show that PMN-CNS are genetically differents from cutaneous melanomas and closer to uveal melanomas (UM), although differences exist. Similar to UM, PMN-CNS frequently harbour activating GNAQ or GNA11 mutations [13], but the other mutations described in UM (BAP1, SF3B1 and EIF1AX) [14, 15] are rare in PMN-CNS [9, 11]. In a series of 19 melanocytic tumours of the CNS, Van de Nes et al. described 71% of GNAQ mutations and 12% of GNA11 mutations. In this cohort, the GNA11 mutated tumours were intermediate prognostic MM and relapsed. Twenty nine genes known to be frequently mutated in UM or cutaneous melanomas were analyzed [11]. Other recurrent mutations were not identified.
We present here the genetic sequencing results of 2 patients with MM, one of which presented an aggressive behaviour with leptomeningeal seeding. We used the FoundationOne® test, a next-generation sequencing (NGS) based assay, which identifies genomic alterations within hundreds of cancer-related genes.
1. Method of Sequencing
FoundationOne CDxTM is a NGS assay that allows in-vitro detection of genomic alterations and signatures in more than 300 cancer-related genes. DNA is extracted from formalin-fixed paraffin embedded tumour tissue samples. Whole-genome shotgun library construction and hybridization-based capture of DNA is then processed. Genomic variants are finally detected using custom software. FoundationOne CDxTM assay is routinely relevant in five solid malignant neoplasms: non-small cell lung cancer, breast cancer, melanoma, colorectal cancer and ovarian cancer. Results can help physicians to identify patients likely to benefit from targeted treatments. A list of clinical trials is provided for patients with potentially actionable alterations without therapeutic agent available. Tumour mutation profiling is achievable in all types of cancer.
2. Results
3.1 Case report 1
A 48 years old man consulted in November 2013 for a one-year history of cervicalgia and recent salivation increase appeared progressively. He had neither history of removed melanocytic tumours nor family history of cancer. MRI at baseline revealed a large retro-bulbar lesion with mass effect on medulla oblongata. This lesion was homogenously enhanced after gadolinium injection on T1-weighted imaging (Figure 1a) and hypointense on T2-weighted sequence (Figure 1b). Radiological assessment was compatible with meningioma despite atypical features. Patient underwent surgery to perform total tumour resection. The intraoperative aspect of the tumour, being fibrotic and moderately haemorrhagic was initially supporting the diagnostic of meningioma. Partial surgical resection was performed due to a significant risk of bleeding. Histopathologic finding revealed a highly cellular tumoral proliferation of meningotheliomatous cells (Figure 2a, Figure 2b) with low mitotic activity and a 10% proliferation index (Ki67) (Figure 2c). Immunohistochemistry (IHC) staining for HBM45 and Melan A were both positive in tumour cells cytoplasm (Figure 2d) and heterogenic for S100 protein (Figure 2e). Other melanoma IHC markers assessed were negative. The diagnosis of intermediate-grade MM was made according to the 2016 WHO classification of tumours of the CNS. Adjuvant radiotherapy was discussed in a neuro-oncology multi-disciplinary team, with a total dose of 50.4 Grays (Gy) (28 fractions of 1.8 Gy). Local tumour control was good for 3 years, and the patient got back to work with a good quality of life.
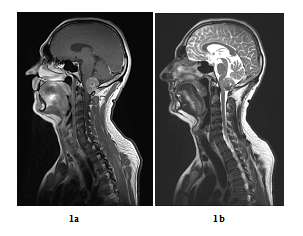
Figure 1: Baseline MRI scan in sagittal plane (case report 1): Gadolinium-enhanced T1-weighted MRI showing a large contrast-enhanced retro-bulbar lesion with mass effect on medulla oblongata (sagittal plane, October 2013) (a) and T2-weighted MRI showing a large hypointense retro-bulbar lesion with mass effect on medulla oblongata (sagittal plane, October 2013) (b).
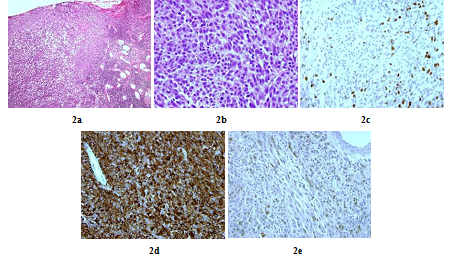
Figure 2: Histopathologic and immunohistochemistry assessment of tumor cells (case repot 1): Meningeal infiltration by a highly vascularized melanocytic tumoral proliferation (x10) (a); Meningeal melanocytoma cells with oval and fusiform nuclei (x40) (b); Tumour cells nuclear staining showed a 10% Ki-67 proliferation index (x40) (c); Intense tumour cells cytoplasmic staining for HMB45 and Melan A (x40) (d) and Heterogenic tumour cells cytoplasmic staining for S100 protein (x40) (e).
In August 2016, bilateral and symmetric lower extremities weakness and sphincter disorders appeared progressively. Whole spine MRI revealed a T2-T3 intramedullary nodular lesion compatible with MM metastasis and a progressive retro-bulbar lesion (Figure 3a, Figure 3b). T2-T3 decompressive radiotherapy was performed with a total dose of 30 Gy (10 fractions of 3 Gy). Follow-up MRI in October 2016 revealed a progressive retro-bulbar lesion but a stable intramedullary lesion. In November 2016, after 3 cycles of palliative Temozolomide-based regimen (200 mg/m2), both the retro-bulbar and the intramedullary lesions have progressed. Temozolomide was discontinued and surgical excision was indicated. However, surgery had to be postponed due to patient’s professional constraints. In February 2017, a C7-C8 paraesthesia of the left upper limb appeared progressively. Target lesions were stable on MRI. Surgical excision of the intramedullary lesion was performed.
Six months later, in October 2017, the intramedullary lesion increased leading to a second line of Fotemustin-based chemotherapy regimen (100 mg/m2). A FoundationOne CDxTM NGS based assay, evaluating more than 300 cancer-related genes, was performed. Two genomic alterations were described: GNAQ Q209P and SF3B1 R625H. Microsatellite status was stable and tumor mutation burden (TMB) was low (5 Muts /Mb).
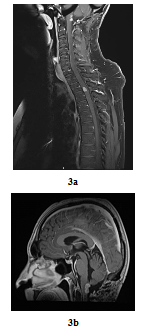
Figure 3: Post-treatment MRI scan in sagittal plan 3 years later (case repot 1): Contrast-enhanced T2-T3 intramedullary lesion on gadolinium-enhanced T1-weighted whole-spine MRI (sagittal plane, August 2016) (a) and Contrast-enhanced retro-bulbar lesion on gadolinium-enhanced T1-weighted MRI (sagittal plane, August 2016) (b).
3.2 Case report 2
A 66 years old woman was referred in 2014 by her general practitioner to the neurosurgery unit for a two years history of progressive left foot elevators deficiency and numbness of the two first left toes. She had neither history of removed melanocytic tumours nor family history of cancer. Physical examination was consistent with grade-2 left foot elevators deficiency, steppage gait at walk and left lower limb pyramidal syndrome. MRI at baseline revealed a contrast-enhanced T11-T12 intramedullary lesion without intracranial lesion. Electro-neuromyogram confirmed a left grade-3 L5 paresis. A tumour biopsy was performed in October 2015 after a T12-L1 laminectomy. Complete surgical resection was not possible due to a highly haemorrhagic tumour. Histological extemporaneous analysis revealed a meningeal melanocytic tumour. Post-operative MRI, performed in November 2015, showed the residual intramedullary lesion with contrast-enhancement on T1-weighted imaging (Figure 4a, Figure 4b), hypo-intense on short inversion recovery (STIR) T1-weighted imaging (Figure 4c). Histological final analysis revealed a highly cellular tumour (Figure 5a) with tumour cells harbouring elongated, fusiform nuclei and condensed nucleolus (Figure 5b). Two mitoses were spotted. Melanic pigment was highly concentrated in tumour cells cytoplasm (Figure 5c). IHC staining was strongly positive for HMB45 and Melan A (Figure 5d, Figure 5e), finally confirming the diagnostic of MM. Initial extension assessment looking for distant locations was negative. Urinary and faecal incontinence rapidly appeared without saddle block anaesthesia. The patient was treated with IMRT spinal stereotactic radiosurgery with a total dose of 43.369 Gy (25 fractions of 1.735 Gy) in November 2015.
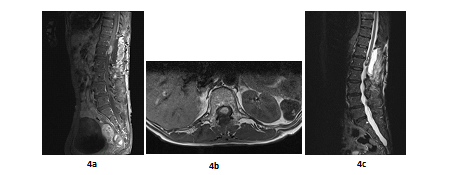
Figure 4: Post-operative MRI scan in sagittal plan (case report 2): T11-T12 contrast-enhanced intramedullary lesion on gadolinium-enhanced T1-weighted MRI (sagittal plane, November 2015) (a); Post-operative T11-T12 contrast-enhanced intramedullary lesion on gadolinium-enhanced T1-weighted MRI (axial plane, November 2015) (b) and Post-operative T11-T12 hypointense lesion on STIR T1-weighted MRI (sagittal plane, November 2015) (c) .
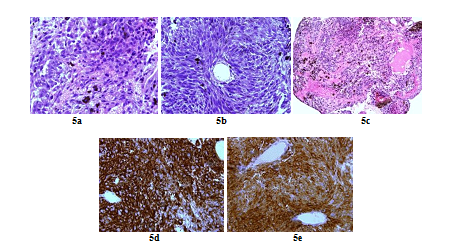
Figure 5: Histopathologic and immunohistochemistry assessment of tumor cells (case repot 2): Highly cellular meningeal melanocytoma (x 60) (a); Meningeal melanocytoma cells with elongated nuclei (x 40) (b); Highly pigmented meningeal melanocytoma (X 20) (c); Intense tumour cells cytoplasmic staining for HMB45 (x 60) (d) and Tumour cells cytoplasmic staining for Melan A (x 60) (e).
Six months later, in July 2016, the follow-up MRI showed progression. Total surgical resection was performed In October 2016 with stable disease for 2 years. In January 2018, the patient presented a cauda equina syndrome with progressive left L5 radiculopathy. In February 2018, cognitive impairment and global lower limbs weakness appeared. MRI revealed a meningeal contrast enhancement compatible with carcinomatous leptomeningitis. A FoundationOne CDxTM NGS based assay was performed. However, only best supportive care could be provided due to fast performance status alteration.
Results were positive for three genomic aberrations: GNA11 Q209L, CDKN2A p16INK4a M52fs*1 and p14ARF H66fs*106 and FANCD2 Q802H. Microsatellite status was stable, and TMB was low (1 Mut /Mb).
3.3 Sequencing results
The results of sequencing of both patients are presented in Table 1.
|
Patients |
Genomic alterations |
Microsatellite status |
Tumor Mutation Burden (TMB) |
|
Patient 1 |
GNAQ Q209P SF3B1 R625H |
Stable |
Low (5 Mutations /Mb) |
|
Patient 2 |
GNA11 Q209L CDKN2A: - p16INK4a M52fs*1 - p14ARF H66fs*106 FANCD2 Q802H |
Stable |
Low (1 Mutations /Mb) |
Table 1: Sequencing results of patients realised by FoundationOne CDxTM.
4. Discussion
4.1 Microstallite status
Microsatellites are small, non-coding DNA sequences that are randomly distributed in the genome. They are often characterized by repetitions of identical bi-nucleotides. Microsatellites show cellular dysfunction during DNA replication. During DNA replication, base matching errors frequently occur, leading to sequence errors within the replicated DNA. During the normal process, these errors are automatically corrected by a coordinated set of proteins: the proteins of the Mismatch Repair (MMR) system. Because of their repetitive sequence, microsatellites are particularly exposed to replication errors. It most often results in a repetition number of nucleotides not retained (either higher or lower than the original sequence of the microsatellite considered). The protein system of repair errors consists of a family of proteins (MMR system proteins), the main ones being MSH2, MLH1, MSH6, PMS1 and PMS2. The inactivation of MMR protein expression leads to a lack of DNA repair and can contribute to oncogenesis [16].
In this study, both tumour microsatellite statuses were stable. High microsatellite instability (MSI) was found in over 25% metastatic lesions of melanoma [17]. It has been proposed that the immunogenicity of some tumors might be determined by mutational heterogeneity, because any mutation could possibly lead to production of neo-antigens [18]. Then microsatellite status might be a predictor for immunotherapy. Indeed, tumors with the highest amount of somatic mutations, such as malignant melanoma and non-small cell lung cancer have showed positive results with immune checkpoint inhibitors. Therefore, larger studies are necessary to understand the role of MSI in immunotherapy response.
4.2 Tumour mutational burden
TMB represents the number of somatic protein-coding base substitution and insertion/deletion mutations in a tumor specimen. TMB is influenced by exposure to mutagens [19-21] or MSI [22-25]. Both tumours in this study harbour a low TMB. Higher TMB predicts favorable outcome to PD-1/PD-L1 blockade across diverse tumors [26]. In contrast, patients with low TMB levels, less benefit from immune checkpoint inhibitors such as anti-CTLA-4 therapy in melanoma [27] or anti-PD-1 therapy in non-small cell lung cancer [22, 28].
4.3 GNAQ/GNA11
Oncogenic mutations in GNAQ and GNA11 genes are found in all grades PMN-CNS [9, 29-34]. GNA11 and GNAQ encode the protein guanine nucleotide-binding protein G(q) subunit alpha, which is a protein that acts as a modulator of transmembrane signaling and activates phospholipase C [35-37]. Activating mutations in GNAQ and GNA11 in PMN-CNS mainly affect codon 209, known to be a mutational hotspot, and consist of substitution of glutamine to leucine [c.626 A>T (p.(Gln209Leu))] or, less frequently, by substitution to proline [c.626 A>C (p.(Gln209Pro))] [12, 31-33]. Only a single case of spinal melanocytoma with a codon 183 GNAQ mutation {c.548 G>A [p.(Arg183Gln)]} has been described[34]. These mutations result in constitutive activation of the protein guanine nucleotide-binding protein G(q), which permanently activates the MAP kinase pathway via protein kinases C (PKC), thus promoting carcinogenicity [38-40]. Based on published series, GNAQ mutations are present in about 39% of melanocytomas and 17% of primary leptomeningeal melanomas, while GNA11 mutations are present in approximately 17% of melanocytomas (including intermediate-grade melanocytic neoplasms) and 29% of primary leptomeningeal melanomas [9].
The prognostic significance of GNAQ and GNA11 mutations in PMN-CNS is unclear but some studies suggest that GNA11 mutations are correlated with poor prognosis [8]. The rapid pejorative evolution of the second patient is consistent with these data. There are no approved therapies targeting GNAQ activating mutations. Preclinical studies have reported that inhibitors of PKC or MEK may have anti-tumor activity on GNAQ mutated tumors with other targeted agents [41-44].
4.4 SF3B1
SF3B1 mutation has been reported in about 20% of UM and meningeal melanoma [15], adenoid cystic carcinomas of the salivary gland and breast [45, 46], pancreatic carcinoma [47], glioblastoma and renal clear cell carcinoma [48]. Mutated SF3B1 tumours have deregulated splice pattern. This leads to a neomorphic activity that upregulates aberrant mRNA splicing [49]. Small molecules such as spliceostatin A, meamycin and sudemycins can modulate aberrant splicing via SF3B1 inhibition[50]. Maguire et al. showed that spliceostatin A inhibits the growth of SF3B1-mutant breast cancer cell lines compared to wild-type cell lines [51]. It is the first time that a mutation of SF3B1 is described in MM.
In UM, SF3B1 mutations are associated with a good prognosis and primarily occur in tumors which do not metastasize [15]. In this case, the SF3B1 mutation was associated with poor prognosis. Inhibition of SF3B1 could be a future strategy in different types of cancers affected by spliceosome-related genes mutations.
4.5 CDKN2A
CDKN2A encodes two distinct tumor suppressor proteins, p16INK4a and p14ARF [52, 53]. The protein p16INK4a is an inhibitor of the cyclin D-dependent kinases CDK4 and CDK6 that prevents them from phosphorylating the retinoblastoma protein (pRB) and thus stop exit from the G1 phase of the cell cycle [52]. The protein p14ARF is involved in the regulation of p53 pathway [53]. p16-cyclin D-CDK4/6-retinoblastoma pathway amplification is a common early event in primary melanoma. A study to compare the genetic alterations of primary melanomas with their adjacent precursor lesions showed biallel inactivation of CDKN2A exclusively in invasive melanomas [54].
In a series of 143 patients with primary invasive melanoma, Young et al. reported a 56% rate of hemizygous or homozygous loss of CDKN2A [55]. Melanoma cell lines with protein p16INK4a loss through CDK2NA gene deletion or methylation were shown to be more sensitive to Palbociclib (PD0332991) compared to wild-type melanoma cell lines. p16INK4a expression loss already had been shown to predict sensitivity to Palbociclib (PD0332991) in ovarian [56], breast [57] and renal cell carcinoma [58] cell lines. A future clinical trial (NCT03454919) will assess Palbociclib in advanced acral melanoma patients with p16INK4a loss. Another clinical trial (NCT02645149) will assess the efficacy of NGS-guided targeted therapy matched to genetic aberration in patients with BRAF and NRAS wild-type unresectable stage III or IV progressive metastatic melanoma after standard therapies. These studies may provide evidence toward the use of CDK4 inhibitors in treating melanoma based on CDKN2A gene status.
According to the literature, this is the first time that this molecular alteration has been described in MM. The role of p16INK4a expression loss as a predictive response biomarker for Palbociclib in patients with MM will have to be explored in future studies.
4.6 FANCD2
FANCD2 is mutated in 5% of Fanconi anemia (FA) patients. FA is an inherited genomic instability disorder, of autosomal recessive transmission, clinically heterogeneous, involving various developmental abnormalities, early-onset bone marrow failure, and a predisposition to cancer [59]. The disease is manifested by defects in DNA repair, hypersensitivity to DNA crosslinking agents, and a high degree of chromosomal aberrations. The FA pathway comprises 13 disease-causing genes involved in maintaining genomic stability.
The FA pathway employs a unique nuclear protein complex, the FA core complex, whose function is to mono-ubiquitinate the FancD2 and FanCI proteins. The FancD2 ubiquitination is indispensable to formation of DNA repair structures that allow the integrity of the genome to be maintained [60]. FancD2 plays a role in preventing chromatin rupture and loss at the end of cell division by interacting with BRCA1 [61] and BRCA2 [62] and with the ATR signalling pathway [63].
It has been shown that the FA pathway is strongly expressed in metastatic melanoma [64] and that gene expression of fanconi anemia was significantly higher in metastatic melanoma than in normal skin [65]. The FA pathway may be a promising new therapeutic target for the treatment of melanoma. The phase II trial NCT03375307 explores the role of Olaparib in metastatic urothelial cancer with DNA-repair defects, including FANCD2 mutation. Talazoparib is being tested in patients with progressive homologous recombination repair deficiency (HRRD) positive stage IV squamous cell lung cancer in a different phase II trial (NCT03377556). To our knowledge, it is the first case of mutation of the FANCD2 gene reported for a MM.
5. Conclusion
MM is a rare CNS tumour with unaddressed issues. In this report, we discussed two cases of MM for whom a FoundationOne CDxTM assay was performed. GNAQ and GNA11 mutations have already been described in several studies. It is however the first time that SF3B1, CDKN2A and FANCD2 mutations are described in MM. The first patient was diagnosed with intermediate-grade MM based on histological assessment, which is consistent with the clinical evolution. The FoundationOne CDxTM assay detected a SF3B1 mutation, usually found in 15% of UM. This mutation is correlated with improved progression-free survival in UM [66].
The second patient’s evolution wasn’t expected based on histology only; her history didn’t match with rather benign histological features microscopically. The FoundationOne CDxTM assay provided two interesting elements: first, the tumour was positive for CDKN2A p16INK4a M52fs*1 and p14ARF H66fs*106 mutations, most commonly found in cutaneous melanomas. Secondly, a GNA11 Q209L mutation, known to be a marker of aggressiveness in UM, was detected. The identification of molecular alterations in PMN-CNS, in addition to immunohistochemical analysis could provide a better understanding of the biological and clinical behaviour of these tumours, a better identification of melanocytic melanomas with poor prognosis and a high risk of relapse or metastasis, and the development of novel therapeutic options for these patients.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest
We have no conflict of interest to disclose.
References
- Douarin NL, Kalcheim C. The Neural Crest. Cambridge University Press (1999).
- Goldgeier MH, Klein LE, Klein-Angerer S, et al. The distribution of melanocytes in the leptomeninges of the human brain, J Invest Dermatol 82 (1984): 235-238.
- Clarke DB, Leblanc R, Bertrand G, et al. Meningeal melanocytoma Report of a case and a historical comparison, J Neurosurg 88 (1998): 116-121.
- Brat DJ, Giannini C, Scheithauer BW, et al. Primary Melanocytic Neoplasms of the Central Nervous System, Am J Surg Pathol 23 (1999): 745.
- O’Brien DF, Crooks D, Mallucci C, et al. Meningeal melanocytoma, Childs Nerv Syst ChNS Off J Int Soc Pediatr Neurosurg 22 (2006): 556-561.
- Rahimi-Movaghar V, Karimi M, Meningeal melanocytoma of the brain and oculodermal melanocytosis (nevus of Ota) case report and literature review, Surg Neurol 59 (2003): 200-210.
- Kutzner H, Schärer L, Requena L, Epithelioid and hyperpigmented melanocytic tumors An overview Pathol 28 (2007): 411-421.
- Roser F, Nakamura M, Brandis A, et al. Transition from meningeal melanocytoma to primary cerebral melanoma Case report, J Neurosurg 101 (2004): 528-531.
- Küsters-Vandevelde HVN, Küsters B, van Engen-van Grunsven ACH, et al. Primary melanocytic tumors of the central nervous system: a review with focus on molecular aspects, Brain Pathol Zurich Switz 25 (2015): 209-226.
- Rades D, Schild SE, Tatagiba M, et al. Therapy of meningeal melanocytomas, Cancer 100 (2004): 2442-2447.
- van de Nes J, Gessi M, Sucker A, et al. Targeted next generation sequencing reveals unique mutation profile of primary melanocytic tumors of the central nervous system, J Neurooncol 127 (2016): 435-444.
- Küsters-Vandevelde HVN, van Engen-van Grunsven IACH, Coupland SE, et al. Mutations in g protein encoding genes and chromosomal alterations in primary leptomeningeal melanocytic neoplasms, Pathol Oncol Res POR 21 (2015): 439-447.
- Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma, N Engl J Med 363 (2010): 2191-2199.
- Harbour JW, Roberson EDO, Anbunathan H, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma, Nat Genet 45 (2013): 133-135.
- Martin M, Maßhöfer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3, Nat Genet 45 (2013): 933-936.
- Pfeifer GP, You Y-H, Besaratinia A, Mutations induced by ultraviolet light, Mutat Res 571 (2005): 19-31.
- Kroiss MM, Vogt TM, Schlegel J, et al. Microsatellite instability in malignant melanomas, Acta Derm Venereol 81 (2001): 242-245.
- Champiat S, Ferté C, Lebel-Binay S, et al. Exomics and immunogenics, Oncoimmunology 3 (2014).
- Pfeifer GP, You Y-H, Besaratinia A, Mutations induced by ultraviolet light, Mutat Res Mol Mech Mutagen 571 (2005): 19-31.
- Hill VK, Gartner JJ, Samuels Y, et al. The genetics of melanoma: recent advances, Annu Rev Genomics Hum Genet 14 (2013): 257-279.
- Pfeifer GP, Denissenko MF, Olivier M, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers, Oncogene 21 (2002): 7435-7451.
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer, Science 348 (2015): 124-128.
- Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma, Nature 497 (2013): 67-73.
- Cancer Genome Atlas Network, Comprehensive molecular characterization of human colon and rectal cancer, Nature 487 (2012): 330-337.
- Roberts SA, Gordenin DA, Hypermutation in human cancer genomes: footprints and mechanisms, Nat Rev Cancer 14 (2014): 786-800.
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers, Mol Cancer Ther 16 (2017): 2598-2608
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma, N Engl J Med 371 (2014): 2189-2199.
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency, N Engl J Med 372 (2015): 2509-2520.
- Cornejo KM, Hutchinson L, Cosar EF, et al. Is it a primary or metastatic melanocytic neoplasm of the central nervous system? A molecular based approach, Pathol Int 63 (2013): 559-564.
- Fuld AD, Speck ME, Harris BT, et al. Primary melanoma of the spinal cord: a case report, molecular footprint, and review of the literature, J Clin Oncol Off J Am Soc Clin Oncol 29 (2011): e499-502.
- Gessi M, Hammes J, Lauriola L, et al. GNA11 and N-RAS mutations: alternatives for MAPK pathway activating GNAQ mutations in primary melanocytic tumours of the central nervous system, Neuropathol Appl Neurobiol 39 (2013): 417-425.
- Koelsche C, Hovestadt V, Jones DTW, et al. Melanotic tumors of the nervous system are characterized by distinct mutational, chromosomal and epigenomic profiles, Brain Pathol Zurich Switz 25 (2015): 202-208.
- Küsters-Vandevelde HVN, Klaasen A, Küsters B, et al. Activating mutations of the GNAQ gene: a frequent event in primary melanocytic neoplasms of the central nervous system, Acta Neuropathol (Berl): 119 (2010): 317-323.
- Murali R, Wiesner T, Rosenblum MK, et al. GNAQ and GNA11 mutations in melanocytomas of the central nervous system, Acta Neuropathol (Berl): 123 (2012): 457-459.
- Jiang M, Pandey S, Tran VT, et al. Guanine nucleotide-binding regulatory proteins in retinal pigment epithelial cells, Proc Natl Acad Sci U S A 88 (1991): 3907-3911.
- Davignon I, Barnard M, Gavrilova O, et al. Gene structure of murine Gna11 and Gna15: tandemly duplicated Gq class G protein alpha subunit genes, Genomics 31 (1996): 359-366 .
- Dong Q, Shenker A, Way J, et al. Molecular cloning of human G alpha q cDNA and chromosomal localization of the G alpha q gene (GNAQ): and a processed pseudogene, Genomics 30 (1995): 470-475.
- Ross EM, Wilkie TM GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS): and RGS-like proteins, Annu Rev Biochem 69 (2000): 795-827.
- Landis CA, Masters SB, Spada A, et al. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours, Nature 340 (1989): 692-696.
- Lyons J, Landis CA, Harsh G, et al. Two G protein oncogenes in human endocrine tumors, Science 249 (1990): 655-659.
- Mitsiades N, Chew SA, He B, et al. Genotype-dependent sensitivity of uveal melanoma cell lines to inhibition of B-Raf, MEK, and Akt kinases: rationale for personalized therapy, Invest Ophthalmol Vis Sci 52 (2011): 7248-7255.
- von Euw E, Atefi M, Attar N, et al. Antitumor effects of the investigational selective MEK inhibitor TAK733 against cutaneous and uveal melanoma cell lines, Mol Cancer 11 (2012): 22.
- Wu X, Zhu M, Fletcher JA, et al. The protein kinase C inhibitor enzastaurin exhibits antitumor activity against uveal melanoma, PloS One 7 (2012): e29622.
- Khalili JS, Yu X, Wang J, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner, Clin Cancer Res Off J Am Assoc Cancer Res 18 (2012): 4345-4355.
- Stephens PJ, Davies HR, Mitani Y, et al. Whole exome sequencing of adenoid cystic carcinoma, J Clin Invest 123 (2013): 2965-2968.
- Martelotto LG, De Filippo MR, Ng CKY, et al. Genomic landscape of adenoid cystic carcinoma of the breast, J Pathol 237 (2015): 179-189.
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes, Nature 491 (2012): 399-405.
- Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies, Nat Med 20 (2014): 1472-1478.
- Alsafadi S, Houy A, Battistella A, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage, Nat Commun 7 (2016).
- Wu G, Fan L, Edmonson MN, et al. Inhibition of SF3B1 by molecules targeting the spliceosome results in massive aberrant exon skipping, RNA N Y N 24 (2018): 1056-1066.
- Maguire SL, Leonidou A, Wai P, et al. SF3B1 mutations constitute a novel therapeutic target in breast cancer, J Pathol 235 (2015): 571-580.
- Quelle DE, Zindy F, Ashmun RA, et al. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest, Cell 83 (1995): 993-1000.
- Sharpless NE INK4a/ARF: a multifunctional tumor suppressor locus, Mutat Res 576 (2005): 22-38.
- Shain AH, Yeh I, Kovalyshyn I, et al. The Genetic Evolution of Melanoma from Precursor Lesions (2015).
- Young RJ, Waldeck K, Martin C, et al. Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines, Pigment Cell Melanoma Res 27 (2014): 590-600.
- Konecny GE, Winterhoff B, Kolarova T, et al. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer, Clin Cancer Res Off J Am Assoc Cancer Res 17 (2011): 1591-1602.
- Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro, Breast Cancer Res BCR 11 (2009): R77.
- Logan JE, Mostofizadeh N, Desai AJ, et al. PD-0332991, a potent and selective inhibitor of cyclin-dependent kinase 4/6, demonstrates inhibition of proliferation in renal cell carcinoma at nanomolar concentrations and molecular markers predict for sensitivity, Anticancer Res 33 (2013): 2997-3004.
- Deakyne JS, Mazin AV Fanconi anemia: at the crossroads of DNA repair, Biochem Biokhimiia 76 (2011): 36-48.
- Moldovan G-L, D’Andrea AD How the fanconi anemia pathway guards the genome, Annu Rev Genet 43 (2009): 223-249.
- Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway, Mol Cell 7 (2001): 249-262.
- Taniguchi T, Garcia-Higuera I, Andreassen PR, et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51, Blood 100 (2002): 2414-2420.
- Taniguchi T, Garcia-Higuera I, Xu B, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways, Cell 109 (2002): 459-472.
- Bourseguin J, Bonet C, Renaud E, et al. FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells, Sci Rep 6 (2016): 36539.
- Kao WH, Riker AI, Kushwaha DS, et al. Upregulation of Fanconi Anemia DNA Repair Genes in Melanoma Compared to Non-Melanoma Skin Cancer, J Invest Dermatol 131 (2011): 2139-2142.
- Furney SJ, Pedersen M, Gentien D, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma, Cancer Discov 3 (2013): 1122-1129.
