Involvement of the Activation Loop of ERK in the Detachment from Cytosolic Anchoring
Article Information
Ido Wolf#, Hadara Rubinfeld#, Seunghee Yoon, Goldie Marmor, Tamar Hanoch, Rony Seger*
Department of Immunology and Regenerative Biology, The Weizmann Institute of Science, Rehovot, 7610001, Israel
# equal contribution
*Corresponding author: Rony Seger, Department of Immunology and Regenerative Biology, The Weizmann Institute of Science, Rehovot, 7610001, Israel.
Received: 29 March 2023; Accepted: 13 April 2023; Published: 02 May 2023
Citation: Ido Wolf, Hadara Rubinfeld, Seunghee Yoon, Goldie Marmor, Tamar Hanoch, Rony Seger. Involvement of the Activation Loop of ERK in the Detachment from Cytosolic Anchoring. Journal of Biotechnology and Biomedicine 6 (2023): 140-150.
View / Download Pdf Share at FacebookAbstract
The Extracellular signal-regulated kinases (ERKs) are translocated into the nucleus in response to mitogenic stimulation. The mechanism of translocation and the residues in ERKs that govern this process were not fully elucidated yet. Here we studied the involvement of residues in the activation loop of ERK2 in determining its subcellular localization. Substitution of residues in the activation loop to alanines indicated that residues 173-181 do not play a significant role in the phosphorylation and activation of ERK2. However, residues 176-181 are responsible for the detachment of ERK2 from MEK1 upon mitogenic stimulation. This dissociation can be mimicked by substitution of residues 176-178 to alanines, and is prevented by deletion of these residues or by substitution of residues 179-181 to alanines. On the other hand, residues 176-181, as well as residues essential for ERK2-dimerization, do not play a role in the shuttle of ERK2 through nuclear pores. Thus, phosphorylation-induced conformational rearrangement of residues in the activation loop of ERK2 plays a major role in the control of subcellular localization of this protein.
Keywords
ERK; MEK; MAPK; Anchor proteins
ERK articles ERK Research articles ERK review articles ERK PubMed articles ERK PubMed Central articles ERK 2023 articles ERK 2024 articles ERK Scopus articles ERK impact factor journals ERK Scopus journals ERK PubMed journals ERK medical journals ERK free journals ERK best journals ERK top journals ERK free medical journals ERK famous journals ERK Google Scholar indexed journals MEK articles MEK Research articles MEK review articles MEK PubMed articles MEK PubMed Central articles MEK 2023 articles MEK 2024 articles MEK Scopus articles MEK impact factor journals MEK Scopus journals MEK PubMed journals MEK medical journals MEK free journals MEK best journals MEK top journals MEK free medical journals MEK famous journals MEK Google Scholar indexed journals MAPK articles MAPK Research articles MAPK review articles MAPK PubMed articles MAPK PubMed Central articles MAPK 2023 articles MAPK 2024 articles MAPK Scopus articles MAPK impact factor journals MAPK Scopus journals MAPK PubMed journals MAPK medical journals MAPK free journals MAPK best journals MAPK top journals MAPK free medical journals MAPK famous journals MAPK Google Scholar indexed journals Anchor proteins articles Anchor proteins Research articles Anchor proteins review articles Anchor proteins PubMed articles Anchor proteins PubMed Central articles Anchor proteins 2023 articles Anchor proteins 2024 articles Anchor proteins Scopus articles Anchor proteins impact factor journals Anchor proteins Scopus journals Anchor proteins PubMed journals Anchor proteins medical journals Anchor proteins free journals Anchor proteins best journals Anchor proteins top journals Anchor proteins free medical journals Anchor proteins famous journals Anchor proteins Google Scholar indexed journals Cytosolic retention sequence articles Cytosolic retention sequence Research articles Cytosolic retention sequence review articles Cytosolic retention sequence PubMed articles Cytosolic retention sequence PubMed Central articles Cytosolic retention sequence 2023 articles Cytosolic retention sequence 2024 articles Cytosolic retention sequence Scopus articles Cytosolic retention sequence impact factor journals Cytosolic retention sequence Scopus journals Cytosolic retention sequence PubMed journals Cytosolic retention sequence medical journals Cytosolic retention sequence free journals Cytosolic retention sequence best journals Cytosolic retention sequence top journals Cytosolic retention sequence free medical journals Cytosolic retention sequence famous journals Cytosolic retention sequence Google Scholar indexed journals Epidermal growth factor articles Epidermal growth factor Research articles Epidermal growth factor review articles Epidermal growth factor PubMed articles Epidermal growth factor PubMed Central articles Epidermal growth factor 2023 articles Epidermal growth factor 2024 articles Epidermal growth factor Scopus articles Epidermal growth factor impact factor journals Epidermal growth factor Scopus journals Epidermal growth factor PubMed journals Epidermal growth factor medical journals Epidermal growth factor free journals Epidermal growth factor best journals Epidermal growth factor top journals Epidermal growth factor free medical journals Epidermal growth factor famous journals Epidermal growth factor Google Scholar indexed journals basic protein articles basic protein Research articles basic protein review articles basic protein PubMed articles basic protein PubMed Central articles basic protein 2023 articles basic protein 2024 articles basic protein Scopus articles basic protein impact factor journals basic protein Scopus journals basic protein PubMed journals basic protein medical journals basic protein free journals basic protein best journals basic protein top journals basic protein free medical journals basic protein famous journals basic protein Google Scholar indexed journals Nuclear export signal articles Nuclear export signal Research articles Nuclear export signal review articles Nuclear export signal PubMed articles Nuclear export signal PubMed Central articles Nuclear export signal 2023 articles Nuclear export signal 2024 articles Nuclear export signal Scopus articles Nuclear export signal impact factor journals Nuclear export signal Scopus journals Nuclear export signal PubMed journals Nuclear export signal medical journals Nuclear export signal free journals Nuclear export signal best journals Nuclear export signal top journals Nuclear export signal free medical journals Nuclear export signal famous journals Nuclear export signal Google Scholar indexed journals Mitogen-Activated articles Mitogen-Activated Research articles Mitogen-Activated review articles Mitogen-Activated PubMed articles Mitogen-Activated PubMed Central articles Mitogen-Activated 2023 articles Mitogen-Activated 2024 articles Mitogen-Activated Scopus articles Mitogen-Activated impact factor journals Mitogen-Activated Scopus journals Mitogen-Activated PubMed journals Mitogen-Activated medical journals Mitogen-Activated free journals Mitogen-Activated best journals Mitogen-Activated top journals Mitogen-Activated free medical journals Mitogen-Activated famous journals Mitogen-Activated Google Scholar indexed journals mechanism articles mechanism Research articles mechanism review articles mechanism PubMed articles mechanism PubMed Central articles mechanism 2023 articles mechanism 2024 articles mechanism Scopus articles mechanism impact factor journals mechanism Scopus journals mechanism PubMed journals mechanism medical journals mechanism free journals mechanism best journals mechanism top journals mechanism free medical journals mechanism famous journals mechanism Google Scholar indexed journals
Article Details
Abbreviations:
CD: Common docking; CRS: Cytosolic retention sequence; EGF: Epidermal growth factor; ERK: Extracellular signal-regulated kinase; MAPK: Mitogen-activated protein kinase; MBP: Myelin basic protein; MEK: MAPK/ERK kinase; NES: Nuclear export signal; TPA: 12-O-tetradecanoylphorbol-13-acetate
Introduction
Key steps in the signaling mechanism of Mitogen-Activated Protein Kinase (MAPK) cascades are the changes in subcellular localization of their components upon extracellular stimulation (reviewed in [1-4]). These relocalization processes are best characterized for protein kinases in the extracellular signal-regulated kinase (ERK) cascade, which seem to be localized primarily in the cytoplasm of resting cells [5]. Upon mitogenic stimulation, Raf is recruited to the plasma membrane, where it is activated [6]. On the other hand, most of the protein kinases downstream of Raf in the cascade, namely MEK1/2 [7], ERK1/2 (ERK; [8]) and RSK [8] translocate into the nucleus upon stimulation. The translocation of MEK1/2 may involve more than one mechanism [9], and it is clear that these kinases are rapidly exported from the nucleus soon after their translocation. This rapid export from the nucleus is mediated by a CRM1-dependent nuclear export signal (NES; [10]), which results in an apparent cytoplasmic localization of MEK1/2 before and after stimulation [11]. ERK and RSK on the other hand are retained in the nucleus for longer times after stimulation. Interestingly, the proper subcellular localization of the various components has been shown to play an important role in regulating the physiological functions of the ERK cascade [2]. Thus, prevention of nuclear localization of constitutively active MEK reduced its oncogenic potential [12], and nuclear localization of ERK1/2 has been correlated with mitogenesis and neurite outgrowth in PC12 cells [13]. Moreover, prevention of the nuclear translocation of ERK strongly inhibited gene transcription [14] and oncogenic transformation [15]. Moreover, RSK2 activity in the nucleus was found necessary for EGF-induced transcription of c-fos gene [16]. In resting cells, both ERK1 and ERK2 are localized in the cytoplasm due to interaction with various anchoring proteins, including microtubules, phosphatases and a MEK-dependent retention [14,17-19]. The association with some of these proteins such as inactive MKP3 [14] or microtubules [17] is not reversible upon activation. However, most of ERK1/2 molecules (70-85%) appear to dissociate from their cytosolic anchors upon stimulation and translocate into the nucleus. This translocation is rapid (seen in 5 min.), reversible, and may involve several distinct mechanisms of nuclear import [20,21]. The translocated ERK1/2 accumulate in the nucleus, where a large amount of these kinases can be observed even after their activity has declined [22,23]. These observations may suggest that the nucleus is a major site for phosphatase-mediated downregulation of ERK1/2 shortly after translocation. It has been suggested that the inactive ERK1/2 is circulated back to the cytosol by a mechanism that involves nuclear shuttling of MEK [9], but this requires further confirmations.
Since ERK1/2 are retained in the cytoplasm of resting cells by anchoring proteins, it was important to find out what is the mechanism that allows the detachment from these anchors upon stimulation. We have previously identified a primary sequence important for MEK1-induced cytoplasmic retention of ERK1/2 that we termed cytosolic retention sequence (CRS), amino acids 312-320 of ERK2 [19]). This region contains residue Asp319 that is known to play a role in interaction with phosphatases [24,25]. Residues in this region were identified as a common docking (CD) domain for other MAPK-interacting proteins as well [26]. However, the wide variety of functions assigned to this region, and its high degree of similarity with other MAPKs suggest that other sequences might be involved in the association of ERK1/2 with cytoplasmic anchoring proteins. Moreover, the residues and the mechanism responsible for the stimulation- induced dissociation of ERK1/2 from the anchoring proteins are not clear. Here we studied the involvement of residues in the activation loop of ERK2 in the determination of its subcellular localization. We found that this dissociation is dependent on residues 176-181 of ERK2, although these residues do not play a significant role in the phosphorylation and activation of ERK2 upon stimulation. The dissociation was also dependent on the regulatory phosphorylation of ERK2 in its activation loop. However, substitution of residues in this region to alanines did not alter either the stimulated or non-regulated shuttle of ERK2 through nuclear pores. These results indicate that nuclear ERK2 translocation is mainly regulated through a stimulation-induced dissociation from cytosolic anchoring due to conformational changes.
Materials and Methods
Constructs
GFP-ERK2 - The cDNA of rat ERK2 (bases 22-1096) was ligated into ApaI and BamHI sites downstream to the GFP gene of pEGFP-C1 (CLONTECH), thus forming a GFP-ERK2 construct in which the N-terminus of ERK2 is modified [19]. All the subsequent modifications (Figure 1) were performed by PCR mutagenesis [27], incorporated into the GFP-ERK2 construct and were fully sequenced to confirm the mutations.
GFP-173A - Residues 173-175 of ERK2 were modified to alanines using the 3' primer: TACCAACGCGTGGCTACATACTCTGTCAAGAACCCTGTATGATCATGGGCTGCAGCTGCAACACGGGC (containing PflMI site) and a 5' PCR primer containing the ApaI site of ERK2.
GFP-176A- Residues 176-178 of ERK2 were modified to alanines using the 3' primer: TACCAACGCGTGGCTACATACTCTGTCAAGAACCCTGTAGCAGCAGCGTCTGGATCTGCAAC (cont-aining PflMI site) and a 5' PCR primer containing the ApaI site of ERK2.
GFP-D176 - Residues 176-178 of ERK2 were deleted using the 3' PCR primer: TACCAACGCGTGGCTACATACTCTGTCAAGAACCCTGTGTCTGGATCTGCAAC (containing PflMI site) and a 5' PCR primer containing the ApaI site of ERK2.
GFP-179A - Residues 179-181 of ERK2 were modified to alanines using the 3' primer: TACCAACGCGTGGCTACATACTCTGTCAACGCCGCTGCATGATCATGGTCTGG (containing PflMI site) and a 5' PCR primer containing the ApaI site of ERK2.
GFP-183A - Residues 183-185 of ERK2 were modified to alanines using the 3' primer: TACCAACGCGTGGCTACAGCCGCTGCCAAGAACCCTGT (containing PflMI site) and a 5' PCR primer containing the ApaI site of ERK2.
GFP-ALC - Residues 170-190 of ERK2 were replaced with residues 213-228 of MEK1 using the 3' primer: TCTGGAGCTCTGTACGAACGCGTACCTACGAAGGAGTTTGCCATTGAATCGATTAGCTGACCGGCAAGGCCAAAGT (containing SacI site) and a 5' PCR primer containing the SacI site of pEGFP-C1. The PCR product was ligated into SacI sites of GFP- ERK2.
GFP-HL - Leucines 333, 336, 341, 344 of ERK2 were modified to alanines using overlapping PCR (i) the 5' PCR primer: GCACCTAAGGAGAAGGCCAAAGAAGCCATTTTTGAA with a 3' primer containing HpaI, and (ii)3'PCR primer: GGCCTTCTCCTTAGGTGCGTCGTCCCCTCCATGTC with 5' primer containing a PflMI site. The overlapping PCR product was ligated into HpaI and PflMI sites of GFP-ERK2. Histidine 176 in this construct was modified to alanine using the 3' primer: GTACCAAGTACCAATGGCTACAT ACTCTGTCAAGAACCCTGTATGATCAGCGTCTGGATCTGC (containing PflMI site) and a 5' PCR primer containing the ApaI site of ERK2.
WT-MEK1 - MEK1 was prepared as previously described (18).
MKP3-C/S - WT-MKP-3 was cloned by RT-PCR with the primer TACGCTAGCATAGATA CGCTCAGACCCGTG and TCGAGCGGCCGCTCACGTAGATTGCAGAGAGTC. The PCR product was ligated into Nhe I and Not I sites of pcDNA3-HA vector (Invitrogen). The C293S mutant was constructed by PCR with the primer CTTGGTACAT TCCTTGGCTGGC.
Figure 1: Localization of GFP-ERK2 and its inactive mutant in CHO cells. CHO cells were transfected (5 hours with LipofectAMINE) with either GFP-ERK2 or GFP-ERK2 in which residues 183-185 were replaced with alanine (TEY/AAA) either without (A) or with (B) MEK1. After the end of transfection, the cells were either transferred into a starvation medium (A) for 1 hour (total of 6 hours) and for 7 hours (12 hours) or transferred to a growing medium for 32 hours, followed by serum starvation for additional 16 hours (A, 48 hours and B). The cells were then either left untreated (-TPA, 0) or stimulated with TPA (+TPA, 5; 250 nM) for 5 min (A, B) and 15 min (B), after which the cells were washed, fixed, and visualized as described under Materials and Methods.
Buffers
Buffer A: 50 mM b-glycerophosphate, pH 7.3, 1.5 mM EGTA, 1 mM EDTA, 1 mM DTT, and 0.1 mM sodium vanadate. Buffer H: (homogenization buffer): 50 mM b-glycerophosphate, pH 7.3, 1.5 mM EGTA, 1 mM EDTA, 1 mM DTT, 0.1 mM sodium vanadate, 1 mM benzamidine, 10 µg/ml aprotinin, 10 µg/ml leupeptin, and 2 µg/ml pepstatin-A. Buffer R: (reaction mixture at 3-fold final concentration): 30 mM MgCl2, 4.5 mM DTT, 75 mM b-glycerophosphate, pH 7.3, 0.15 mM sodium vanadate, 3.75 mM EGTA, 30 µM calmidazolium, and 2.5 mg/ml bovine serum albumin.
Transfection and localization studies: The various GFP-ERKs plasmids were transfected into CHO cells using LipofectAMINE (Life Technologies, Inc.). When plasmid expressing MEK1 was co-transfected with the various GFP-ERKs, ratio of 2:1 in the amount of DNA transfected was kept, respectively. When plasmid expressing MKP-3 C/S was co-transfected with the various GFP-ERKs, ratio of 4:1 in the amount of DNA transfected was kept, respectively. Visualization was performed essentially as described (7). Cells were stimulated with tetradecanoyl phorbol acetate (TPA; 250 nM, 5 min), fixed (3% paraformaldehyde) and visualized either by fluorescence microscopy (Zeiss Axioscop microscope, HBO 100 W/2; x400 magnification) or by confocal microscopy (BioRad).
Western blotting: Transfected cells were serum starved (0.1% FCS) for 16 hours. After stimulation, the cells were washed twice with ice-cold PBS and once with ice-cold Buffer A. Cells were harvested with Buffer H followed by sonication (2x7 sec, 40 W) and centrifugation (15,000xg, 15 min, 4oC). The supernatants, which contained cytosolic proteins were collected and aliquots from each sample (20 µg) were separated on 10% SDS PAGE followed by Western blotting. Activated ERK was detected by probing blots with anti-activated ERK monoclonal antibody (di-phospho (DP)-ERK, Sigma, Israel) or antibody PT/DP-ERK (Sigma, Israel, clone 115). Total ERK protein was detected by anti-MAPK antibody (Sigma, Israel). MEK1 was detected with anti-MEK1 monoclonal antibody (Transduction Laboratories).
Immunoprecipitation: Transfected CHO cells were serum-starved (24 hours), stimulated (TPA, 5 min), and harvested as described above. The cell extracts were then subjected to immunoprecipitation with anti-GFP monoclonal antibody (Roche Molecular Biochemicals). For MEK1 co-immunoprecipitation studies, the beads
were washed as described (10) with low stringency buffer (20 mM HEPES pH 8.0, 2 mM MgCl2, 2 mM EGTA), and then subjected to immunoblotting with monoclonal anti-MEK and anti-GFP antibodies. For the determination of ERK activity the beads were washed once with radioimmune precipitation buffer, twice with 0.5M LiCl, and twice with buffer A as described (27). The immunoprecipitates were subjected to myelin basic protein (MBP) phosphorylation assay as below.
Determination of ERK activity: The immunoprecipitates of GFP-ERKs proteins obtained from the stimulated transfected CHO cells were mixed with MBP (8.4 µg) and Buffer R with 100 µM [g-32P] ATP (1-2 cpm/fmol) in a final volume of 30 µl. The phosphorylation reaction allowed to proceed at 30oC for 20 min, after which it was terminated by sample buffer and boiling for 5 min. Phosphorylation of MBP was assessed by autoradiography.
Results
Subcellular localization of ERK2 and its inactive mutant. In response to mitogenic stimulation, ERK1/2 are translocated from the cytoplasm into the nucleus [8]. In order to characterize this dynamic change in subcellular localization we have previously [19] fused green fluorescent protein (GFP) to the N-terminal end of ERK2 (GFP-ERK2). We showed that overexpression of GFP-ERK2 in CHO cells results in a nuclear accumulation of the protein both before and after stimulation. Co-expression with MEK1 results in a cytosolic-retention of the GFP-ERK2 in resting cells, and this is mediated by residues 312-320 of ERK2 (CRS). This localization is rapidly changed upon mitogenic stimulation of the cells, which causes a dissociation of ERK2 from its cytosolic anchor, and enables its translocation into the nucleus. To further study the molecular mechanisms that govern the subcellular localization of ERK2 we followed the changes in the subcellular localization of GFP-ERK2 shortly after transfection (six hours after the beginning of the transfection procedure and one hour after its end). Under these conditions, GFP-ERK2 was equally distributed all over the resting cells (Fig. 1A), and TPA stimulation induced a rapid nuclear translocation of the majority of GFP-ERK2 molecules. In later time points after transfection (12 to 48 hours after transfection) the total amount of GFP-ERK2 was increased, and the vast majority of it was detected in the nucleus both before and after TPA stimulation. These results can be explained by saturation of a limited number of endogenous cytosolic anchoring proteins by newly synthesized GFP-ERK2 molecules. A slow rate nuclear-translocation of an excess of synthesized GFP-ERK2 into the nucleus is probably responsible for the gradual accumulation of protein in the nucleus. However, stimulation is able to cause the detachment of the ERK1/2 from their endogenous cytosolic anchor and induce a fast rate translocation that is completed within 5 min after stimulation (Figure 1A). This system enabled us to follow interactions with endogenous anchoring proteins as well as ERK2 translocation into the nucleus.
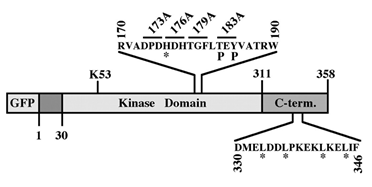
Figure 2. Schematic representation of the activation loop mutants of ERK2. Sites of mutation of GFP-173A, GFP-176A, GFP-D176, GFP-179A and GFP-183A is indicated. The phosphorylated residues 183 and 185 are indicated with P. GFP-ALC was prepared by substitution of residues 170-190 with the homologous residues from MEK. The residues that were substituted to alanines in HL-GFP are indicated by *.
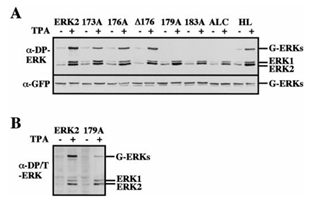
Figure 3: Phosphorylation state of GFP-ERK2 and its activation loop mutants. CHO cells were transfected with the indicated constructs. Thirty-two hours after transfection the cells were serum-starved for 16 hours and then were either left untreated (-) or stimulated with TPA (+; 250 nM, 5 min). A: cell lysates were prepared as described under Materials and Methods and analyzed with anti-DP-ERK antibody (a-DP-ERK, upper panel) or anti-GFP antibody (a-GFP, lower panel). B: cell lysates of GFP-ERK2 and GFP-179A were prepared and analyzed with anti di-phosphorylated/Threonine phosphorylated ERK antibody (clone 115, Sigma, Israel). G-ERKs indicates the position of GFP-ERK2, and ERK1/ERK2 indicates the position of the endogenous ERK1 and ERK2. These results were reproduced five times.
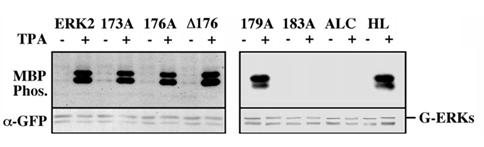
Figure 4: Activity of GFP-ERK2 and its activation loop mutants. CHO cells were transfected with the indicated constructs. Thirty-two hours after transfection the cells were serum-starved for 16 hours and then were either left untreated (-) or stimulated with TPA (+; 250 nM, 5 min). Cell lysates were prepared as described under Materials and Methods and subjected to immunoprecipitation with monoclonal anti-GFP antibody. The catalytic activity of the different GFP-ERKs was assayed by an in-vitro kinase assay using MBP as a substrate, as described under Materials and Methods. Samples were separated by an SDS-PAGE and blotted onto nitrocellulose. Blots were probed with monoclonal anti-GFP antibody (lower panel). Radioactivity was determined by 24 hours exposure to X ray film (MBP Phos.; upper panel). MBP indicates the place of phosphorylated MBP and G-ERKs indicates the position of the immunoprecipitated GFP-ERKs. The results are taken from two distinct gels. These results were reproduced four times.
Construction and biochemical analysis of activation loop mutants of ERK2. As previously reported [18,19], coexpression of GFP-ERK2 and MEK1 resulted in a cytosolic distribution of the GFP-ERK2 (Figure 1B), and this was reversed by TPA stimulation. When the inactive mutant of ERK2, in which the regulatory Thr and Tyr residues were replaced by alanines, was coexpressed with MEK1, TPA failed to induce nuclear translocation of the mutated ERK2, indicating that phosphorylation of ERK2 is essential for its nuclear translocation. Therefore, we undertook to analyze the role of the activation loop of ERK2 in both its stimulated and passive nuclear translocation processes. To do so we constructed sequential mutations in the activation loop, and as previously described for ERK2 [19], the constructs were fused to GFP to enable their easy detection. The mutations introduced (Figure 2) were: (i) substitution of amino acids 173-175 to alanines (GFP-173A); (ii) substitution of amino acids 176-178 to alanines (GFP-176A); (iii) substitution of amino acids 179-181 to alanines (GFP-179A); (iv) substitution of amino acids 183-185 to alanines (GFP-183A). Other mutations introduced (Figure 2) were (i) deletion of amino acids 176-178 (GFP-D176) to further study the role of these residues in the subcellular localization (ii) replacement of the residues required for homodimerization of ERK2 (His 176, Leu333, Leu336, Leu341 and Leu344; [28]) termed GFP-HL; and (iii) replacement of the whole activation loop of ERK2 (residues 170-190) with the activation loop of MEK1, thus forming an activation loop chimera (GFP-ALC). We examined the activation and catalytic activity of the above mutants by transfecting them into CHO cells. Thirty-two hours after transfection the cells were serum-starved (0.1% serum) for 16 hours, and then were either stimulated with TPA or left untreated. All the mutants were expressed to a similar level, as judged by their detection with anti-general ERK antibody at the expected size of ~70 kDa (Figure 3). Western blot with anti-DP-ERK antibody revealed that 173A, 176A, D176, and HL were phosphorylated as much as the wild type ERK2, whereas no phosphorylation was detected with 179A, 183A, and ALC. Staining of the GFP-ALC with anti-phospho MEK antibody did not yield any appreciable staining, indicating that it could not be activated under the conditions used. However, staining with another anti-DP-ERK antibody detected a TPA-induced phosphorylation on GFP-179A (Figure 3B), but not on GFP-183A or GFP-ALC, and similar detection was obtained with anti-phospho Tyr antibody (data not shown). These results indicate that the GFP-179A construct is phosphorylated in response to TPA on both regulatory Thr and Tyr, and the lack of recognition by the initial anti-DP-ERK antibody was due to the specificity of the antibody. The catalytic activity of the constructs was then examined by immunoprecipitation of the proteins expressed in CHO cells with anti-GFP antibody followed by an in-vitro kinase assay towards MBP. The specific catalytic activity observed with GFP-ERK2 upon TPA stimulation was comparable to that of endogenous ERK1/2 (~150 nmol/min/mg, data not shown). This activity was not significantly changed when overexpressed GFP-HL or GFP-179A constructs were examined and was slightly elevated (25%+/- 7%) with GFP-D176 (Figure 4). On the other hand, the activity of GFP-173A and GFP-176A was lower than that of GFP-ERK2 (35%+/- 12% vs 50%+/-10% respectively). GFP-183A, and GFP-ALC, had no detectable activity.
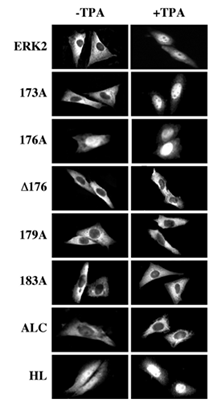
Figure 5: Localization of GFP-ERK2 and its activation loop mutants in CHO cells upon cotransfection with MEK1. CHO cells were transfected with the indicated constructs together with MEK1. Thirty-two hours after transfection the cells were serum starved for 16 hours and then were either left untreated (Basal) or stimulated with TPA (250 nM, 5 min), after which the cells were fixed and visualization using conventional and confocal fluorescence microscopy. Each of the experiments was reproduced at least five times.
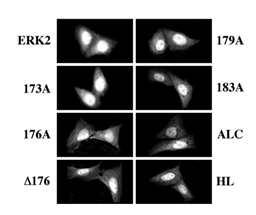
Figure 6: Localization of GFP-ERK2 and its activation loop mutants in CHO cells 48 hours after transfection without MEK1. CHO cells were transfected, with each of the indicated constructs. After transfection the cells were transferred into a growing medium for 32 hours and then to starvation medium for 16 hours. The cells were then washed, fixed, and visualized as described. Each of the experiments was reproduced at least five times.
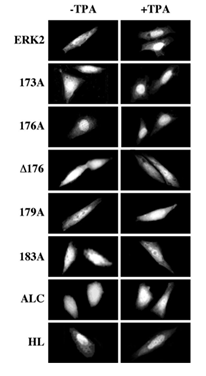
Figure 7: Localization of GFP-ERK2 and its activation loop mutants in CHO cells 6 hours after the beginning of transfection without MEK1. CHO cells were transfected for 5 hours using LipofectAMINE, with each of the indicated constructs. After the end of transfection, the cells were transferred into a starvation medium for 1 hour. The cells were then either left untreated (Basal) or stimulated with TPA (250 nM) for 5 min, after which the cells were washed, fixed, and visualized as described. Each of the experiments was reproduced at least four times.
Subcellular localization of the activation loop-mutants of ERK2. We then undertook to study the subcellular localization of the various activation loop-mutated proteins in the presence of MEK1. Thus, each of the GFP-conjugated ERK2 was cotransfected into CHO cells together with MEK1 and visualized 48 hours after transfection. As expected, GFP-ERK2 was localized in the cytosol of resting cells and translocated into the nucleus shortly after TPA stimulation (see Figure 1). However, the only mutant that behaved similarly to GFP-ERK2 was GFP-173A (Figure 5). The inactive GFP-183A and GFP-ALC were unable to leave the cytosolic compartment of the CHO cells as expected (Figure 1 and [20]). Interestingly, GFP- 176A was localized in the nucleus both before and after TPA stimulation (Figure 5), whereas GFP-D176 was restricted to the cytosol. Other residues that appear to be important in the subcellular localization of ERK2 are residues 179-181, because GFP-179A, which could be phosphorylated and activated upon TPA treatment, failed to translocate into the nucleus after this stimulation. Finally, the dimerization-deficient GFP-HL was distributed all over resting cells and very rapidly translocated into the nucleus upon stimulation, indicating that His 176 and possibly also the four C-terminal Leu residues are important for the association of ERK2 with MEK1 and are probably not essential for TPA-induced translocation. As previously reported [19], overexpression of GFP-ERK2 without MEK1 for 48 hours resulted in nuclear accumulation of most expressed molecules (Figure 6). Similar to GFP-ERK2, all the GFP-conjugated mutants of ERK2 tested here were detected in the nuclei of resting CHO cells both before (Figure 6) and after (data not shown) stimulation. These results may indicate that all mutants have the capability to enter into the nucleus of resting cells, probably due to a passive mechanism [20]. Their accumulation in the nucleus can be explained by attachment to nuclear-anchoring proteins [11], since endogenous MEK1 that can serve as a nuclear export vehicle for ERK2 [9] was not able to induce any appreciable nuclear export of the overexpressed ERK2. In order to examine whether the interaction of the ERK2 mutants with endogenous anchoring proteins is similar to their interaction with overexpressed MEK1, we examined the subcellular localization of the ERK2 mutants six hours after the beginning of transfection without the MEK1 construct. The results obtained with this system correlated nicely with those obtained when the mutants were co-transfected with MEK1 above. Thus, as much as the GFP- ERK2, also GFP173A, GFP-D176, GFP-179A, GFP-183A and GFP-ALC were equally distributed throughout the resting cells, indicating retention of part of the synthesized molecules (Figure 7). On the other hand, GFP-176A and the GFP-HL, that were not retained in the cytosol by MEK1, were localized primarily in the nucleus in this system too, indicating that they lost their ability to interact with endogenous retention proteins. We then treated the cells with TPA and found that this induced nuclear translocation of GFP-ERK2 and GFP-173A, while GFP-176A and GFP-HL remained localized in the nucleus. Again, in agreement with the results obtained with the MEK1-cotransfection system (Fig. 5), GFP-D176, GFP-179A, GFP- 183A and GFP-ALC remained spread all over the cell, and were unable to translocate into the nucleus even in the absence of overexpressed MEK1. Thus, our results indicate that the endogenous retention proteins behave similarly to MEK1. The detachment from these proteins, like that from MEK1, was dependent on residues 176-181, and the substitution of residues 176-178 to alanines mimics this detachment. The subcellular localization of the various ERK mutants was also examined after stimulation with peroxovanadate, and gave similar results to those obtained after TPA stimulation (data not shown). Therefore, the mechanisms that govern ERK dissociation from the cytosolic anchor are probably general and are independent on the type of cellular stimulation.
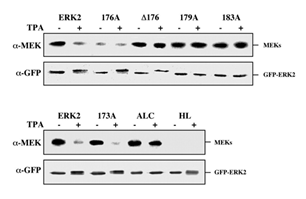
Figure 8: Co-immunoprecipitation of ERK2 and its activation loop mutants with MEK1. CHO cells were transfected with each of the indicated constructs together with a plasmid expressing MEK1. Thirty-two hours after transfection the cells were serum starved for 16 hours and then were either left untreated (-) or stimulated with TPA (+, 250 nM, 5 min). Lysates from all conditions were subjected to immunoprecipitation with monoclonal anti-GFP antibody. After SDS-PAGE, samples were immunoblotted with monoclonal anti-MEK (a-MEK) and monoclonal anti-GFP antibodies (a-GFP). GFP-ERK2 and MEKs represent their respective positions. The amount of MEK in each of the extracts used for co-immunoprecipitation was roughly equal (data not shown). The two panels represent two independent experiments. Each of the experiments was reproduced at least four times.
Co-immunoprecipitation of activation loop mutants with MEK1. We then assayed the activation loop mutants for their ability to interact with MEK1 in a co-immunoprecipitation assay. Thus, GFP-ERK2 and the various mutants were cotransfected into CHO cells together with MEK1, serum-starved for 16 hours and then were either left untreated or stimulated with TPA for 5 min. At this stage, the cells were gently lysed, and GFP-ERKs were immunoprecipitated with anti-GFP antibodies. The washes of the immunocomplexes were extremely mild [18], and those were subjected to SDS-PAGE and Western blot analysis with both anti-MEK1 and anti-GFP antibodies. As expected, immunoprecipitation of GFP-ERK2 was able to pull down some amount of MEK1, which was reduced upon treatment with TPA (Figure 8). As detected in the subcellular localization studies above, the only construct that behaved in a similar way to the GFP-ERK2 was GFP-173A. On the other hand, GFP-D176, 179A, 183A and GFP-ALC did not seem to dissociate from MEK1 upon stimulation, whereas GFP-176A as well as GFP-HL did not associate with MEK1 to any appreciable level even under basal conditions. The correlation between the association with MEK1 (Figure 8) and the nuclear translocation of the ERK-constructs (Figures 5,7) indicates that the cytosolic interaction with MEK1 is the main parameter that determines the subcellular localization of the ERK2.
Interaction of the activation loop mutants with MKP3. Several proteins have been shown to act as irreversible cytosolic anchors for ERK1/2, including cytoskeletal elements [17], and protein Tyr phosphatases [14]. We therefore studied the ability of the various ERK2 constructs to associate with the inactive form of MAP Kinase phosphatase 3 (MKP3-C/S, [14]). Cytosolic localization of GFP-ERK2 was detected when it was coexpressed with MKP3 in CHO cells and this localization was not reversed by TPA stimulation. However, MKP3 failed to anchor GFP-312A (substitution of residues 312-319 of ERK2 to alanines [19]) in the cytosol, indicating that, in similarity to MEKs, the main binding of ERK2 to MKP3 occurs by to residues 316-319 [26]. However, unlike the results with MEK1 above, when coexpressed with MKP3, all the activation-loop mutants of ERK2 behaved in a similar manner to the GFP-ERK2 and localized to the cytosol both before and after TPA stimulation. These results indicate that although the activation loop plays a role in the association of ERK2 with MEK1, it does not contribute to the association of ERK2 to MKP3-C/S. Moreover, the inability of ERK2 to dissociate from MKP3-C/S upon TPA stimulation, support our suggestion that the activation loop is responsible for the stimulation-induced dissociation of ERK2 from some of its cytosolic anchor.
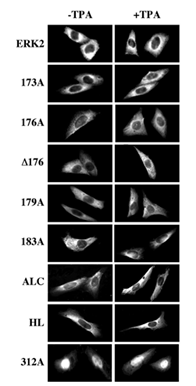
Figure 9: The subcellular localization of GFP-ERK2 and its activation loop mutants upon cotransfection with MKP3 C/S. CHO cells were transfected with each of the indicated constructs together with plasmid containing MKP3-C/S. Thirty-two hours after transfection the cells were serum starved for 16 hours and then were either left untreated (Basal) or stimulated with TPA (250 nM, 5 min), after which the cells were fixed and visualized using conventional and confocal fluorescence microscopy. Each of the experiments was reproduced at least five times.
Discussion
The requirement of catalytic activation for the nuclear translocation of ERK1/2 prompted our studies on the importance of residues in the activation loop of ERK2 for its activation, dissociation from MEK and translocation into the nucleus. Here we designed a series of mutations that covered the activation loop of ERK2. To our surprise, mutations in residues 173-181, which are very close to the regulatory Thr183 and Tyr185 had only a small effect on the phosphorylation and activation of ERK2 (Figures 3 and 4). The only constructs that were inactive were GFP-183A and GFP-ALC that lacked the regulatory Tyr and Thr residues, and thus served as an inactive control in our study. We then studied the MEK1-dependent subcellular localization of the various activation loop mutants of ERK2. It has previously been reported [18] that MEKs may directly interact with ERK1/2 and thus retain them in the cytosol. However, because of the weak nature of the association between the two proteins [19], it is possible that this association is not direct but mediated via a scaffold protein [29]. Our results revealed that, as reported [20], non-phosphorylatable mutants (183A, ALC) did not dissociate form MEK1 (Figure 8) and did not translocate into the nucleus after stimulation (Figure 5).
Beside the inactive constructs, also changes in residues 176-181 modified the MEK1-dependent subcellular localization of ERK2. Thus, when residues 176-178 were substituted to alanines, there was no interaction with MEK1, and this correlated with a nuclear accumulation of the ERK2 construct. However, deletion of the same residues resulted in an irreversible association with MEK1, which caused a cytoplasmic localization both before and after TPA stimulation. These results are best explained by assuming that these residues are important for the dissociation of ERK2 from MEK1. Thus, as for GFP-ERK2, the association of GFP-D176 with MEK1 is mediated by the CRS at residues 312-320 of ERK2 [19]. Hence, the inability of GFP-D176 to dissociate from MEK1 in response to stimulation can be explained by an ability of residues 176-178 to serve as a lever that allows dissociation from MEK1. Substitution of the 176-178 residues to alanines probably mimics the activated conformation of ERK2 and therefore do not allow association between the two proteins even though the CRS region was intact. This suggestion is supported by the fact that substitution of these residues with Gly residues gave a similar response as GFP- D176 and not as GFP-176A (data not shown). Moreover, GFP-176A could interact with MKP3-C/S (through CRS, Figure 9), without interference of the alanines at position 176-178.
As the association with MKP3-C/S is not reversible, there is no influence of residues that prompt dissociation and therefore no dissociation of the construct GFP-176A that mimics the MEK1- dissociated form of ERK2 was observed. Interestingly, substitution of residues 179-181 to alanines also demonstrated an irreversible association with MEK1 and it is likely therefore that residues 176-181 are all involved in the stimulated dissociation. In addition to the involvement of residues 176-181 in the dissociation of ERK2 from MEK1, these residues play a role in determining the specificity of interaction between the proteins. Since the CD domain of ERK1/2 seems to be similar to the same region in other MAPKs, it was predicted [26] that other regions will be responsible for the specificity of binding of MAPKs to their particular MAPKKs. Thus, our findings that residues 176-181 are important for the dissociation from MEK1 but not from MKP3-C/S may indicate that these residues participate also in the determination of specificity of ERK1/2 to MEKs.
The region of the activation loop identified here joins a list of other regions of ERK1/2 that were postulated to be important in the association between ERK and MEKs. These are residues in subdomain III of ERK1/2 [30], multiple regions in the N- and C- termini of ERK1/2 [31], amino acids 19-25 of ERK2 [32] and residues 312-320 [19], among which residues 316 and 319 seem to play the most important role in the interaction with MEKs [26]. It is clear that all these residues cannot interact with one molecule of MEK1 at the same time, because they are located in completely different areas of the ERK2 molecule. It is possible however, that two types of interactions between ERK1/2 and MEKs exist. One is probably required for the immediate activation of ERK1/2 by MEKs and could involve the regions in the same plane of the activation loop [31]. The other interaction may involve the CRS (CD), which does not seem to play a significant role in the activation process of ERK2 [19,26], but rather in its subcellular localization and sensitivity to phosphatases. Although there is accumulating evidence that ERK2 and MEK1 can directly interact with each other [18,33], it is still possible that this interaction occurs via a third protein such as MP1 for ERK1 [34]. In this case the stimulation-dependent dissociation observed in our experiments would not be from MEK1 itself, but from this putative scaffolding protein.
The mechanism of ERK2 translocation into the nucleus can be divided into two stages. In the first step the enzyme is detached from its cytosolic anchor, while the second step comprise of a shuttle through the nuclear envelope. The process of nuclear import is usually accomplished through nuclear pores that are easily accessible to proteins with molecular mass smaller than 40 kDa. Molecules with higher molecular masses usually shuttle through the nuclear envelope by an active mechanism which requires a complex proteins network [35]. In similarity to previous findings [20], we show here that the 70 kDa GFP-ERK2 can translocate into the nucleus in two rates: a basal, non-regulated rate (Fig. 6 and Fig. 7, -TPA) and a TPA-stimulated fast rate (Fig. 5 and Fig. 7 +TPA). We believe that the molecular mass of the GFP- ERK2 proteins requires that both rates of translocation would be mediated by an active mechanism. All mutants used in the current study showed equal nuclear accumulation in resting cells (Fig. 6), indicating that the activation loop of ERK2 does not play a role in its slow-rate nuclear translocation. It is therefore possible that the machinery responsible for the basal state of translocation operates in a low rate in resting cells and is activated by mitogenic stimulation of the cells. However, the stimulated translocation correlated with dissociation of the ERK2 constructs from MEK1.
Our results suggest that the translocation of ERK2 into the nucleus is regulated primarily by its dissociation from MEK1 [20]. Thus, the detachment is the main process that allow translocation to occur. In later studies we found the actual mechanism that allow the nuclear translocation. Thus, upon detachment of ERK1/2 from cytoplasmic anchoring proteins ([19,26] and this study) ERK1/2 are phosphorylated on the two Ser residues within its kinase insert domain (SPS motif also known as nuclear translocation signal (NTS) [36]). This phosphorylation is mediated primarily by CK2 and autophosphorylation [37], and allows interaction with importin 7 (Imp7 [36]). Next, Imp7 escorts ERK1/2 into the nucleus through the nuclear pores, detachment of these two proteins, which is mediated by Ran-GTP and export of Imp7 back to the cytoplasm. The released active ERK1/2 are now free to phosphorylate its nuclear targets including transcription factors and other nuclear proteins [2,38]. We also found that the nuclear translocation of the other MAPKs, p38 and JNK, has some similarities to ERK1/2, but the importins involved are either Imp 3/7 or Imp3/9 dimers [39]. Upon their release from the importin dimers by Ran-GTP, the translocated p38 and JNK phosphorylate their targets in the nucleus to allow specific physiological or pathological functions [3].
It has previously been proposed that dimerization of ERK2 is required for its active nuclear transport and that phosphorylated ERK2 can form dimers with either phosphorylated or unphosphorylated ERK2 partners [28]. Based on crystal structure of phosphorylated ERK2, mutants were constructed at the predicted interface shared by the two monomers. A mutant, H176E and L333,336,341,344A of ERK2 was identified, which lacked the ability to dimerize upon stimulation, and when microinjected into the cytosol of cells, could not translocate into the nucleus. Therefore, it has been suggested that these residues participate in homodimerization formation and are critical for the translocation process of ERK2. However, in our system, the HL mutant in which these five residues were substituted to alanines, accumulted in the nucleus without overexpression of MEK1 (Fig. 6 and 7). When coexpressed with MEK1 the mutant was spread all over the cells, and TPA stimulation induced its rapid nuclear translocation. Thus, our results indicate that these residues or at least the H176 participates neither in the non-regulated nor in the stimulated translocation of ERK2 into the nucleus. Similar results showing that dimerization is not required for translocation were shown by others as well were shown by others as well [40-42]. Rather, it seems that the dimerization may be involved in cytoplasmic ERK signaling due to binding to various anchoring proteins [41,43].
In summary, we identified here six amino acids in the activation loop of ERK2, which appear to be important for the stimulation-induced dissociation of ERK2 from MEK1. Conformational change of these residues upon phosphorylation seems to be involved in the mechanism of release of ERK2 from its cytosolic anchor, and this can be mimicked by substitution of residues 176-178 to alanines. Surprisingly, residues 173-181 did not seem to play a significant role in the activatory phosphorylation of ERK2 upon TPA stimulation. We also show here that ERK2 translocates into the nucleus in two rates, a non-regulated rate in resting cells and fast rate upon mitogenic stimulation. The residues of the activation loop of ERK2, including the dimerization-promoting His 176, do not play a role in these two types of translocations. Therefore, our results suggest that the translocation of ERK2 into the nucleus is mainly regulated through the dissociation of ERK2 from cytosolic anchoring proteins and this is mediated by residues in the activation loop.
Author Contributions
“Conceptualization, R.S.; methodology, I.W., H.R.; investigation, I.W., H.R., S.Y., G.M., T.H; writing: original draft preparation, R.S., I.W, H.R.; writing: review and editing, R.S.; supervision, R.S.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by This work was supported by grants from the Moross Institute for Cancer Research at the Weizmann Institute of Science, the Benozyio Institute for Molecular Medicine at the Weizmann Institute of Science, the Estate of Siegmund Landau, the Israel Academy of Sciences, and by grant number 4949 from the Chief Scientists Office of the Ministry of Health, Israel (to I.W.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tubita A, Lombardi Z, Tusa I, et al. Beyond Kinase Activity: ERK5 Nucleo-Cytoplasmic Shuttling as a Novel Target for Anticancer Int J Mol Sci 21 (2020): 938.
- Maik-Rachline G, Hacohen-Lev-Ran A, Seger R. Nuclear ERK: Mechanism of Translocation, Substrates, and Role in Int J Mol Sci 20 (2019): 1194.
- Maik-Rachline G, Lifshits L, Seger R. Nuclear P38: Roles in Physiological and Pathological Processes and Regulation of Nuclear Int J Mol Sci 21 (2020): 6102.
- Flores K, Yadav SS, Katz AA, et al. The Nuclear Translocation of Mitogen- Activated Protein Kinases: Molecular Mechanisms and Use as Novel Therapeutic Neuroendocrinology 108 (2019): 121-131.
- Wainstein E, Seger R. The dynamic subcellular localization of ERK: mechanisms of translocation and role in various organelles. Curr Opin Cell Biol 39 (2016): 15-20.
- Lavoie H, Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol 16 (2015): 281-298.
- Jaaro H, Rubinfeld H, Hanoch T, et al. Nuclear translocation of mitogen- activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc. Acad. Sci. USA 94 (1997): 3742-3747.
- Chen RH, Sarnecki C, Blenis Nuclear localization and regulation of erk- and rsk- encoded protein kinases. Mol Cell Biol 12 (1992): 915-927.
- Adachi M, Fukuda M, Nishida E. Nuclear export of MAP kinase (ERK) involves a MAP kinase kinase (MEK)-dependent active transport mechanism . J Cell Biol 148 (2000): 849-856.
- Fukuda M, Gotoh I, Gotoh Y, et al. cytoplasmic localization of mitogen- activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem 271 (1996): 20024-
- Lenormand P, Brondello JM, Brunet A, et al. Growth factor-induced p42/p44 MAPK nuclear translocation and retention requires both MAPK activation and neosynthesis of nuclear anchoring J Cell Biol 142 (1998): 625-633.
- Fukuda M, Gotoh I, Adachi M, et al. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role of nuclear export signal of MAP kinase J. Biol. Chem 272 (1997): 32642-32648.
- Traverse S, Gomez N, Paterson H, et al. Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth Biochem J 288 (1992): 351-355.
- Brunet A, Roux D, Lenormand P, et al. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor- induced gene expression and cell cycle EMBO J 18 (1999): 664-674.
- Plotnikov A, Flores K, Maik-Rachline G, et al. The nuclear translocation of ERK1/2 as an anticancer Nat Commun 6 (2015): 6685.
- Swanson KD, Taylor LK, Haung L, et al. Transcription Factor Phosphorylation by pp90(rsk2). Identification of fos kinase and ngfi-b kinase i as pp90 (rsk2). J Biol Chem 274 (1999): 3385-3395.
- Reszka AA, Seger R, Diltz CD, et al. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc. Natl. Acad. USA 92 (1995): 8881-8885.
- Fukuda M, Gotoh Y, Nishida Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J 16 (1997): 1901-1908.
- Rubinfeld H, Hanoch T, Seger Identification of a cytoplasmic-retention sequence in ERK2. J. Biol. Chem 274 (1999): 30349-30352.
- Adachi M, Fukuda M, Nishida E. Two co-existing mechanisms for nuclear import of MAP kinase: passive diffusion of a monomer and active transport of a EMBO J 18 (1999): 5347-5358.
- Lorenzen JA, Baker SE, Denhez F, et al. Nuclear import of activated D-ERK by DIM-7, an importin family member encoded by the gene Development 128 (2001): 1403-1414.
- Yao Z, Dolginov Y, Hanoch T, et al. Detection of partially phosphorylated forms of ERK by monoclonal antibodies reveals spatial regulation of ERK activity by FEBS Lett 468 (2000): 37-42.
- Yao Z, Flash I, Raviv Z, et al. Non-regulated and stimulated mechanisms cooperate in the nuclear accumulation of Oncogene 20 (2001): 7588-7596.
- Bott CM, Thorneycroft SG, Marshall The sevenmaker gain-of-function mutation in p42 MAP kinase leads to enhanced signalling and reduced sensitivity to dual specificity phosphatase action. FEBS Lett 352 (1994): 201-205.
- Camps M, Chabert C, Muda M, et al. Induction of the mitogen-activated protein kinase phosphatase MKP3 by nerve growth factor in differentiating FEBS Lett 425 (1998): 271-276.
- Tanoue T, Adachi M, Moriguchi T, et al. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol 2 (2000): 110-
- Silverman MA, Benard O, Jaaro H, et al. CPG16, a novel protein Serine/Threonine kinase downstream of cAMP- dependent protein kinase. J Biol Chem 274 (1999): 2631-2636.
- Khokhlatchev AV, Canagarajah B, Wilsbacher J, et al. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear Cell 93 (1998): 605-615.
- Whitmarsh AJ, Cavanagh J, Tournier C, et al. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science 281 (1998): 1671-1674.
- Brunet A, Pouyssegur J. Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science 272 (1996): 1652-1655.
- Wilsbacher JL, Goldsmith EJ, Cobb MH. Phosphorylation of MAP kinases by MAP/ERK involves multiple regions of MAP kinases. J Biol Chem 274 (1999): 16988-16994.
- Eblen ST, Catling AD, Assanah MC, et al. Biochemical and biological functions of the N-terminal, noncatalytic domain of extracellular signal-regulated kinase Mol Cell Biol 21 (2001): 249-259.
- Bardwell AJ, Flatauer LJ, Matsukuma K, et al. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal J. Biol. Chem 276 (2001): 10374- 10386.
- Schaeffer HJ, Catling AD, Eblen ST, et al. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase Science 281 (1998): 1668-1671.
- Moore MS, Blobel A G protein involved in nucleocytoplasmic transport: the role of Ran. Trends Biochem Sci 19 (1994): 211-216.
- Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Cell 31 (2008): 850-861.
- Plotnikov A, Chuderland D, Karamansha Y, et al. Nuclear ERK Translocation is Mediated by Protein Kinase CK2 and Accelerated by Cell. Physiol. Biochem., 53 (2019): 366-387.
- Unal EB, Uhlitz F, Bluthgen A Compendium of ERK Targets. FEBS Lett 591 (2017): 2607-2615.
- Zehorai E, Seger R. Beta-Like Importins Mediate the Nuclear Translocation of Cell Physiol Biochem 52 (2019): 802-821.
- Lidke DS Huang F, Post JN, et al. ERK nuclear translocation is dimerization- independent but controlled by the rate of phosphorylation. J Biol Chem 285 (2010): 3092-3102.
- Casar B, Pinto A, Crespo P. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol Cell 31 (2008): 708-721.
- Burack WR, Shaw AS. Live Cell Imaging of ERK and MEK: simple binding equilibrium explains the regulated nucleocytoplasmic distribution of ERK. J Biol Chem 280 (2005): 3832-3837.
- Casar B, Pinto A, Crespo ERK dimers and scaffold proteins: unexpected partners for a forgotten (cytoplasmic) task. Cell Cycle 8 (2009): 1007-1013.
