Lung Adenocarcinoma: Next Generation Sequencing and Outcome in Regional Community Oncology Setting
Article Information
Laura E Skacel1, Michael J Babcock1, Antoine Harb2, Adam D Curtis2, Carter L Liou1, Kathleen T Brawn1, Frantisek Sandor3, Marek Skacel1*
1Dahl-Chase Diagnostic Services, Bangor, Maine, USA
2Northern Light Cancer Institute, Brewer, Maine, USA
3Penobscot Respiratory & Critical Care, Bangor, Maine, USA
*Corresponding author: Marek Skacel, MD, Dahl-Chase Pathology Associates, 417 State St, Suite 540, Bangor, ME 04401, USA
Received: 08 December 2021; Accepted: 14 December 2021; Published: 23 December 2021
Citation:
Laura E Skacel, Michael J Babcock, Antoine Harb, Adam D Curtis, Carter L Liou, Kathleen T Brawn, Frantisek Sandor, Marek Skacel. Lung Adenocarcinoma: Next Generation Sequencing and Outcome in Regional Community Oncology Setting. Journal of Surgery and Research 4 (2021): 804-819.
View / Download Pdf Share at FacebookAbstract
Purpose
To analyze lung adenocarcinomas (LUAD) from a geographically unique population of rural Maine by next generation sequencing (NGS), correlate mutational findings with clinical features, patient outcomes and published data from other populations.
Methods
210 consecutive LUADs diagnosed in 2017-2018 were analyzed for 50 oncogene/tumor suppressor gene hot spots by NGS. ALK, ROS-1, RET and MET were assessed by FISH, PD-L1 by immunohistochemistry. Findings were correlated with age, gender, smoking history, stage, overall (OS) and progression-free survival (PFS) and compared to published literature.
Results
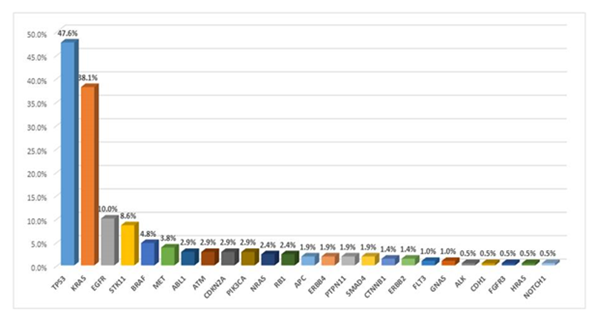
The cohort included 113 (54%) women and 97 (46%) men, ages 33 to 91 (mean: 67.4 years), 52% active and 41% former smokers, 79 (38%) of advanced stage (stage IV). Most frequently detected mutations included TP53 (47.6%), KRAS (38.1%), EGFR (10%), STK11 (8.6%), BRAF (4.8%), MET (3.8%), ABL-1, ATM, CDKN2A, PIK3CA, (all 2.9%), RB-1 and NRAS (2.4%), APC, ERBB4, PTPN11, SMAD4, (all 1.9%), CTNNB1 and ERBB2 (both 1.4%). MET amplification occurred in 3.3%, RET and ALK/ROS-1 rearrangements in 1.4% and 0.5%, high PD-L1 expression in 35.2%. Treatment included surgery/radiation/adjuvant chemotherapy for stages I-II, definitive chemo/radiation therapy and immunotherapy for stage III, immunotherapy, chemo-immunotherapy, targeted therapy, palliative radiation for stage IV. At median of 26 months (minimum 21 month for surviving patients), OS/PFS were 44.3%/39.5%. Stage, male gender, TP53 mutation and KRAS/STK11 co-mutations correlated with adverse OS. In stages I-II, KRAS/TP53 co-mutation was unfavorable.
Conclusion
NGS testing
Keywords
<p>Lung adenocarcinoma, TP53, KRAS, STK11, Gene mutation, Survival</p>
Article Details
1. Introduction
Lung cancer is the second most common cancer and the leading cause of cancer-related death in the U.S. [1]. Non-small cell carcinoma (NCSLC) accounts for approximately 85% of cases, with lung adenocarcinoma (LUAD) being the most common subtype, accounting for approximately 40% of NSCLC cases [2]. There is a known urban-rural disparity in lung cancer incidence within the United States, with the most rural counties having an annual lung cancer incidence almost twice that of the largest metropolitan areas [3]. The cause for this disparity is largely attributed to higher prevalence of cigarette smoking, as the increase in lung cancer incidence tracks closely rural smoking rates [4]. Such urban/rural smoking rate differences were found to be most significant in the New England region [5]. The state of Maine with its quintessential New England rural communities, has rates of lung cancer astonishingly 30% higher than the national average [6]. The lung cancer incidence in Maine is even higher than what would be expected for its smoking rate, suggesting exposure to other risk factors besides smoking, such as residential radon exposure or air pollution [7]. In fact, 12 of 16 Maine counties are classified as zones with predicted average indoor radon levels greater than those recommended as safe by the EPA [8]. Whether such potential factors may contribute to the molecular genetic makeup of lung cancer from this region and its mutational profiles differ from those reported in other parts of the country is unknown, as to date, no molecular profiling studies from this geographically unique region have been published. The recent standardization of next-generation sequencing (NGS) methods allowed its gradual implementation in molecular pathology laboratories outside of larger academic institutions [9]. This trend provides an opportunity to profile tumors diagnosed locally in more homogeneous rural populations. In addition to markers endorsed by current clinical practice guidelines, NGS provides molecular data that can be utilized for investigational and clinical trial-based treatment purposes [10]. Routine local NGS testing of lung cancer samples with short turn-around time can improve the efficiency of regional oncology care by providing timely results for genomic-based therapies without delay, which often results from sending samples to remote reference centers [11]. Our single-center institution and laboratory collects and analyzes samples and treats patients from the entire central and northern Maine region, thus concentrating the vast majority of lung carcinomas from the rural parts of the state. As a result, the cases in our series represent a geographically homogeneous lung cancer patient population from an area with its prevalence ranking among the highest in the nation. Our retrospective cohort consists of consecutive LUAD cases analyzed by NGS, fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC). In addition to the standard markers endorsed by the National Comprehensive Cancer Network (NCCN) [12], our analysis includes a panel of tumor suppressor genes and oncogenes, as well as correlation with clinical features and outcomes. The relative geographic isolation and ethnic uniformity of our patient population provided us with an opportunity to compare the mutational profiles of “rural” LUAD to the data in the literature, generated predominantly by academic centers and reflecting patients from predominantly larger metropolitan areas. Our work also highlights the benefits of "reflexive testing" in community oncology practice. We describe the utilization of "standing order" for molecular pathology and integration of upfront sample preparation for molecular testing as part of routine diagnostic process, and assess their impact on tissue adequacy and turnaround time as important contributors to efficient oncology care in a regional setting.
2. Methods
2.1 Patient selection
210 consecutive LUAD cases from patients with available clinical follow up tested by NGS during the period of January 2017- June 2018 were retrieved from the NGS database at Dahl-Chase Pathology Laboratory and Northern Light Oncology Institute. This work was performed in concordance with an Institutional Review Board approval (19-1-A-001). LUADs arising in the lung or involving metastatic sites in patients where primary tumor was unavailable were utilized for testing. A molecular pathologist confirmed the diagnosis ensured minimum of 20% tumor content in each sample. Staging of patients was performed according to the 8th edition of the tumor, node and metastasis (TNM) classification for lung cancer [13].
2.2 Reflexive molecular testing utilizing a standing order
In order to perform molecular testing in a short turnaround time, avoid the need for sending individual test requests to pathology and to preserve tissue, our institution utilizes “reflexive” molecular standing order. Updated regularly to ensure it includes all NCCN-endorsed biomarkers and meets the current standards of care, this standing order is approved by the institutional molecular tumor board in conjunction with the hospital medical staff and initiated automatically by the pathologist, as soon as the diagnosis of LUAD is established.
2.3 DNA extraction and sample preparation
A pathologist confirmed the presence of sufficient tumor tissue for NGS analysis by direct microscopic visualization of tissue on glass slides stained with hematoxylin-eosin. Ten 4-5 μm formalin-fixed, paraffin-embedded sections were marked for manual microdissection of genomic DNA. 15 ng of DNA was amplified, fragmented, ligated to adapters, barcoded, clonally amplified onto beads to create DNA libraries using the Ion AmpliSeq™ Cancer HotSpot Panel v2.0 (CHPv2.0; Life Technologies, ThermoFisher, Carlsbad, CA, USA) and subjected to NGS on the Ion Torrent Personal Genome Machine (PGM) or S5 instrument.
2.4 Mutational analysis by NGS
The DNA primer pool in the AmpliSeq™ CHPv2.0 panel is designed to examine 2800 mutation hotspots previously described as somatic mutations occurring in human cancers. The genes assessed by the Cancer Hotspot Panel v2 include: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, and VHL.
2.5 Variant interpretation
The resulting DNA sequencing reads were aligned to the hg19 (GRCh37) reference genome and data analysis was performed using the Ion Torrent Suite (5.8.0) and Golden Helix VarSeq (2.0.2) software. The lower limit of detection of this assay has been established at a 5% variant allele frequency (VAF), with normal range (mutation not detected) being a VAF <5%, and mutational status of VAF ≥ 5% indicating a mutation present. Previously established minimum read depth cut off was set at 100X coverage with a minimum of 25 variant allele observations, with a 250X read depth minimum for a 5% sensitivity (based on prior validation studies, data not shown). Previously reported false-positive mutations and other potential artifacts due to nonspecific mispriming events were removed according to McCall et al [14]. Genomic alterations were reviewed by a molecular pathologist, tiered according to their clinical relevance, and reported with accompanying NCCN therapeutic guidelines.
2.6 Additional testing
Fluorescence in situ hybridization (FISH) was performed according to an established protocol, using probes for ALK, ROS-1, RET and MET. Immunohistochemistry (IHC) for PD-L1 was performed according to manufacturer’s protocol using 22C3 antibody (Dako, Carpinteria, CA) and FDA-approved scoring approach, reporting the percentage of tumor cells with membranous positivity (tumor proportion score; TPS) [15]. MET Exon 14 skipping mutation was evaluated via send-out testing, since this test was not available in-house at the time of this study period.
2.7 Statistical analysis
All statistical analyses were performed using SPSS for Windows (v19.0, IBM, Armonk, NY). Comparisons of proportions across different groups were evaluated using Fisher’s exact test or Chi-square test where appropriate. The presence of molecular abnormalities was correlated with patients’ age, gender, smoking history (including pack-years) and tumor stage. Progression-free survival (PFS) was measured from the date of diagnosis to tumor recurrence or death, while overall survival (OS) was measured from the date of diagnosis to the date of death. Patients were censored on September 30, 2020 if alive and disease-free for OS and PFS, respectively. A log-rank test was used to compare survival curves in Kaplan-Meier analysis. Cox regression analysis was performed to evaluate the significance and calculate relative risks in a time-dependent multivariate model, with risk expressed as hazard ratios (HR) with 95% confidence intervals (CI). All statistical analyses were two-sided with p-values at significance level of 0.05.
3. Results
3.1 Specimen types, clinical features and treatment modalities
The specimens included 89 (42.3%) core needle biopsies, 53 (25.2%) cytology specimens with cell blocks (endobronchial ultrasound-guided fine-needle biopsies (EBUS) or pleural fluid aspirates) and 68 (32.4%) excisional specimens. 140 cases (66.7%) were samples of primary lung tumor and 70 (33.3%) were metastases (mediastinal or distant). Among the excisional specimens, 55 were primary lung tumor excisions, 18 (33%) of which were wedge excisions and 37 (67%) lobectomies. Of the 55 excised tumors, 8 (14%) were grade 1, 34 (62%) grade 2 and 13 (24%) grade 3. Histologic subtypes included 34 (60%) acinar, 12 (22%) solid, 2 (4%) lepidic, 3 (5%) micropapillary, 1 (2%) mucinous and 4 (7%) not otherwise specified types. The patient cohort consisted of 97 (46.2%) men and 113 (53.8%) women, ages 33 to 91 years (mean: 67.4 years), living in central/eastern and northern regions of the state of Maine. 208 of the 210 patients (99%) were Caucasian, 1 (0.5%) Asian and 1 (0.5%) of Native American (Penobscot) ancestry. 14 patients (6.7%) were never smokers, 109 (51.9%) were active smokers at the time of diagnosis and 87 (41.4%) former smokers (quit 1 or more years prior to diagnosis). The mean number of pack years among the smokers was 47 (range: 1-122), with 79 (42.9%) having at least 50-pack year history. 131 (62.4%) patients had limited stage: 71 (33.8%) stage I (51 (72%) IA and 20 (28%) IB), 25 (11.9%) stage II (14 (56%) IIA and 11 (44%) IIB and 35 (16.6%) stage III (21 (60%) IIIA, 12 (34%) IIIB and 2 (6%) IIIC). 79 (37.6%) patients had advanced (stage IV) disease. Treatment was administered according to the NCCN guidelines16, dictated by stage, presence of predictive/therapeutic biomarkers and including: surgery/radiation plus/minus adjuvant chemotherapy for stages I-II, definitive chemo/radiation therapy followed by immunotherapy for stage III, and immunotherapy, combination chemo-immunotherapy, targeted therapy or palliative radiation for stage IV. At a median follow-up of 26 months (range: 1-46 months, SD 13.9), with a minimum of 21 months follow-up for surviving patients, the overall survival (OS) and progression-free survival (PFS) were 44.3% and 39.5%, respectively.
3.2 NGS Performance and mutations detected in the studied cases
The tissue sections used for analysis contained a minimum of 500 cancer cells in a micro-dissected area from which 10 consecutive unstained sections, which were obtained during initial sectioning of the tissue for diagnosis. The mean tumor cell content per sample was 40% (range 20-80%). Overall, the variant allelic frequency (VAF) of the mutations detected in this study ranged from 6% to 48% (mean: 32%). Read depths ranged from a minimum of 350X to over 12,000X (mean 2,800X), indicating a satisfactory level of coverage for the regions of interest (ROI). Overall frequencies of gene mutations detected in at least 1% of the studied cases were (Figure 1): TP53 (44.80%), KRAS (38.1%), EGFR (10%), STK11 (8.6%), BRAF (4.8%), MET (3.8%), ABL-1, ATM, CDKN2A, PIK3CA, (all 2.9%), RB-1 and NRAS (2.4%), APC, ERBB4, PTPN11, SMAD4, (all 1.9%), CTNNB1 and ERBB2 (both 1.4%). Detailed lists of all detected mutations with numbers of patients harboring each mutation are included in the supplemental table 1.
|
TP53 Mutation |
KRAS Mutation |
EGFR Mutation |
STK11 Mutation |
|||||
|
N (%) |
P |
N (%) |
P |
N (%) |
P |
N (%) |
P |
|
|
Gender |
||||||||
|
Male |
40 (41%) |
26 (27%) |
7 (7%) |
11 (11%) |
||||
|
Female |
54 (48%) |
0.4 |
54 (48%) |
0.003 |
14 (12%) |
0.25 |
7 (6%) |
0.22 |
|
Age |
||||||||
|
>70 years |
38 (38%) |
35 (35%) |
10 (10%) |
8 (8%) |
||||
|
<70 years |
56 (50%) |
0.09 |
45 (41%) |
0.48 |
11 (10%) |
1 |
10 (9%) |
1 |
|
Stage |
||||||||
|
I-III |
61 (47%) |
46 (35%) |
13 (10%) |
10 (8%) |
||||
|
IV |
33 (42%) |
0.56 |
34 (43%) |
0.31 |
8 (10%) |
1 |
8 (10%) |
0.6 |
|
Smoking History |
||||||||
|
Smoker |
90 (46%) |
77 (39%) |
14 (7%) |
18 (9%) |
||||
|
Never smoker |
4 (29%) |
0.27 |
3 (21%) |
0.26 |
7 (50%) |
<0.001 |
0 (0%) |
0.6 |
|
Smoking Status |
||||||||
|
Current Smoker |
58 (53%) |
45 (41%) |
3 (3%) |
10 (9%) |
||||
|
Former Smoker |
36 (36%) |
0.018 |
35 (35%) |
0.39 |
18 (18%) |
<0.001 |
8 (8%) |
0.8 |
|
Pack Years |
||||||||
|
50 or more |
35 (44%) |
26 (33%) |
2 (3%) |
9 (11%) |
||||
|
Less than 50 |
45 (43%) |
0.88 |
44 (42%) |
0.22 |
18 (17%) |
0.001 |
5 (5%) |
0.16 |
|
PD-L1 Expression |
||||||||
|
Low/High |
64 (48%) |
56 (42%) |
11 (8%) |
5 (4%) |
||||
|
Absent |
30 (39%) |
0.25 |
24 (31%) |
0.14 |
10 (13%) |
0.34 |
13 (17%) |
0.002 |
|
High |
33 (45%) |
39 (53%) |
5 (7%) |
2 (3%) |
||||
|
Low/Absent |
61 (45%) |
1 |
41 (23%) |
0.002 |
16 (12%) |
0.34 |
16 (12%) |
0.04 |
Table 1: Clinicopathological parameters and the four most commonly detected mutations
3.3 Most commonly mutated genes (occurring in greater than 5% of cases)
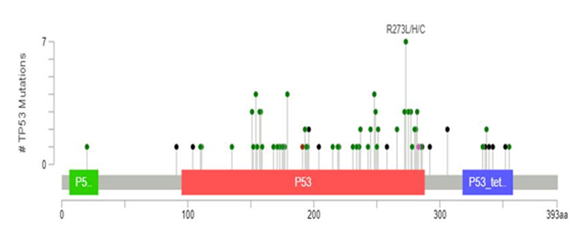
3.3.1 TP53 mutations: TP53 mutation was the most commonly detected, occurring in 100 (47.6%) cases. The detected mutations were highly diverse; 77% occurred only in a single case in this cohort. The most common recurring mutations affected residue 273 (p.R273L, p.R273C or p.R273H) seen in 7% of the mutated cases, followed by residue 179 (p.H179R or H179Y), accounting for 5.5% of cases, 248 (p.R248L or R248W; 4.4% of cases), 154 (p.G154V; 3.3% of cases) and 282 (p.R282G; 2.2% of cases). Most missense, frameshift, and nonsense mutations were located in known “hot spots”, annotated as functional protein domains for DNA-binding (amino acid residues 100-292) and tetramerization (residues 325-356). 1718 Such functional domains mutations were seen in 94 (44.8%) cases. The remaining rare mutations were seen outside of these functional domains, including variants located in the transactivation domain (residues 6-29) or splice site variants. Distribution of TP53 mutations in studied cases across protein domains is depicted in figure 2.
Figure 2: Distribution of TP53 mutations in the studied cases. Mutations occurred most commonly in the DNA binding (red) and tetramerization (purple) domains of the p53 protein. The individual mutations are depicted as a “lollipops” on the x-axis and their frequency on the y-axis. The plot was constructed using the MutationMapper method described by Cerami et al. [17]. Missense mutations are depicted as green, truncating black and splice variants pink circles. The most commonly detected mutation at residue 273 is annotated.
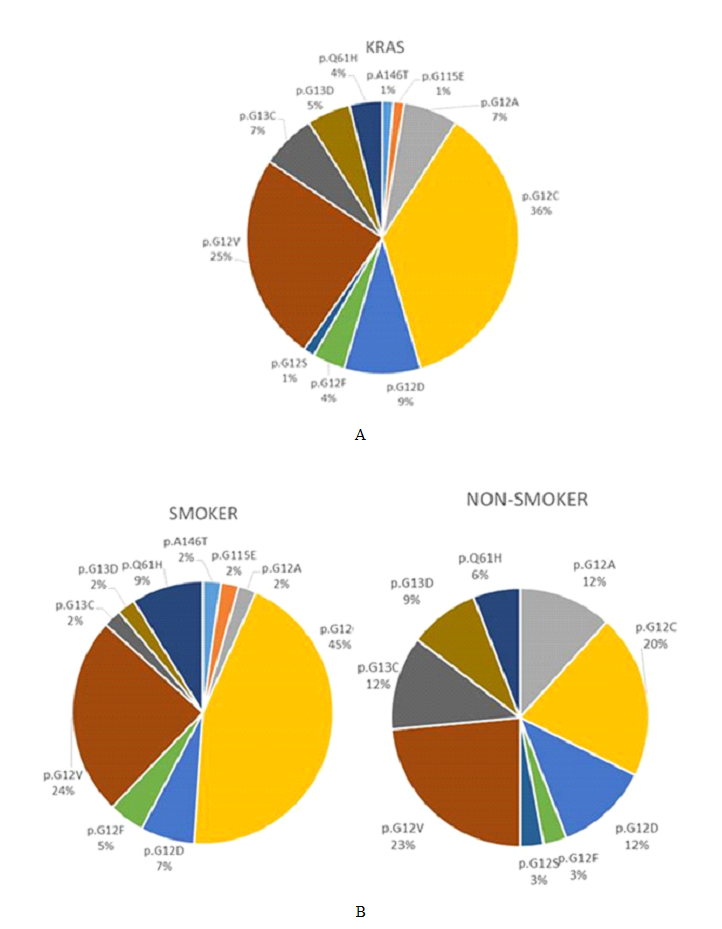
3.3.2 KRAS mutations: KRAS mutations were the second most commonly detected, seen in 80 (38.1%) cases. The vast majority of KRAS mutations affected codon 12 (63 cases, 78.8%), with the remaining mutations occurring in codon 13 (9 cases, 11.3%), codon 61 (6 cases, 7.5%), and codons 115 or 146 (one case, 1.3% each). The proportion of KRAS mutations were as follows: p.G12C (28 cases, 35%), p.G12V (19 cases, 23.8%), p.G12D (7 cases, 8.8%), p.Q61H (6 cases, 7.5%), p.G12A and p.G13C (5 cases each, 6.3%), p.G13D (4 cases, 5%), p.G12F (3 cases, 3.8%) and one case each of p.G12S, p.G155E and p.A146T (1.3% each). The spectrum of KRAS mutations detected in the cohort overall is depicted in figure 3A.
3.3.3 EGFR mutations: EGFR mutations were seen in 21 (10%) of cases studied. The most common were exon 19 deletion/insertions and exon 21 p.L858R mutations accounting for 47.6% and 38% of the EGFR mutations, respectively. T790M mutation in exon 20 was identified as a second mutation with a lower allelic frequency in 3 (10.3%) of the EGFR-mutated patients that previously received EGFR inhibitors. STK11 mutations. STK11 mutations occurred in 18 (8.6%) cases. The most prevalent mutations were p.Y60Ter (2), p.H168R (2) and p.E256Ter (2) and splice donors (3). The remaining 9 mutations were each identified in single cases (5.6%).
3.3.4 Overall mutational rates per tumor and co-mutation rates of the most commonly mutated genes: The mean number of mutations per case was 1.41 (range: 0-10, 95% confidence interval: 1.27-1.56). No mutations were detected in 31 (14.7%) cases, 94 (44.8%) had a single mutation, 63 (30%) had two concurrent mutations, 17 (9%) had three and 5 (2.4%) had four or more mutations. TP53, KRAS and STK11 mutations accounted for the majority of mutations, either one of them detected in 152 (72.4%) of cases. TP53 mutation co-occurred with KRAS, STK11 or both of these mutations in 29 (13.8%), 9 (4.3%) and 5 (2.4%) of cases, while KRAS co-occurred with STK11 mutation in 12 (5.7%) cases, respectively.
3.4 FISH and IHC results
MET amplification, ALK and ROS1 rearrangement. MET amplification was seen in 7 cases (3%), with MET/chromosome 7 ratios ranging from 2.17 to 6.78 in these amplified cases (mean 3.8). Only one case had ratio greater than 5.0 (high-level amplification). RET rearrangements were seen in 3 cases (1.4%), ROS1 and ALK rearrangements in 1 case (0.5%) each, respectively.
3.5 PD-L1 expression and its correlation with molecular genetic abnormalities
High PD-L1 expression (>50% TPS) was seen in 74 cases (35%), low PD-L1 expression (1-49% TPS) in additional 59 cases (29%). The remaining 77 patients (37%) were negative for PD-L1 expression (TPS<1). High PD-L1 expression occurred more commonly in tumors with KRAS (p=0.002) or BRAF (p=0.04) mutations, as well as in tumors with MET gene amplification (p=0.011). Both low/high and high PD-L1 expression were less common in tumors with STK11 mutation (p=0.009 and p=0.04, respectively).
3.6 Reflexive molecular standing order experience
Utilizing a reflexive standing order for molecular testing ensured completion of testing within 10 working days after the biopsy/excision date in all of the cases studied (mean: 8.5 days; range 7-10 working days after the biopsy date). Obtaining unstained sections for molecular testing at the time of cutting the initial hematoxylin-eosin sections for diagnosis allowed performing molecular studies on tissue from small biopsies and cytology cell blocks, by obtaining unstained ribbons of tissue during the initial diagnostic evaluation, avoiding “refacing” the paraffin blocks, which is necessary when the blocks need to be cut for molecular testing at a later date. During the study period, only two cases (less than 1%) had no residual tumor tissue at diagnosis and could thus not be included in this cohort (never progressed to molecular testing). Prior to introducing the reflexive testing in 2017, our turn-around time exceeded this by up to 5 working days, plus not obtaining sections for molecular testing at diagnosis resulted in an approximately 8% testing failure rate due to lack of residual tissue in the paraffin blocks (data not shown).
3.7 Molecular abnormalities, clinical features and survival
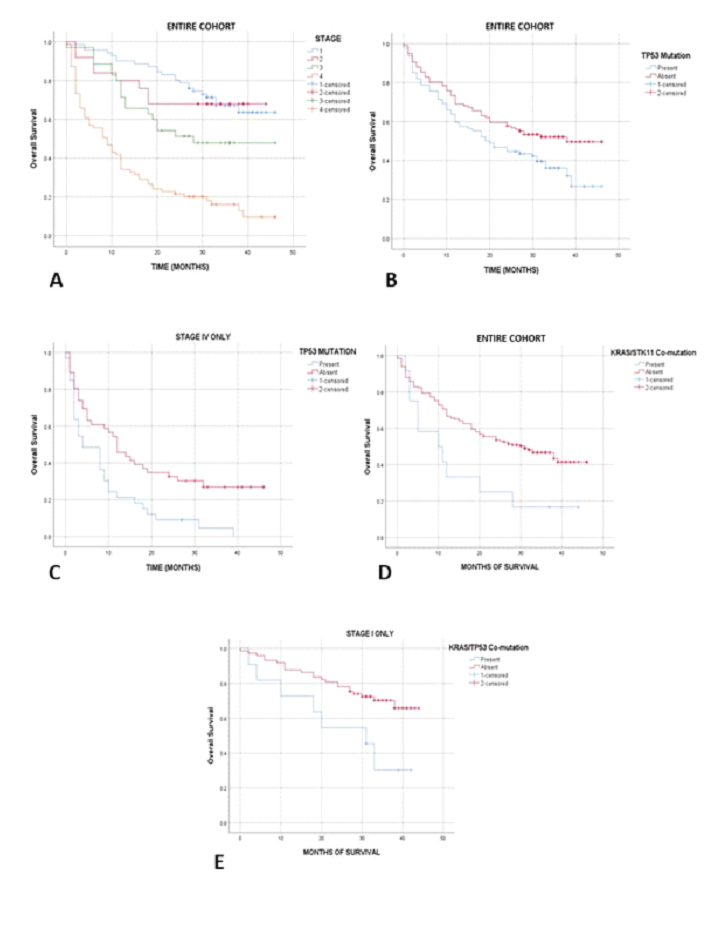
3.7.1 Overall survival/progression-free survival and clinical stage: The overall survival and progression-free survival rates across all stages at the mean follow up of 26 months (with a minimum of 21 months for surviving patients) were 44.3% and 39.5%, respectively. Among patients with stages I, II and III disease (lower stage), 24/71 (33.8%), 8/25 (32%) and 17/34 (50%) died of the disease within the studied time period, resulting in 62.2% overall survival rate in this lower stage category. In contrast, 68/80 (85%) of patients in the advanced stage group (stage IV) died, resulting in a 15% overall survival rate (p<0.001, Kaplan-Meier analysis with log rank; Figure 4A). Similar findings were seen for progression-free survival. In stages I, II and III, 27/71 (38%), 11/25 (44%) and 16/34 (47%) of patients experienced disease recurrence or progression, with overall 58.5% progression-free survival rate in the lower stage group. This contrasted with 73/80 (91.3%) of patients with stage IV disease, who recurred/progressed, resulting in 8.7% progression-free survival rate (p<0.001, Kaplan-Meier analysis with log rank). The clinical and molecular findings are summarized in table 1.
3.7.2 TP53 mutation and survival: Among the patients with TP53 mutations affecting the regions encoding the two principal functional domains (DNA-binding and tetramerization), 44% died of disease, compared to 24% of patients without a "hot spot” TP53 gene mutation (p=0.01). By Kaplan-Meier analysis, such TP53-mutated patients showed significantly worse overall survival (p=0.023, Figure 4B). Controlled for gender, age, stage, grade, histologic subtype and smoking history in a multivariate analysis, a stepwise backward elimination Cox regression model showed that clinical stage (p<0.001, HR=1.85, 95% CI 1.57-2.2), TP53 mutation status (p=0.004, HR=1.71, 95% CI 1.18-2.47), male gender (p=0.04; HR 1.47, 95% CI 1.01-2.12) and KRAS/STK11 co-mutation (p=0.05; HR 1.92, 95% CI 1.00-3.69) independently correlated with overall survival (Table 2). Male gender was no longer significant for overall survival when EGFR mutated cases were excluded from analysis, reflecting the higher prevalence of the prognostically favorable EGFR mutation in non-smoking women (p=0.007). Multivariate Cox regression model showed only advanced stage as significant variable for progression-free survival (PFS) (p<0.001, HR=2.62, 95% CI 1.66-4.11). A separate stage IV survival analysis confirmed the adverse effect of TP53 mutation on both OS and PFS in patients with advanced disease (p=0.007 and p=0.003, respectively; Figure 4C).
|
P |
HR |
95% CI |
|
|
UNIVARIATE ANALYSIS: |
|||
|
Stage I (N=64)*: |
|||
|
KRAS/TP53 co-mutation |
0.009 |
3.78 |
1.39 - 10.27 |
|
Stages I-II (N=84)*: |
|||
|
KRAS/TP53 co-mutation |
0.03 |
2.6 |
1.11 - 6.1 |
|
Stage IV (N=71)*: |
|||
|
TP53 mutation |
0.004 |
2.07 |
1.26 – 3.4 |
|
MULTIVARIATE ANALYSIS: |
|||
|
All stages (N=210): |
|||
|
Stage |
<0.001 |
1.86 |
1.57 - 2.2 |
|
TP53 mutation |
0.004 |
1.71 |
1.19 - 2.47 |
|
Male gender |
0.04 |
1.47 |
1.01 - 2.12 |
|
KRAS/STK11 co-mutation |
0.05 |
1.92 |
1.0 – 3.69 |
|
All stages without EGFR-mutated cases (N=189): |
|||
|
Stage |
<0.001 |
1.82 |
1.53 – 2.16 |
|
TP53 mutation |
0.009 |
1.67 |
1.13 – 2.44 |
|
KRAS/STK11 co-mutation |
0.03 |
2.1 |
1.08 – 4.07 |
*Without EGFR-mutated cases
Table 2: Results of univariate and multivariate analyses: P = P- value, HR = Hazard Risk, CI = Confidence Interval
3.7.3 KRAS mutations and clinical features: Any KRAS mutation, codon 12 mutations (both p=0.003) and most specifically G12C mutation (p<0.001) tended to occur in women, smokers (both p=0.04) and most prominently in women smokers (p<0.001). No associations of KRAS mutations with outcome were noted.
3.7.4 EGFR mutations and survival: Among the EGFR mutated cases, shortened survival was seen by Kaplan Meier analysis in advanced stage (p=0.02) and male gender (p=0.025); both independent in multivariate Cox Regression analysis (p=0.005 and 0.01; HR 2.9/95% CI 1.4-6.2 and 8.9, 95% CI 1.7-46.6, respectively). TP53/KRAS, KRAS/STK11 co-mutations and survival. KRAS/STK11 co-mutation was associated with worse OS (p=0.018; Figure 4D). This was independent of stage, gender, or TP53 mutation in the above multivariate model. In stage I, as well as stages I-II, after excluding cases with favorable prognostic effect of EGFR mutation and controlled for type of excision of the primary tumor, KRAS/TP53 co-mutation was predictive of adverse OS (p=0.005 and p=0.02; Figure 4E).
3.7.5 Mutually exclusive mutations: EGFR mutations were entirely mutually exclusive with BRAF mutations, as well as nearly mutually exclusive with KRAS and NRAS mutations (in only one case, EGFR mutation co-occurred with mutation in either KRAS or NRAS; both p=0.001).
3.7.6 PD-L1 status and survival: High PD-L1 expression appeared as an adverse factor in both OS and PFS on univariate analysis (p=0.03 and 0.04, respectively) but this was shown to be due to a correlation with advanced stage, where PD-L1 was more often expressed (p=0.012). A multivariate Cox regression model controlled for age, gender, stage and smoking history, showed that PD-L1 status was not independently associated with OS or PFS.
3.7.7 Gene mutations and smoking: As mentioned earlier, KRAS G12C mutation was more commonly seen in smokers (p=0.04), with G12V being second most common. In non-smokers this order was reversed (Figure 3B). An inverse association was seen between EGFR mutation and smoking, with 50% of non-smokers harboring an EGFR mutation, in contrast to only 7% of smokers (p<0.001). Among patients with positive smoking history, EGFR mutation occurred in 18% of prior smokers, in contrast to only 2.7% of current smokers (p<0.001), and in 17% of patients with lower than 50 pack-year history, in contrast to only 2.5% of smokers with 50 or greater pack-year history (p=0.001). Considering the positive correlation of male gender with heavy smoking (greater than 50 pack-years; p=0.007) and a female non-smoker status with EGFR mutation (p<0.001), a relative survival advantage of women non-smokers was seen by Kaplan-Meier survival analysis (p=0.05; log rank). However, unlike male gender, such “female non-smoker” status was not an independent survival factor in a multivariate Cox Regression model with stage, TP53 mutation and KRAS/STK11 co-mutation.
3.7.8 PD-L1 expression, gene mutations and survival: High PD-L1 expression occurred significantly more commonly in tumors with mutations in the MAPK pathway (KRAS, BRAF or NRAS; p<0.001) as well as those with MET gene amplification (p=0.009). Similar trend was seen for tumors with KRAS/TP53 co-mutation, but did not reach significance (p=0.08). In contrast, PD-L1 expression was more commonly absent in tumors with STK11 mutations (p=0.009), even more so in those with KRAS/STK11 co-mutations (p=0.002).
4. Discussion
In the present study, we report mutational profiles and clinical outcomes of lung adenocarcinomas from patients residing in the rural parts of the state of Maine- a region of the US with a particularly high incidence of this disease. As could be expected from the clinical experience and established knowledge [19], clinical stage at presentation was a strong factor determining the outcome in our patients. In addition, several mutations and co-mutations were found to strongly affect survival independently of stage, both in the entire cohort overall, as well as in select stage subgroups. The 44.3% overall survival rate of our cohort within the studied time frame is comparable to the approximately 40% 2-year survival rate across all stages of LUAD in the national cancer databases [20]. TP53 mutations affecting DNA-binding and tetramerization domains are known to dysregulate the transactivation of TP53-dependent genes and promote tumorigenesis [21-22]. TP53 mutations in these two domains occurred in 41% of our patients and showed a strong, stage-independent prognostic effect on OS with a hazard ratio of 1.7. The stage-independent prognostic significance of these TP53 mutations was further demonstrated in a separate analysis of stage IV tumors, showing a similar hazard ratio for adverse OS. A high prevalence of TP53 mutations in our lung cancer population was not unexpected. TP53 mutation has been identified in a number of prior studies as consistently the most frequent mutation in LUAD - with a prevalence reported around 45%, ranging from mid-30% range to 50% of cases [23-24]. The adverse prognostic effect of TP53 mutations has been reported less consistently among studies, but has likewise not been an infrequent finding [25]. The TP53 mutations identified in our tumors were very diverse, with the same mutation only rarely seen in more than a single patient. This is also a common occurrence, as there is typically no consistent pattern in the spectrum of TP53 mutations, even in cancers of the same type [26]. Six point mutations (R175, G245, R248, R249, R273 and R282), seen in 14% of our cases, typically account for up to 30% of all mutations in human cancers [22]. Since they are located in the DNA binding region, these mutations disrupt the tumor suppressive activity of the p53 protein. Finding these mutations in our study is similar to that of Zahra et al. who utilized the same NGS platform used in our study, detecting 11% of TP53 mutations in their lung cancers in these “hotspots” [23]. TP53 mutations at seven residues (R157, R158, R179, G245, R248, R249 and R273) known to be susceptible to damage caused by benzopyrene diol epoxide, a potent cigarette smoke carcinogen, occurred in 25% of our mutated patients (all smokers), similar to what has been reported in lung cancer patients previously [27]. A TP53 transversion mutation at residue 249, which is rarely seen outside of radon exposure, only occurred in one of our patients, suggesting that radon exposure may not have played a major role in our cohort, although other studies have questioned the sensitivity of this mutation as a marker of radon exposure in LUAD [28]. The spectrum of KRAS mutations detected in our study was similar to previously reported lung cancer studies [29,30]. The most common mutations affected codon 12 (predominantly G12C and G12V), followed by codons 13, 61 and rarely others. The most common mutation types differed between smokers and non-smokers, similar to what was reported previously by Yu et al. [31]. While by themselves, KRAS mutations didn’t show an impact on prognosis, when co-occurring with other mutations, KRAS had a very significant prognostic impact. Co-mutation KRAS/STK11 identified an adverse group with 18% OS at 40 months, which contrasted with a 40% OS rate seen in patients without such co-mutation over the same timeframe. Similarly, KRAS/TP53 co-mutations delineated an adverse disease subset within the low-stage (I-II) cancers, when excluding the prognostically positive effect of EGFR mutation [32]. This difference was marked, with 65% of low-stage patients without such co-mutation being alive at 40 months, in contrast to only 30% of those with KRAS/TP53 co-mutation. In other words, less than half of the patients with these co-mutations achieved the overall survival seen in patients with the same stage of disease, who lacked these adverse mutational signatures. Prior reports have suggested existence of such molecular LUAD subsets, defined by the presence of co-existing mutations in the tumor, such as: the wild-type group, isolated TP53 group, KRAS group, KRAS/TP53 group and KRAS/STK11 group [33]. Others however, could not confirm the effect of such genomic co-alterations on survival [34]. Among the remaining mutations detected most frequently in our patients, of note is the relatively lower prevalence of EGFR mutations, compared to the literature typically reporting it to range anywhere between 17% to 52% [35]. These cumulative averages reflect data influenced by extrapolations from East Asian data, as well as that derived from populations with larger contribution of women and non-smokers [36]. In contrast, lung cancer patient populations more similar in composition to that of rural Maine (majority Caucasian of predominantly northern European/often German or Scandinavian ancestry, with a large proportion of smokers), have shown the prevalence of EGFR mutations closer to 10%, similar to our study [37,38]. In fact, even in The Cancer Genome Atlas (TCGA), the overall rate of EGFR mutations is only 14% [39]. The prevalence of other mutations in our cohort was similar to previous reports [40]. We observed that tumors with mutations affecting the MAPK pathway (KRAS, BRAF and NRAS), as well as those with MET gene amplification, more frequently show high expression of PD-L1, with a trend for KRAS/TP53 co-mutated tumors to be overexpressed. In contrast, tumors with STK11 mutations, as well as with KRAS/STK11 co-mutations were significantly less commonly PD-L1 positive in our experience. This supports the notion that the previously proposed co-mutation subtypes not only exist and vary in terms of biologic aggressiveness, but may also differ in their responsiveness to immunotherapy, as has been recently suggested [41,42]. From methodological standpoint, utilizing reflexive standing order for local molecular testing, initiated by the pathologist at the time when the diagnosis is established, resulted in the testing being completed in a short enough turn-around time for the results to be available at the time of the patient’s first encounter with the oncologist (generally 2 weeks after diagnosis). Such completion times are not only out of the reach of most send-out tests, but the reflexive procedure, which also includes obtaining sections for molecular testing during the initial sectioning, prevents loss of material typically associated with later procurement of sections for any subsequently requested testing. This in turn leads to a very low unsatisfactory rate for molecular testing (generally within single percentage range in our experience across many different tumor types).
In conclusion, our analysis of oncogenes and tumor suppressor genes in LUAD showed a distribution of mutations in rural Maine tumors to be similar to what has been reported from other regions of related ancestry, supporting the recently proposed subclassification of LUAD into different co-mutational subsets. Importantly, in this study we show such subclassification to be possible to accomplish by using a smaller gene panel in a regional oncology care setting, with the only pre-requisite being a sufficient coverage of the TP53, KRAS and STK11 genes. In particular, our study adds to the recently emerged data emphasizing the importance of detecting therapeutically and prognostically significant mutations in early stage tumors [43]. The recent FDA-approval of targeted therapies such as EGFR inhibitors for lung carcinomas of stages IB-IIIA provides a strong support for mutational testing of early stage lung tumors [44], and may constitute a tipping point for laboratories and hospitals to adopt similar reflexive molecular testing strategies as described herein. Our results contribute to the so far elusive efforts to develop risk stratification models for early stage lung cancer utilizing their molecular characteristics [45]. In comparison to previously published studies derived predominantly from academic institutions or large commercial laboratories, we show that NGS performed in a regional medical center setting yields molecular genetic information of equal value for patient risk stratification and management. While utilizing a smaller panel can prove to be a technical advantage in a regional community oncology setting due to its lower complexity, it also represents a limitation of our study. For example we could not assess the recently reported KEAP1/NFE2L2 pathway alterations, predicted to associate with therapy resistance and rapid progression [46]. Despite this limitation, the results we report complement the predominantly more urban data in the literature by providing region-specific mutational profiles from a geographically unique lung cancer population with high disease prevalence and a strong known risk factor, addressing what is often referred to as an “urban-rural disparity” in oncology. Identification of high-risk groups amenable to targeted intervention available in regional setting (such as targeting the specific G12C KRAS mutation most recently) [47] is essential for achieving sustainable improvement to rural cancer survival, which is especially true for lung cancer [48]. Our future work utilizing a larger mutation panel will allow further expansion of the current profiles and continue our efforts of mapping the molecular lung cancer landscape in our region.
Declarations
Funding
The authors did not receive any specific grant funding or financial support from funding agencies or organizations for the submitted work
Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose
Availability of data and material (Data Transparency)
All data is available per request
Code availability
Standard software for next generation sequencing available for review
Authors contributions
All authors contributed to the study conception and design. Material preparation, data collection, formal analysis and original draft preparation were performed by Laura Skacel, Michael Babcock and Marek Skacel. Data curation, conceptualization and editing were performed by Antoine Harb, Adam Curtis, Carter Liou and Kathleen Brown. Editing and supervision were performed by Frantisek Sandor. All authors read and approved the final manuscript.
Ethics approval
This study was approved by the Institutional Review Board of Northern Light Eastern Maine Medical Center, filed under number 19-1-A-001. All work was performed in accordance with the ethical standards as laid down by the 1964 Declaration of Helsinki and its later amendments. This study was retrospective in nature and was conducted on already available data and biologic materials obtained for routine diagnostic purposes.
References
- Lung and Bronchus Cancer- Cancer Stat Facts. Surveillance, Epidemiology, and End Results (SEER) Program, National Cancer Institute (NCI). Cancer stat facts: lung and bronchus cancer. Bethesda, MD: NCI, National Institutes of Health (2016).
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6 (2011): 244-285.
- Singh GK, Siahpush M, Williams SD. Changing urbanization patterns in US lung cancer mortality 1950-2007. J Community Health 37 (2012): 412-420.
- Atkins GT, Kim T, Munson J. Residence in rural areas of the United States and lung cancer mortality: Disease incidence, treatment disparities, and stage-specific survival. Ann Am Thorac Soc 14 (2017): 403-411.
- Roberts ME, Doogan NJ, Kurti AN, et al. Rural tobacco use across the United States: How rural and urban areas differ, broken down by census regions and divisions. Health Place 39 (2016): 153-159.
- State Cancer Profiles (2018).
- Maine | American Lung Association (2018).
- Farah C, Beard K, Hess CT, et al. Analyzing spatial and temporal 222Rn trends in Maine. Health Phys 102 (2012): 115-123.
- Ivanov M, Laktionov K, Breder V, et al. Towards standardization of next-generation sequencing of FFPE samples for clinical oncology: Intrinsic obstacles and possible solutions. J Transl Med 15 (2017): 22.
- Sholl L. Molecular diagnostics of lung cancer in the clinic. Transl Lung Cancer Res (2017).
- Miller TE, Yang M, Bajor D, et al. Clinical utility of reflex testing using focused next-generation sequencing for management of patients with advanced lung adenocarcinoma. J Clin Pathol (2018).
- Saslow D, Solomon D, Lawson HW, et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology Screening Guidelines for the Prevention and Early Detection of Cervical Cancer. Am J Clin Pathol 137 (2012): 516-542.
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (8th) edtn, TNM Classification for Lung Cancer. J Thorac Oncol 11 (2016): 39-51.
- McCall CM, Mosier S, Thiess M, et al. False positives in multiplex PCR-based next-generation sequencing have unique signatures. J Mol Diagn 16 (2014): 541-549.
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of Non-Small-Cell Lung Cancer. N Engl J Med 372 (2015): 2018-2028.
- Org N. NCCN Clinical Practice guidelines in Oncology (NCCN Guidelines®) NCCN evidence blocks TM Non-Small Cell Lung Cancer (2020).
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2 (2012): 401-404.
- Bouaoun L, Sonkin D, Ardin M, et al. TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum Mutat 37 (2016): 865-876.
- Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Heal 85 (2019).
- SEER* Explorer Application (2021).
- Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem 77 (2008): 557-582.
- Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev (1993): 2556-2564.
- Shajani-Yi Z, de Abreu FB, Peterson JD, et al. Frequency of somatic TP53 mutations in combination with known pathogenic mutations in colon adenocarcinoma, non-small cell lung carcinoma, and gliomas as identified by next-generation sequencing. Neoplasia 20 (2018): 256-262.
- Jao K, Tomasini P, Kamel-Reid S, et al. The prognostic effect of single and multiple cancer-related somatic mutations in resected non-small-cell lung cancer. Lung Cancer 123 (2018): 22-29.
- Ludovini V, Pistola L, Gregorc V, et al. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: A study of the perugia multidisciplinary team for thoracic oncology. J Thorac Oncol 3 (2008): 365-373.
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2(2010): a001008.
- Menzies GE, Reed SH, Brancale A, et al. Base damage, local sequence context and TP53 mutation hotspots: A molecular dynamics study of benzo[a]pyrene induced DNA distortion and mutability. Nucleic Acids Res (2015).
- McDonald JW, Taylor JA, Watson MA, et al. p53 and K-ras in radon-associated lung adenocarcinoma. Cancer Epidemiol Biomarkers Prev 4 (1995): 791-793.
- Román M, Baraibar I, López I, et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 17 (2018): 33.
- Yu, Helena A, Planchard D, et al. Sequencing therapy for gentically defined subgroups of non-small cell lung cancer 12 (2020): 726-739.
- Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol 10 (2015): 431.
- Stanic K, Turnsek N, Vrankar M. Incorporation of EGFR mutation status into M descriptor of new TNM classification influences survival curves in non-small cell lung cancer patients. Radiol Oncol 53 (2019): 453-458.
- La Fleur L, Falk-Sörqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 130 (2019): 50-58.
- Gibert J, Clavé S, Hardy-Werbin M, et al. Concomitant genomic alterations in KRAS mutant advanced lung adenocarcinoma. Lung Cancer 140 (2020): 42-45.
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists. International association for the study of lung cancer, and association for molecular pathology. Arch Pathol Lab Med 137 (2013): 828-860.
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in Lung Cancers. J Natl Cancer Inst 97(2005): 339-346.
- Gahr S, Stoehr R, Geissinger E, et al. EGFR mutational status in a large series of Caucasian European NSCLC patients: data from daily practice. Br J Cancer 109 (2013): 1821-1828.
- Helland Å, Skaug HM, Kleinberg L, et al. EGFR gene alterations in a Norwegian cohort of lung cancer patients selected for surgery. J Thorac Oncol 6 (2011): 947-950.
- Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45 (2013): 1113-1120.
- Mäki-Nevala S, Sarhadi VK, Rönty M, et al. Hot spot mutations in Finnish non-small cell lung cancers. Lung Cancer 99 (2016): 102-110.
- Serra P, Petat A, Maury J-M, et al. Programmed cell death-ligand 1 (PD-L1) expression is associated with RAS/TP53 mutations in lung adenocarcinoma. Lung Cancer 118 (2018): 62-68.
- Bange E, Marmarelis ME, Hwang WT, et al. Impact of KRAS and TP53 co-mutations on outcomes after first-line systemic therapy among patients with STK11-mutated advanced non-small-cell lung cancer. JCO Precis Oncol 3 (2019).
- Kneuertz PJ, Carbone DP, D’Souza DM, et al. Prognostic value and therapeutic implications of expanded molecular testing for resected early stage lung adenocarcinoma. Lung Cancer 143 (2020): 60-66.
- Herbst RS, Tsuboi M, John T, et al. Osimertinib as adjuvant therapy in patients (pts) with stage IB-IIIA EGFR mutation positive (EGFRm) NSCLC after complete tumor resection: ADAURA. J Clin Oncol 38 (2020): LBA5-LBA5.
- Kratz JR, Haro GJ, Cook NR, et al. Incorporation of a molecular prognostic classifier improves conventional Non-Small Cell Lung Cancer staging. J Thorac Oncol 14 (2019): 1223-1232.
- Goeman F, De Nicola F, Scalera S, et al. Mutations in the KEAP1-NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J Thorac Oncol 14 (2019): 1924-1934.
- Molina-Arcas M, Moore C, Rana S, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci Transl Med 11 (2019): 11-21.
- Dalwadi S, Teh BS, Bernicker E, et al. Community-based disparities in the treatment and outcomes of early-stage Non-small-cell Carcinoma. Cureus 11 (2019): e5889.