LDBPR: Latest Database of Protein Research
Article Information
Shahid Ullah2*#, Tianshun Gao1#, Wajeeha Rahman2, Farhan Ullah2, Riffat Jahan2, Anees Ullah3, Gulzar Ahmad2, Muhammad Ijaz2, Yihang Pan1*
1Research Center, The Seventh Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangzhou, China
2S Khan Lab Mardan, Khyber Pakhtunkhwa, Pakistan
3Kyrgyz State Medical University, Bishkek, Kyrgyzstan
*Corresponding author: Yihang Pan, Research Center, The Seventh Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangzhou, China; Shahid Ullah, S Khan Lab Mardan, Khyber Pakhtunkhwa, Pakistan.
Received: 03 February 2022; Accepted: 15 February 2022; Published: 21 February 2022
Citation: Shahid Ullah, Tianshun Gao, Wajeeha Rahman, Farhan Ullah, Riffat Jahan, Anees Ullah, Gulzar Ahmad, Muhammad Ijaz, Yihang Pan. LDBPR: Latest Database of Protein, Research. Journal of Bioinformatics and Systems Biology 5 (2022): 34-44.
View / Download Pdf Share at FacebookAbstract
With the vast and rapid growth of protein research data, a large number of databases are produced to annotate proteins. How to use these databases is becoming a crucial part of modern biology. Database research is usually the first step in the analysis of a new protein. The combined utilization of multiple databases could help researchers to understand the evolution, structure, and function of proteins. Therefore, a well comprehensive and large-scale resource integrated with most of databases is urgently desirable for systematic and precise studies of proteins. Here we designed a platform LDBPR with a collection of 564 latest scientific protein databases. It fully covered physical, chemical, and biological information of Protein sequence, structure, and model, domain, function, and protein-protein interactions. Furthermore, The LDBPR can be explored by three ways: (i) single database can be browsed by typing the name in the given search bar; (ii) all protein categories can be browsed by clicking on the name of the category; (iii) the image icon, could give all categorized protein databases on single click. Moreover, the programming languages including PHP, HTML, CSS, and MySQL were used to construct LDBPR for the protein scientific community that can be freely searched by clicking http://www.habdsk.org/ldbpr.php and will be updated timely.
Keywords
HABDSK; LDBPR; MySQL; Protein; Protein‐protein interactions; Sequence
HABDSK articles; LDBPR; MySQL articles; Protein articles; Protein?protein interactions articles; Sequence articles
HABDSK articles HABDSK Research articles HABDSK review articles HABDSK PubMed articles HABDSK PubMed Central articles HABDSK 2023 articles HABDSK 2024 articles HABDSK Scopus articles HABDSK impact factor journals HABDSK Scopus journals HABDSK PubMed journals HABDSK medical journals HABDSK free journals HABDSK best journals HABDSK top journals HABDSK free medical journals HABDSK famous journals HABDSK Google Scholar indexed journals LDBPR articles LDBPR Research articles LDBPR review articles LDBPR PubMed articles LDBPR PubMed Central articles LDBPR 2023 articles LDBPR 2024 articles LDBPR Scopus articles LDBPR impact factor journals LDBPR Scopus journals LDBPR PubMed journals LDBPR medical journals LDBPR free journals LDBPR best journals LDBPR top journals LDBPR free medical journals LDBPR famous journals LDBPR Google Scholar indexed journals MySQL articles MySQL Research articles MySQL review articles MySQL PubMed articles MySQL PubMed Central articles MySQL 2023 articles MySQL 2024 articles MySQL Scopus articles MySQL impact factor journals MySQL Scopus journals MySQL PubMed journals MySQL medical journals MySQL free journals MySQL best journals MySQL top journals MySQL free medical journals MySQL famous journals MySQL Google Scholar indexed journals Protein articles Protein Research articles Protein review articles Protein PubMed articles Protein PubMed Central articles Protein 2023 articles Protein 2024 articles Protein Scopus articles Protein impact factor journals Protein Scopus journals Protein PubMed journals Protein medical journals Protein free journals Protein best journals Protein top journals Protein free medical journals Protein famous journals Protein Google Scholar indexed journals Protein?protein interactions articles Protein?protein interactions Research articles Protein?protein interactions review articles Protein?protein interactions PubMed articles Protein?protein interactions PubMed Central articles Protein?protein interactions 2023 articles Protein?protein interactions 2024 articles Protein?protein interactions Scopus articles Protein?protein interactions impact factor journals Protein?protein interactions Scopus journals Protein?protein interactions PubMed journals Protein?protein interactions medical journals Protein?protein interactions free journals Protein?protein interactions best journals Protein?protein interactions top journals Protein?protein interactions free medical journals Protein?protein interactions famous journals Protein?protein interactions Google Scholar indexed journals Sequence articles Sequence Research articles Sequence review articles Sequence PubMed articles Sequence PubMed Central articles Sequence 2023 articles Sequence 2024 articles Sequence Scopus articles Sequence impact factor journals Sequence Scopus journals Sequence PubMed journals Sequence medical journals Sequence free journals Sequence best journals Sequence top journals Sequence free medical journals Sequence famous journals Sequence Google Scholar indexed journals biological information articles biological information Research articles biological information review articles biological information PubMed articles biological information PubMed Central articles biological information 2023 articles biological information 2024 articles biological information Scopus articles biological information impact factor journals biological information Scopus journals biological information PubMed journals biological information medical journals biological information free journals biological information best journals biological information top journals biological information free medical journals biological information famous journals biological information Google Scholar indexed journals biology articles biology Research articles biology review articles biology PubMed articles biology PubMed Central articles biology 2023 articles biology 2024 articles biology Scopus articles biology impact factor journals biology Scopus journals biology PubMed journals biology medical journals biology free journals biology best journals biology top journals biology free medical journals biology famous journals biology Google Scholar indexed journals
Article Details
1. Introduction
To deposit the precious protein information for easy retrieving, a handful of databases such as such as “SCOP” [1], “HAMAP” [2], “Plprot” [3], “AHD” [4], “STRING” [5], and “PRIDE” [6] have been designed. These databases were mainly focused on structure, sequence, model, pathway, Protein-Protein and other Interaction (PP&OI), and expression respectively and provided comprehensive information of the proteins for the protein research community. Meanwhile, there are also a lot of well-known animal and plant databases [7] including “HMDB” [8], “P3DB” [9], “PhytAMP” [10], “Nextprot” [11], “TSTMP” [12], and “dbPAF” [13], which focused on special species. Especially, a number of articles are published based on a collection of a small number of databases simply listed in a table and didn’t construct an online website to display the compensative features for the research object (Table 1). These studies showed low coverage of category, and some of them are specific for special organisms (e.g., mouse. human, or plant). Thus, a well comprehensive and large-scale database is needed for further studies of proteins. Here we integrate a collection of the latest scientific protein data raised from physical, chemical and biological information of Protein sequence, structure, Modal, domain, function, and protein-protein interactions. These data cannot be managed without computational databases [14, 15], which become a crucial part of modern biology. Some widely known protein databases are far from being fully used by the protein scientific community. Therefore, we provided a starting point to explore the potential of all protein databases on the internet by presenting a friendly and easy searching platform. We will also update the protein information with the passage of time.
|
PMID |
YEAR |
CATEGORY |
FORM OF |
DB. NO |
JOURNAL NAME |
|
LDBPR |
2022 |
Protein |
DB+Table |
564 |
|
|
25712261 |
2015 |
Human |
Table |
74 |
Genomic, Proteomic Bioinformatics (GPB) |
|
18265344 |
2012 |
Protein |
Table |
121 |
Current Protocols in Molecular Biology |
|
16381921 |
2006 |
Pathway |
Database |
190 |
Nucleic Acids Research (NAR) |
|
7764641 |
1994 |
DNA+ Protein |
Table |
50 |
Current Opinion in Biotechnology |
|
31906604 |
2020 |
Nucleic acid |
Table |
70 |
Nucleic Acids Research (NAR) |
Table 1: Comparison table of the LDBPR with other published work.
2. Material and Method
2.1. Database construction and content
2.1.1. Construction of LDBPR
We integrated the data from four well-known resources including PubMed, Google, Google Scholar, and Web of Science. Multiple keywords such as “Protein database”, “Protein databases”, “protein database list”, “database of protein”, “databases of protein”, and “list of protein databases” were searched to retrieve published protein related databases with PubMed ids (http://www.ncbi.nlm.nih.gov/pubmed). To circumvent missing data, we have manually collected the latest protein databases from Nucleic Acids Research journal (NAR) (https://academic.oup.com/nar), and journal of Genomics, Proteomics & Bioinformatics (GPB) (https://www.journals.elsevier.com/genomics-proteomics-and-bioinformatics), which are the leading edge research journals on database issue. We only collected all available protein databases and removed all broken links. Programming languages such as PHP, MYSQL, HTML, CSS, and JavaScript were used to construct LDBPR. Finally, our database is easy for operation and updating (Figure 1).
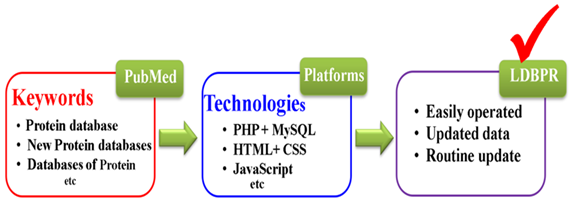
Figure 1: Procedure for the collection of protein databases in LDBPR.
2.2. Content of the LDBPR
2.2.1. Proteins databases classification
several projects [16-18] made their own special classifications of protein databases on the base of the function, application, some technical features, or a special organism such as human, mouse [17], or drosophila [19] and so on. According to the classifications in these projects, we classified all the protein databases into 6 categories, which are protein model database, protein structure database, protein sequence database, protein-protein interaction database, protein expression database, and protein pathway database in LDBPR. We have previously provided some databases of different research area like DBHR: database for human research [20] (Figure 2A), Co-19 PDB: About COVID-19 [21] (Figure 2B), and DBPR: database of plant research [7] (Figure 2C).
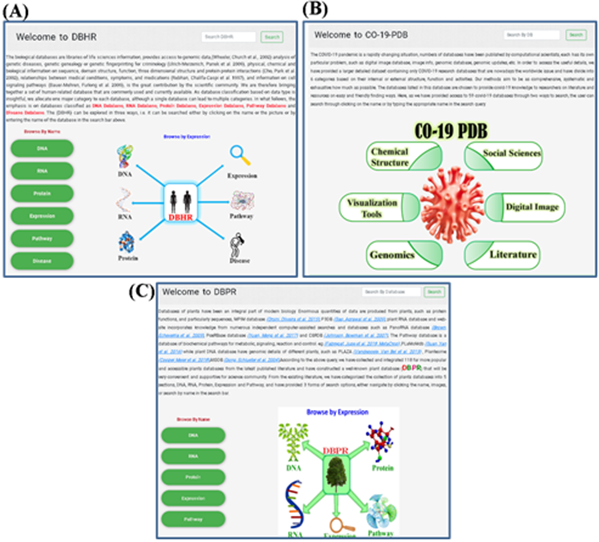
Figure 2: The screenshots of some relevant databases. (A) Database relevant to human research. (B) Covid-19 relevant database. (C) Plant related database.
2.2.2. Protein model databases
Protein model databases provided the protein three-dimensional structure on the base of predication from its amino acids or primary structure [22], which could help discover the most important targets soughed by bioinformatics and theoretical chemistry [23]. In addition, the protein model is of great significance in the field of medicine (e.g., drug design), while it is the development of novel enzymes in the field of biotechnology [24]. A lot of well-known databases were built in this field, such as “PMDB” [25], and “MODELLER” [26]. we have collected totally 27 protein model databases.
2.2.3. Protein structure databases
In this classification, the databases contained a large number of experimental determinations for protein structures, and aimed to organize and annotate useful protein information [16] including unit cell dimensions and angles for structures determined by x-ray crystallography, and structure-based drug design that is the deep study about the function of the proteins [27], e.g., “PDB” [28], “PDBTM” [29], “P3DB” [9], etc.
2.2.4. Protein sequence databases
Protein sequence databases were developed for a large collection of mass-spectrometry based proteomics data [30] including protein sequences [31], post-translation modification [32], and sequence alignment [33]. They are not only the simple sequence databases but also provide a rich annotation from other known research results for proteins. As far as we know, different databases (e.g., “Proteome db” [34], “Uniprot” [35], “dbPSP” [36]) annotated sequences of proteins with different levels [16].
2.2.5. Protein–protein interaction (PPI) databases
Protein-Protein Interactions (PPIs) usually involve two or more protein molecules and could be considered as high-specific physical contacts induced by a result of biochemical events including electrostatic forces, hydrogen bonding, and hydrophobic influence occurring in a cell or a living organism [37]. Since PPIs annotate proteins in a large-scale level, much more specialized databases in this classification are designed to provide complete interactomes [38]. Some typical examples like “DIP” [39], “Biogrid” [40], and “STRING-db” [41] have been widely used as reliable references for PPI analysis.
2.2.6. Protein Expression databases
The protein expression databases integrated data of protein expression from microarray and allowed users to search proteins by gene name, splice variant, protein attribute, disease, treatment, or organism part that is a form of metadata manually curated and analyzed through standard analysis pipelines [42]. For example, the database Expression Atlas provides information about protein expression in animal and plant samples of different cell types, organism parts, developmental stages, diseases and other conditions [43].
2.2.7. Protein pathway database
Pathway diagrams are the roadmaps for molecular biology and could illustrate the connections between genes, proteins, and metabolites [44]. A well-illustrated pathway database should provide a biological context to complex molecular processes in an easily understood and highly visual manner. In this regard, the pathway databases we collected provide remarkably useful information for scientists to share, integrate, interpret, and visualize “omics data” and “omics measurements”. In this category, two databases “PathBank” [44] and “KEEG” [45] have been widely used in analysis of biological pathways.
3. Result and Discussion
3.1. Database statistics
In the current work, we have provided almost all protein databases (Table S1) and shown the category-wise, chronological order, and percentile development in LDBPR. Figure 3A showed the percentile of the proteins databases. Figure 3B displayed the chronological order of the category, while Figure 3C presented the category-wise growth distribution of protein database, indicating the tremendous growth and achievement for the protein scientific community. Furthermore, we have deleted all the broken and non-accessible database links and provided a new and updated protein database in the form of a database named LDBPR as well as a table (Table S1).
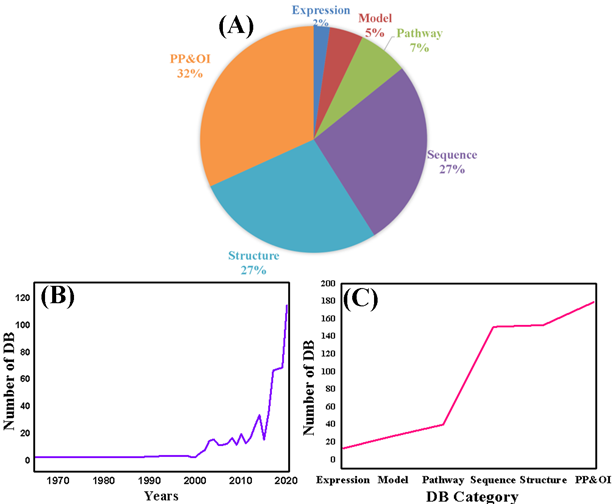
Figure 3: The statistics data of LDBPR. (A) Distribution of the database category. (B) Chronological order of the LDBPR. (C) Category-wise growth of the DBHR.
3.2. Usage of the LDBPR database
The (LDBPR) is developed in an easy and friendly searching way. For easier and faster search, three options are provided for accessing protein databases. First, users can browse LDBPR by clicking on the name of the category (Figure 4A), or image expression (Figure 4B) linking to the category list page (Figure 4C), or a brief overview with the original link. Users can access the database of interest by simply clicking the database name. Furthermore, to advance specific database search, users can also type the name of the database in the search bar (Figure 4D). Here, we used the “PathBank” database as an example from the Disease Databases to display the search process.
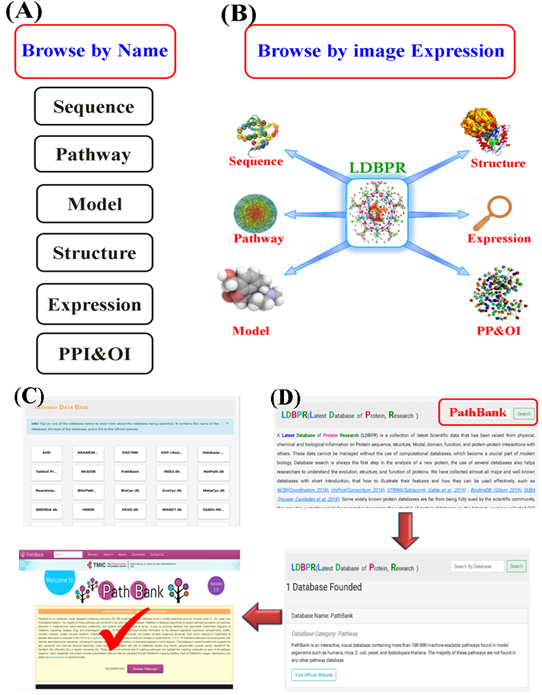
Figure 4: The browse options of the LDBPR. (A) Browsing by clicking the name. (B) Browsing by image expression. (C) Browsing database name in the search bar. (D) A browsing example of the final result.
4. Conclusion
A useful biological database should provide facilities for storing, organizing, and retrieving biological data such as DNR, RNA, carbohydrate, protein, and so on. It should also be easily viewed, managed, and modified. Although hundreds of databases have been constructed in protein research field, and have their own classifications of protein features like sequence, function, structure and pathway, there is still a lack of research community to effectively manage these resources and give a comprehensive annotation for all proteins. Hence, we collected 564 protein related databases and divided them into 6 categories based on protein model, structure, sequence, protein-protein interaction, expression, and pathway. Furthermore, we added a short introduction for nearly each protein database and kept updating for them. Our database can be searched in an easy and friendly way by clicking on category name, image expression, or database name in the given search bar.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Availability of data and materials
These data will be available under the journal rule and regulation
Competing interests
The authors don’t have any compete of interests.
Funding
This project is supported by National Natural Science Foundation of China (32100434) and Research Start-up Fund of the Seventh Affiliated Hospital of Sun Yat-sen University (ZSQYBRJH0020).
Authors' contributions
Dr. Shahid Ullah designed and supervised the project with Prof. Yihang Pan's assistance. Dr. Tianshun Gao worked as a co-first author performed data analysis. Farhan Ullah, Wajeeha Rahman, Muhmmad Ijaz Gulzar Ahmad, Riffat Jahan and Dr. Anees Ullah contributed to data analysis. Shahid Ullah wrote the manuscript. All authors reviewed the manuscript.
Acknowledgement
To avoid future conflict and plagiarism issue, LDBPR database is uploaded on https://habdsk.org/ldbpr.phpso that we have provided some contents in this article.
Supplementary Information
Download the supplementary information from the below link
https://www.fortunejournals.com/supply/JBSB_5004s.pdf
References
- Andreeva A, Howorth D, Brenner SE, et al. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic acids research 32 (2004): 226-229.
- Pedruzzi I, Rivoire C, Andre HA, et al. HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic acids research 43 (2015): 1064-1070.
- Kleffmann T, Matthias HH, Gruissem W, et al. plprot: A comprehensive proteome database for different plastid types. Plant and cell physiology 47 (2006): 432-436.
- Jiang, Z, Liu X, Peng Z, et al. AHD2. 0: an update version of Arabidopsis Hormone Database for plant systematic studies. Nucleic acids research 39 (2010): 1123-1129.
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic acids research 45 (2016): 362-368.
- Vizcaíno JA, Csordas A, Del-Toro N, et al., 2016 update of the PRIDE database and its related tools. Nucleic acids research 44 (2016): 447-456.
- Ullah S, Rahman W, Ullah F, et al. DBPR: Data Base of Plant Research (2021).
- Wishart DS, Feunang YD, Marcy A, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic acids research 46 (2018): 608-617.
- Ya Q, Ge H, Wu S, et al. P3DB 3.0: from plant phosphorylation sites to protein networks. Nucleic Acids Research 42 (2014): 1206-1213.
- Hammami R, Hamida JB, Vergoten G, et al. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic acids research 37 (2009): 963-968.
- Gaudet P, Michel PA, Monique ZZ, et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic acids research 45 (2017): 177-182.
- Varga J, Dobson L, Remenyi I, et al. TSTMP: target selection for structural genomics of human transmembrane proteins. Nucleic acids research 45 (2017): 325-330.
- Ullah S, Lin S, Xu Y, et al. dbPAF: an integrative database of protein phosphorylation in animals and fungi. Scientific reports 6 (2016): 23534.
- KhanSA JM. Tremendous Contribution of Dr. Shahid Ullah to Scientific Community during COVID-19 Pandemic in the Form of Scientific Research. J Clin Med Res 2 (2020): 1-7.
- Ullah S, Ullah F, Rahaman W, et al. EDBCO-19: Emergency Data Base of COVID-19. J Clin Med Res 2 (2020): 1-4.
- Xu D. Protein databases on the internet. Current protocols in protein science 70 (2012).
- Celis JE, Ostergaard M, Jensen NA, et al. Human and mouse proteomic databases: novel resources in the protein universe. FEBS letters 430 (1998): 64-72.
- Harper R. Access to DNA and protein databases on the Internet. Current Opinion in Biotechnology 5 (1994): 4-18.
- Sanchez C, Lachaize C, Janody F, et al. Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database. Nucleic acids research 27 (1999): 89-94.
- Ullah S, Rahman W, Ullah F, et al. DBHR: a collection of databases relevant to human research. Future Science OA (2022): 780.
- Ullah S, Ullah A, Rahman W, et al, An innovative user-friendly platform for Covid-19 pandemic databases and resources. Computer Methods and Programs in Biomedicine Update 1 (2021): 100031.
- Deng H, Jia Y, Zhang Y. Protein structure prediction. International Journal of Modern Physics B 32 (2018): 1840009.
- Sliwoski G, Kothiwale S, Meiler J, et al. Computational methods in drug discovery. Pharmacological reviews 66 (2014): 334-395.
- Khan S, Ullah MW, Siddique R, et al. Role of recombinant DNA technology to improve life. International journal of genomics 2016 (2016): 2405954.
- Tiziana C, Cozetto D, Talamo IG, et al. The PMDB protein model database. Nuclic Acid Research 34 (2005): 306-309.
- Webb B, A Sali. Comparative protein structure modeling using MODELLER. Current protocols in bioinformatics 54 (2016): 1-5.
- Smyth M, Martin J. X Ray crystallography. Molecular Pathology 53 (2000): 8.
- Burley SK, Berman HM, Kleywegt GJ, et al. Protein Data Bank (PDB): the single global macromolecular structure archive, in Protein Crystallography. Springer 2017 (2017): 627-641.
- Kozma D, Simon I, Tusnady GE. PDBTM: Protein Data Bank of transmembrane proteins after 8 years. Nucleic acids research 41 (2012): 524-529.
- Chen C, Hou J, Tanner JJ, et al. Bioinformatics methods for mass spectrometry-based proteomics data analysis. International Journal of Molecular Sciences 21 (2020): 2873.
- Dayhoff MO. Atlas of protein sequence and structure. National Biomedical Research Foundation 4 (1966): 262-263.
- Bond A, Row P, Dudley E. Post-translation modification of proteins; methodologies and applications in plant sciences. Phytochemistry 72 (2011): 975-996.
- Rost B. Twilight zone of protein sequence alignments. Protein engineering 12 (1999): 85-94.
- Lippert D, Yuen M, Bohlmann J. Spruce proteome DB: a resource for conifer proteomics research. Tree genetics & genomes 5 (2009): 723-727.
- Consortium U. UniProt: the universal protein knowledgebase. Nucleic acids research 46 (2018): 2699.
- Shi Y, Zhang Y, Lin S, et al. dbPSP 2.0, An updated database of protein phosphorylation sites in prokaryotes. Scientific Data 7 (2020): 1-9.
- De Las Rivas J, Fontanillo C. Protein–protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol 6 (2010): 1000807.
- Zahiri J, Hannon Bozorgmehr J, Masoudi-Nejad A. Computational prediction of protein–protein interaction networks: algorithms and resources. Current genomics 14 (2013): 397-414.
- Salwinski L, Miller CS, Smith AJ, et al. The database of interacting proteins: 2004 update. Nucleic acids research 32 (2004): 449-451.
- Chatr-Aryamontri A, Rose O, Boucher L, et al. The BioGRID interaction database: 2017 update. Nucleic acids research 45 (2017): 369-379.
- Szklarczyk D, Gabble AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research 47 (2019): 607-613.
- Chen C, Huang H, Wu CH. Protein bioinformatics databases and resources, in Protein Bioinformatics. Springer 2017 (2017): 3-39.
- Petryszak R, Burdett T, Fiorelli B, et al. Expression Atlas update-a database of gene and transcript expression from microarray-and sequencing-based functional genomics experiments. Nucleic acids research 42 (2014): 926-932.
- Wishart DS, Li C, Marcu A, et al. PathBank: a comprehensive pathway database for model organisms. Nucleic acids research 48 (2020): 470-478.
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research 45 (2017): 353-361.
