In vivo-like Culture of Monophagous Animal Organ using Dietary Components
Article Information
Norichika Ogata1*, Shogo Konishi1, Takeshi Yokoyama2
1Nihon BioData Corporation, 3-2-1 Sakado, Takatsu-ku, Kawasaki, Kanagawa 213-0012, Japan
2Laboratory of Sericultural Science, Faculty of Agriculture, Tokyo University of Agriculture and Technology, 3-5-8, Saiwai-cho, Fuchu, Tokyo, 183-8501, Japan
*Corresponding author: Norichika Ogata, Nihon BioData Corporation, 3-2-1 Sakado, Takatsu-ku, Kawasaki, Kanagawa 213-0012, Japan.
Received: 24 January 2023; Accepted: 31 January 2023; Published: 10 February 2023
Citation: Norichika Ogata, Shogo Konishi, Takeshi Yokoyama. In vivo-like Culture of Monophagous Animal Organ using Dietary Components. Journal of Biotechnology and Biomedicine 6 (2023): 42-48.
View / Download Pdf Share at FacebookAbstract
Animals depend on other species to live, with monophagy being an extreme mode. Monophagous animals depend on their diet not only for nutritients but also for developmental and reproductive controls. Thus, dietary components may be useful in culturing tissues from monophagous animals. We hypothesized that a dedifferentiated tissue from the monophagous silkworm, Bombyx mori, would re-differentiate when cultured in a medium containing an extract of mulberry (Morus alba) leaves, the only food of B. mori. Over 40 fat-body transcriptomes were sequenced, and we concluded that it is possible to establish in vivo-like silkworm tissue cultures using their diet.
Keywords
Dedifferentiation; Monophagy; Primary Explant Culture; RNA-seq; Silkworm
Dedifferentiation articles Dedifferentiation Research articles Dedifferentiation review articles Dedifferentiation PubMed articles Dedifferentiation PubMed Central articles Dedifferentiation 2023 articles Dedifferentiation 2024 articles Dedifferentiation Scopus articles Dedifferentiation impact factor journals Dedifferentiation Scopus journals Dedifferentiation PubMed journals Dedifferentiation medical journals Dedifferentiation free journals Dedifferentiation best journals Dedifferentiation top journals Dedifferentiation free medical journals Dedifferentiation famous journals Dedifferentiation Google Scholar indexed journals Monophagy articles Monophagy Research articles Monophagy review articles Monophagy PubMed articles Monophagy PubMed Central articles Monophagy 2023 articles Monophagy 2024 articles Monophagy Scopus articles Monophagy impact factor journals Monophagy Scopus journals Monophagy PubMed journals Monophagy medical journals Monophagy free journals Monophagy best journals Monophagy top journals Monophagy free medical journals Monophagy famous journals Monophagy Google Scholar indexed journals Primary Explant Culture articles Primary Explant Culture Research articles Primary Explant Culture review articles Primary Explant Culture PubMed articles Primary Explant Culture PubMed Central articles Primary Explant Culture 2023 articles Primary Explant Culture 2024 articles Primary Explant Culture Scopus articles Primary Explant Culture impact factor journals Primary Explant Culture Scopus journals Primary Explant Culture PubMed journals Primary Explant Culture medical journals Primary Explant Culture free journals Primary Explant Culture best journals Primary Explant Culture top journals Primary Explant Culture free medical journals Primary Explant Culture famous journals Primary Explant Culture Google Scholar indexed journals RNA-seq articles RNA-seq Research articles RNA-seq review articles RNA-seq PubMed articles RNA-seq PubMed Central articles RNA-seq 2023 articles RNA-seq 2024 articles RNA-seq Scopus articles RNA-seq impact factor journals RNA-seq Scopus journals RNA-seq PubMed journals RNA-seq medical journals RNA-seq free journals RNA-seq best journals RNA-seq top journals RNA-seq free medical journals RNA-seq famous journals RNA-seq Google Scholar indexed journals Silkworm articles Silkworm Research articles Silkworm review articles Silkworm PubMed articles Silkworm PubMed Central articles Silkworm 2023 articles Silkworm 2024 articles Silkworm Scopus articles Silkworm impact factor journals Silkworm Scopus journals Silkworm PubMed journals Silkworm medical journals Silkworm free journals Silkworm best journals Silkworm top journals Silkworm free medical journals Silkworm famous journals Silkworm Google Scholar indexed journals isolated articles isolated Research articles isolated review articles isolated PubMed articles isolated PubMed Central articles isolated 2023 articles isolated 2024 articles isolated Scopus articles isolated impact factor journals isolated Scopus journals isolated PubMed journals isolated medical journals isolated free journals isolated best journals isolated top journals isolated free medical journals isolated famous journals isolated Google Scholar indexed journals pharmaceutical† articles pharmaceutical† Research articles pharmaceutical† review articles pharmaceutical† PubMed articles pharmaceutical† PubMed Central articles pharmaceutical† 2023 articles pharmaceutical† 2024 articles pharmaceutical† Scopus articles pharmaceutical† impact factor journals pharmaceutical† Scopus journals pharmaceutical† PubMed journals pharmaceutical† medical journals pharmaceutical† free journals pharmaceutical† best journals pharmaceutical† top journals pharmaceutical† free medical journals pharmaceutical† famous journals pharmaceutical† Google Scholar indexed journals monophagous articles monophagous Research articles monophagous review articles monophagous PubMed articles monophagous PubMed Central articles monophagous 2023 articles monophagous 2024 articles monophagous Scopus articles monophagous impact factor journals monophagous Scopus journals monophagous PubMed journals monophagous medical journals monophagous free journals monophagous best journals monophagous top journals monophagous free medical journals monophagous famous journals monophagous Google Scholar indexed journals hydrophobic permethrin articles hydrophobic permethrin Research articles hydrophobic permethrin review articles hydrophobic permethrin PubMed articles hydrophobic permethrin PubMed Central articles hydrophobic permethrin 2023 articles hydrophobic permethrin 2024 articles hydrophobic permethrin Scopus articles hydrophobic permethrin impact factor journals hydrophobic permethrin Scopus journals hydrophobic permethrin PubMed journals hydrophobic permethrin medical journals hydrophobic permethrin free journals hydrophobic permethrin best journals hydrophobic permethrin top journals hydrophobic permethrin free medical journals hydrophobic permethrin famous journals hydrophobic permethrin Google Scholar indexed journals mulberry leaf articles mulberry leaf Research articles mulberry leaf review articles mulberry leaf PubMed articles mulberry leaf PubMed Central articles mulberry leaf 2023 articles mulberry leaf 2024 articles mulberry leaf Scopus articles mulberry leaf impact factor journals mulberry leaf Scopus journals mulberry leaf PubMed journals mulberry leaf medical journals mulberry leaf free journals mulberry leaf best journals mulberry leaf top journals mulberry leaf free medical journals mulberry leaf famous journals mulberry leaf Google Scholar indexed journals
Article Details
1. Introduction
Tissue culturing is performed to maintain isolated portions of multicellular organisms in an artificial milieu that is outside the individual organism over considerable periods of time. The technique is used not only to produce materials for bio-manufacturing [1] but also for studying plant and animal cell behaviors in environments free from systemic variations that might arise in vivo during both normal homeostasis and experimental stress. Conventionally, when cells are used to produce pharmaceutical proteins, a culture method that achieves a greater target productivity is required, but owing to the diversification of modalities, the cells themselves have become therapeutic agents. As a result, there is an increasing demand for culturing methods that provide mature cells [2]. How can we establish in vivo-like culture environments that provide mature cells? Tissues in the culture environment easily dedifferentiate and lose their mature characteristics [3,4]. To suppress dedifferentiation [5,6], methods of devising cell scaffolds [7,8] and the inclusion of gases in incubators [9], as well as methods using components derived from other species, have been proposed. Methods for suppressing dedifferentiation using components derived from a microorganism [10] and from tea [11] have been reported. Why are components derived from other species effective in suppressing cell culture environment-dependent dedifferentiation? In a relationship between a certain species and another species, the latter may be indispensable for the former’s establishment. Monophagous animals are extreme examples of such interspecies relationships. For example, Koalas eat only eucalyptus leaves, and silkworms eat only mulberry leaves [12]. When an animal is completely dependent on a forage plant for its diet, its cells alone are not sufficient to form a normal individual. In these relationships, the forage plant provides more than just nutrients, proteins, lipids, sugars, vitamins, minerals and water resources to the animal. For example, components derived from the leaves may promote the actions of animal organs or function like hormones. Therefore, we investigated the effects of adding diet-derived components to the medium of the primary cultured tissues of a monophagous animal, the silkworm Bombyx mori. In previous studies, the fat bodies of B. mori, a monophagous insect, were dedifferentiated in a primary culture environment [13]. A method to quantitatively evaluate the dedifferentiation of primary cultured tissues using a transcriptome has been established [13,14]. The previous work of the author-imposed significance of utilizing entropy in an excellent way in 2010, and its piercing insight lead to other fine articles [15,16]. Specifically, the information entropy of the transcriptome data can be used as an index of transcriptome diversity. The value decreases when a certain cell is in a differentiated state in the living body and expresses intensively a specific gene. The value increases when a certain cell tends to dedifferentiate in a culture environment and the expression of the specific gene is not promoted. A system that has lost control has increased entropy. The dedifferentiated cultured tissues may be re-differentiated by the addition of a high concentration of hydrophilic phenobarbital or hydrophobic permethrin [14]. The addition of these molecules, with their opposite affinities for water, result in similar responses. Thus, the tissue responses were not interpreted as being caused by a compound-specific bioactivity, but by the stimulation of a control system that exists in vivo. Thus, we aimed to identify the stimuli in this in vivo control system. In this monophagous insect, the fat body is located adjacent to the midgut, which is filled with chewed mulberry leaves. The fat body appears as a thin worm-like tissue wrapped around a mulberry-leaf sausage. Thus, we hypothesized that the mulberry leaf inside the midgut, or compounds from the leaves, contributes to the in vivo environment. Therefore, in this study, we investigated the effects of mulberry leaf-derived components on the transcriptomes of cultured silkworm fat bodies. Dedifferentiated ‘p50’ fat bodies were primary cultured in a medium containing a silkworm midgut extract and a medium containing a mulberry leaf extract, and changes in the information entropy of the transcriptomes were examined. In addition, fat bodies of ‘Sawa-J’, a broad-eating silkworms strain obtained by breeding, were used. The dedifferentiated ‘Sawa-J’ fat bodies were cultured in media containing silkworm midgut, mulberry leaf, or cabbage leaf extracts, and changes in the information entropy of the transcriptomes were examined. In addition, we examined the information entropies of the transcriptomes of the ‘Sawa-J’ fat bodies 12, 24, and 48 h after the start of culturing and compared them to those of in vivo ‘Sawa-J’ fat bodies. Silkworm fat bodies dedifferentiate 80 h after culturing [13,14], but it was unclear how many hours were sufficient for dedifferentiation.
2. Materials and Methods
2.1 Insects and Plants
The ‘p50’ and ‘Sawa-J’ strains of B. mori were reared on fresh leaves from the mulberry Morus alba. At 3 days after the 4th ecdysis, 5th-instar larvae were harvested. Female insects were used. Morus bombycis was cultivated on a farm at the Tokyo University of Agriculture and Technology, Fuchu-shi, Tokyo, Japan. The cabbage Brassica oleracea var. capitata was obtained at a supermarket (Maruetsu, Kawasaki, Kanagawa, Japan).
2.2 Media Preparation
The MGM-450 insect medium (10% fetal bovine serum) was used in this study [17]. A 2-mL stock solution of silkworm midgut extract was prepared culturing isolated the midgut for 10 min. For culturing, the silkworm midgut extract’s stock solution was diluted 10-fold in the medium. The mulberry leaf extract’s stock solution was prepared by homogenizing 0.25 g of the leaves in 2 mL of medium using scissors (Dissecting Scissors Iris 11 cm D340-476; FRIGZ, Chiba, Japan). For culturing, the mulberry leaf extract’s stock solution was diluted 100-fold and 1,000-fold independently in the medium. The cabbage leaf extract’s stock solution was prepared by homogenizing 0.25 g of leaves in 2 mL of medium using scissors. For culturing, the cabbage leaf extract’s stock solution was used.
2.3 Culturing of ‘p50’ Fat Bodies in Mulberry Leaf Extract
The silkworm larvae were surface-sterilized by submersion in 70% ethyl alcohol, and the fat bodies were removed without injuring the digestive system. The intact fat bodies of four ‘p50’ larvae (Samples 1, 2, 3, and 4) were dissected, soaked in 0.35 mL of TRIzol LS (Invitrogen, CA, USA), and homogenized using a BioMasher II (Nippi, Tokyo, Japan) and a PowerMasher II (Nippi). Then, the tissues were maintained at −60°C until used. More than 100 fat body tissue samples with intact lobes (approx. 2 mm3) were excised from fat bodies dissected from 21 larvae. Tweezers (KFI K-23 No. 3?120 m/m 18-8 Stainless 230001; Kowa Pinset Industry, Tokyo, Japan) and scissors (FRIGZ) were used. These tissue samples were incubated in cell culture dishes (ø=35 mm; AS ONE, Osaka, Japan) with 2 mL of MGM-450 insect medium and no gas change. The tissue samples were cultured without antibiotics for 80 h at 25°C. We examined the morphology of cultured fat body cells using a phase contrast microscope (ECLIPSE TS100; Nikon, Tokyo, Japan). The presence or absence of microbial contamination in the culture was monitored by microscopic inspection. Whole cultured tissues were mixed in a cell culture dish (ø= 100 mm; AS ONE) to eliminate the effects of inter-individual variation observed in primary cultures [18]. The original medium in each dish was replaced with an assay medium. In ‘p50’ controls, the original medium was replaced with fresh medium (Samples 5, 6, and 7). The primary cultured ‘p50’ tissues were incubated in two concentrations of mulberry leaf extract (mulberry 1/1,000: Samples 8, 9, and 10; and mulberry 1/100: Samples 11, 12, and 13). The primary cultured ‘p50’ tissues were incubated in medium containing silkworm midgut extract (Sample 14). After 10 h, the cultures were terminated by soaking the tissues in 0.35 mL of TRIzol LS, and tissues were then homogenized using a BioMasher II (Nippi) and a PowerMasher II (Nippi). Samples were maintained at −60°C until used.
2.4 Culturing of ‘Sawa-J’ Fat Bodies in Mulberry Leaf Extract
The larvae were surface-sterilized as described previously. The intact fat bodies of ‘Sawa-J’ larvae (Sample 15) were dissected and maintained at −60°C until used as described previously. More than 50 fat body tissue samples with intact lobes (approx. 2 mm3) were excised from fat bodies dissected from 12 larvae. These tissue samples were incubated 80 h and observed as described previously. Whole cultured tissues were mixed in a cell culture dish as described previously. The primary cultured ‘Sawa-J’ control tissues were incubated in fresh medium for 10 h (Samples 16, 17, and 18). The primary cultured ‘Sawa-J’ tissues were incubated in two concentrations of mulberry leaf extract for 10 h (mulberry 1/1,000: Samples 19, 20, and 21; and mulberry 1/100: Samples 22, 23, and 24). The primary cultured ‘Sawa-J’ fat bodies were incubated in a medium containing silkworm midgut extract for 10 h (Samples 25, 26, and 27) and in a medium containing cabbage leaf extract for 10 h (Samples 28, 29, and 30). The cultures were terminated as described previously.
2.5 Time Course of the ‘Sawa-J’ Tissue Cultures
The larvae were surface-sterilized as described previously. The intact fat bodies of three ‘Sawa-J’ larvae (0 h; Samples 31, 32, and 33) were dissected and maintained at −60°C until used as described previously. More than 30 fat body tissue samples with intact lobes were excised from fat bodies dissected from 9 larvae. These ‘Sawa-J’ tissue samples were incubated for 12 (Samples 34, 35, 36, 37, and 38), 24 (Samples 39, 40, and 41), and 48 (Samples 42, 43, and 44) h. The cultures were terminated as described previously.
2.6 RNA Extraction and Sequencing
Total RNA was extracted using TRIzol LS and a Direct-zol RNA Kit (Zymo Research, CA, USA) following the manufacturers’ instructions. The homogenates were incubated for 5 min at 25°C, and the samples were subjected to centrifugation at 12,000 ×g for 10 min at 5°C. The supernatants were then transferred to new tubes and incubated at 25°C for 5 min. An equal volume (approximately 0.35 mL) of ethanol was added to each sample homogenate and mixed thoroughly. The mixtures were then transferred into Zymo-Spin IIICG Columns in collection tubes and centrifuged. The flow-through was discarded, and columns were transferred into new collection tubes. Then, 400 µL Direct-zol RNA PreWash was added to each column, and the columns were centrifuged. The flow-through was discarded, and the prewash and centrifugation were repeated. Afterward, 700 µL RNA Wash Buffer was added to each column, and they were centrifuge for 1 min. The columns were carefully transferred into RNase-free tubes. To elute the RNA, 100 µL of DNase/RNase-free water was added directly to each column matrix, and the columns were centrifuged. The integrity of the rRNA in each sample was checked using an Agilent TapeStation 4200 (Agilent Technologies, CA, and USA). mRNA libraries were prepared for RNA sequencing using a TruSeq RNA Sample kit (Illumina, San Diego, CA, USA) in accordance with the manufacturer’s protocols. These libraries were sequenced using a NextSeq 500 sequencer (Illumina) in accordance with the manufacturer’s protocols. Short-read data were deposited in the Short Read Archive (project ID, DRA011291) of the DNA Data Bank of Japan. Short-read sequences were mapped to an annotated silkworm transcript sequence obtained from SilkDB [19] using hisat2 [20]. We did not use other full-length cDNA data set from National Agriculture and Food Research Organisation in 2011 (AK377185 - AK388575) since over 30% multi-hits were found.
2.7 Data Analyses
The Shannon entropies of the transcriptome data were calculated as described previously [14] using R (Supplemental Material 1). A t-test was performed on the null hypothesis, which stated that the mulberry leaf extract’s concentration did not affect the information entropy of the transcriptomes of the ‘p50’ fat bodies, using R [21] and Fitting Linear Models (lm) (Samples 5, 6, 7, 8, 9, 10, 11, 12, and 13). A t-test was performed on the null hypothesis, which stated that the mulberry leaf extract’s concentration did not affect the information entropy of the transcriptomes of the ‘Sawa-J’ fat bodies, using R and lm (Samples 16, 17, 18, 19, 20, 21, 22, and 23). A t-test was performed on the null hypothesis, which stated that the hours after dissection did not affect the information entropy of the transcriptomes of the ‘Sawa-J’ fat bodies, using R and lm (Samples 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, and 44). Data were visualized using R.
|
Sample ID |
Shannon Entropy |
Culture Condition |
Strain |
|
1 |
6.865143 |
Not cultured |
p50 |
|
2 |
5.914764 |
Not cultured |
p50 |
|
3 |
6.605657 |
Not cultured |
p50 |
|
4 |
6.454235 |
Not cultured |
p50 |
|
5 |
10.03083 |
Control |
p50 |
|
6 |
10.03309 |
Control |
p50 |
|
7 |
9.586332 |
Control |
p50 |
|
8 |
9.587584 |
MulberryLeaf1/1000 |
p50 |
|
9 |
9.915635 |
MulberryLeaf1/1000 |
p50 |
|
10 |
9.313175 |
MulberryLeaf1/1000 |
p50 |
|
11 |
9.1602 |
MulberryLeaf1/100 |
p50 |
|
12 |
9.480333 |
MulberryLeaf1/100 |
p50 |
|
13 |
8.539101 |
MulberryLeaf1/100 |
p50 |
|
14 |
8.255311 |
Midgut |
p50 |
|
15 |
6.532235 |
Not cultured |
Sawa-J |
|
16 |
10.09469 |
Control |
Sawa-J |
|
17 |
9.414184 |
Control |
Sawa-J |
|
18 |
9.33448 |
Control |
Sawa-J |
|
19 |
8.774243 |
MulberryLeaf1/1000 |
Sawa-J |
|
20 |
8.881261 |
MulberryLeaf1/1000 |
Sawa-J |
|
21 |
7.642716 |
MulberryLeaf1/1000 |
Sawa-J |
|
22 |
8.665842 |
MulberryLeaf1/100 |
Sawa-J |
|
23 |
8.931488 |
MulberryLeaf1/100 |
Sawa-J |
|
24 |
7.911349 |
MulberryLeaf1/100 |
Sawa-J |
|
25 |
8.412092 |
Midgut |
Sawa-J |
|
26 |
8.295697 |
Midgut |
Sawa-J |
|
27 |
8.95734 |
Midgut |
Sawa-J |
|
28 |
9.434283 |
CabbageLeaf |
Sawa-J |
|
29 |
9.661787 |
CabbageLeaf |
Sawa-J |
|
30 |
9.491179 |
CabbageLeaf |
Sawa-J |
|
31 |
6.859353 |
Not cultured |
Sawa-J |
|
32 |
6.202299 |
Not cultured |
Sawa-J |
|
33 |
6.239494 |
Not cultured |
Sawa-J |
|
34 |
5.89826 |
Culture12hours |
Sawa-J |
|
35 |
6.813056 |
Culture12hours |
Sawa-J |
|
36 |
6.593159 |
Culture12hours |
Sawa-J |
|
37 |
6.432197 |
Culture12hours |
Sawa-J |
|
38 |
7.37291 |
Culture12hours |
Sawa-J |
|
39 |
6.774276 |
Culture24hours |
Sawa-J |
|
40 |
7.083945 |
Culture24hours |
Sawa-J |
|
41 |
6.835882 |
Culture24hours |
Sawa-J |
|
42 |
6.537758 |
Culture48hours |
Sawa-J |
|
43 |
6.895498 |
Culture48hours |
Sawa-J |
|
44 |
6.266251 |
Culture48hours |
Sawa-J |
Supplementary Material 1: Information entropy of transcriptomes. In total, 44 transcriptomes were sequenced. Sample IDs, information entropies, culture conditions, and strains are summarized.
3. Results
The mulberry leaf extract decreased the information entropies of the transcriptomes of the ‘p50’ fat bodies (p = 0.025) (Figures 1A–D and 2). The silkworm midgut extract decreased the transcriptome diversity levels in the ‘p50’ fat bodies (Figure 1). The decrease in the information entropies of the transcriptomes of the ‘p50’ fat bodies cultured in the mulberry leaf extract were reproduced in another strain, ‘Sawa-J’ (p = 0.0457) (Figures 3A–E and 4). The silkworm midgut extract also decreased the transcriptome diversity levels in the ‘Sawa-J’ fat bodies (Figure 3). Fat-body dedifferentiation was not observed in the primary cultures until 48 h (Figures 5A–D and 6).
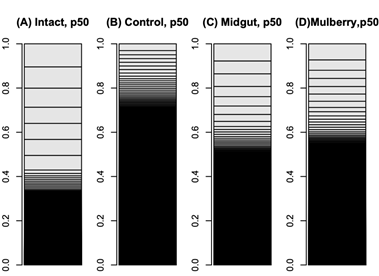
Figure 1: Bar charts of ‘p50’ silkworm fat-body transcriptomes. The occupation rates of genes in transcriptomes were expressed as bar charts. Heights of boxes in the bar charts indicate the occupation rates of the genes in the transcriptomes. Although more than 14,000 genes are included in these bar charts, most are invisible and are included in the black regions. (A) Transcriptome of intact silkworm fat-body tissue. (B) Transcriptome of fat-body tissues cultured for 90 h in MGM-450 insect medium. (C, D) Transcriptomes of fat-body tissues cultured for 80 h in MGM-450 insect medium and then an additional 10 h in medium supplemented with silkworm midgut (C) or mulberry leaf (D) extract.
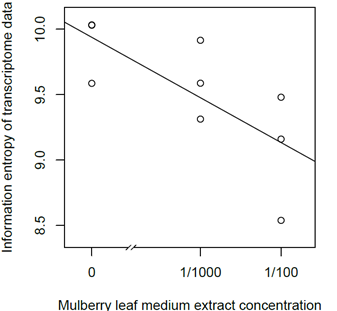
Figure 2: Scatter plot of the Mulberry leaf extract concentration vs the information entropy of the transcriptome data. Transcriptomes of ‘p50’ silkworm fat-body tissues that were cultured for 80 h in MGM-450 insect medium followed by 10 h in MGM-450 insect medium supplemented with Mulberry leaf extract.
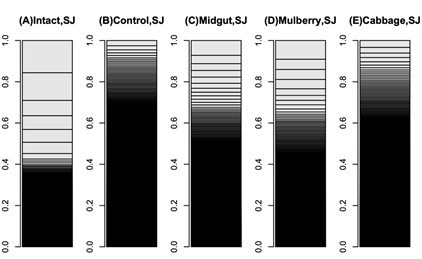
Figure 3: Bar charts of ‘Sawa-J’ silkworm fat-body transcriptomes. The occupation rates of genes in the transcriptomes were expressed as bar charts. Heights of boxes in the bar charts indicate the occupation rates of the genes in the transcriptomes. Although more than 14,000 genes are included in these bar charts, most are invisible and are included in the black regions. (A) Transcriptome of intact silkworm fat-body tissue. (B) Transcriptome of fat-body tissues cultured for 90 h in MGM-450 insect medium. (C–E) Transcriptomes of fat-body tissues cultured for 80 h in MGM-450 insect medium and then an additional 10 h in medium supplemented with silkworm midgut (C), mulberry leaf (D), or cabbage leaf (E) extract.
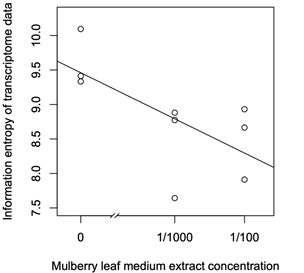
Figure 4: Scatter plot of Mulberry leaf extract concentrations vs the information entropy of the transcriptome data. Transcriptomes of ‘Sawa-J’ silkworm fat-body tissues that were cultured for 80 h in MGM-450 insect medium followed by 10 h in MGM-450 insect medium supplemented with Mulberry leaf extract.
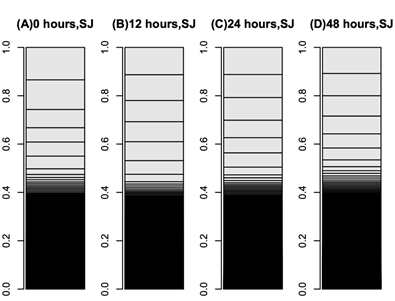
Figure 5: Bar charts of ‘Sawa-J’ silkworm fat-body transcriptomes. The occupation rates of genes in the transcriptomes were expressed as bar charts. Heights of boxes in the bar charts indicate the occupation rates of the genes in the transcriptomes. Although more than 14,000 genes are included in these bar charts, most are invisible and are included in the black regions. (A–D) Transcriptomes of fat-body tissues cultured for 0 (A), 12 (B), 24 (C), and 48 (D) h in MGM-450 insect medium.
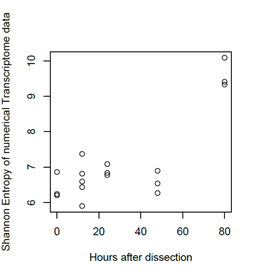
Figure 6: Scatter plot of culture time vs the information entropy of the transcriptome data. Transcriptomes of ‘Sawa-J’ silkworm fat-body tissues that were cultured for 0, 12, 24, 48 and 80 hours in MGM-450 insect medium.
4. Discussion
The medium containing the midgut extract showed a bioactivity similar to that of the medium containing the mulberry leaf extract (Figures 1 and 3). This suggested that the bioactivity found in the midgut extract is derived from mulberry leaves. Thus, the midgut in living silkworms may release compounds from the mulberry leaves. Therefore, we hypothesized that components of mulberry leaves affect the fat bodies in the living organisms. The information entropies of the transcriptomes of the fat bodies decreased when primary culture-dependent dedifferentiated fat bodies were cultured in the mulberry leaf extract (Figures 2 and 4). The mulberry leaf extract-dependent tissue re-differentiation was shown in two silkworm strains (‘p50’ and ‘Sawa-J’). Thus, the previously discovered tissue responses to phenobarbital and permethrin may not have been caused by compound-specific bioactivity but by stimulating the in vivo control system, which originally functioned using components of mulberry leaves. This indicates that monophagous animals use components of their special feed not only as nutrients but, like hormones, to control homeostasis. Because maintaining a system that ensures a constant output, regardless of the input, requires a cost, there may be situations where a system that performs only the fixed processing of the input is selected. As a result of choosing the latter, monophasic animals may have lost the system that allows any food to be used in growth and development. In fact, silkworms grow poorly when raised on artificial diets that do not contain mulberry. In other insects, dietary changes result in changed outputs, such as pheromone secretions. One lepidopteran insect changes its secreted sex pheromone components in response to being fed artificial diets [22]. ‘Sawa-J’ is an abnormal feeding-habit strain that feeds on an artificial diet lacking mulberry leaf components [23,24]. It was expected that the fat bodies from members of this strain would differentiate in response to components derived from plants other than mulberry, but the cabbage extract had no effect (Figure 3). Micro-particles of a uniform shape were observed in ‘Sawa-J’ tissue cultures. We revealed that the cultures were infected with NPV, and we identified many NPV sequences (GenBank: JQ071499.1) using RNA-seq. The NPV infection did not interrupt the mulberry leaf-dependent re-differentiation of the fat bodies. In this experiment, mulberry leaf extract was used in the medium to develop a culture environment, and thus, fat-body transcriptomes, that was closer to the in vivo environment. However, the information entropy of the in vivo fat-body transcriptome is clearly less than that of the transcriptome of a fat body cultured in a medium supplemented with the mulberry leaf extract. Thus, the in vivo-like culture techniques for silkworm fat bodies can be improved. Here, we revealed that cultured fat bodies did not immediately dedifferentiate (< 48 h). This contrasts the cell re-differentiation seen within 10 h when cells were transferred from a medium supplemented with Phenobarbital and permethrin to a drug-deficient medium [14]. By clarifying this difference, we hope to obtain information that will allow us to mimic the in vivo environment in an in vitro culture system. In this decade, several studies [15,25-27] following our research [14] repeatedly measured the degree of cellular dedifferentiation and differentiation as a transcriptome Shannon entropy. The Shannon entropy is a kind of alpha diversity in ecology [28], and the transcriptome Shannon entropy is simply transcriptome diversity [14,29]. It is not incorrect to call it the alpha diversity of the transcriptome, but that would leave its biological significance undefined, as would each principal component that came up in the principal component analysis. Since we can quantitatively assess, judge, and define that dedifferentiation is an increase in the Shannon entropy of the transcriptome and differentiation is a decrease in the Shannon entropy of the transcriptome, it is more accurate to position the value not as a mere bioinformatics measure; however, as a number with obvious biological and bioengineering significance, such as viable cell rate, cell density, specific growth rate, or pcd (pg/cell/day). Here we call the quantitative value of cellular dedifferentiation and differentiation “liberality,” since a previous study explained the changes were happening to cultured cells as “libère” [30-33].
5. Conclusions
To investigate the possibility that feed-derived components are essential to the normal lives of monophagous silkworm, mulberry leaf components were added to the dedifferentiating primary cultures of silkworm fat bodies. Using mulberry leaf extract, we were able to differentiate silkworm fat bodies in primary cultures.
Acknowledgements
We would like to thank Ms. Haruna Nagata at the Tokyo University of Agriculture and Technology for maintaining the silkworm lines. We thank Lesley Benyon, PhD, from Edanz Group (https://en-author-services.edanz.com/) for editing a draft of this manuscript.
References
- Ogata N, Nishimura A, Matsuda T, et al. Single-cell transcriptome analyses reveal heterogeneity in suspension cultures and clonal markers of CHO-K1 cells. Biotechnology and Bioengineering, bit (2020): 27624.
- Matsumoto E, Koide N, Hanzawa H, et al. Fabricating retinal pigment epithelial cell sheets derived from human induced pluripotent stem cells in an automated closed culture system for regenerative medicine. PLOS ONE 14 (2019): e0212369.
- Champy C. Quelques résultats de la méthode de culture de tissus., IV. La Rétine. Arch, de Zool. Exp 66 (1914).
- Sato G, Zaroff L, Mills SE. Tissue Culture Populations and their Relation to the Tissue of Origin. Proc Natl Acad Sci 46 (1960): 963-972.
- Bolleyn J, Fraczek J, Rogiers V, et al. Epigenetic Modifications as Antidedifferentiation Strategy for Primary Hepatocytes in Culture (2015): 203-211.
- Fraczek J, Bolleyn J, Vanhaecke T, et al. Primary hepatocyte cultures for pharmaco-toxicological studies: at the busy crossroad of various anti-dedifferentiation strategies. Archives of Toxicology 87 (2013): 577-610.
- Burkhardt B, Martinez-Sanchez JJ, Bachmann A, et al. Long-term culture of primary hepatocytes: New matrices and microfluidic devices. Hepatology International 8 (2014): 14-22.
- Mao Y, Block T, Singh-Varma A, et al. Extracellular matrix derived from chondrocytes promotes rapid expansion of human primary chondrocytes in vitro with reduced dedifferentiation. Acta Biomaterialia 85 (2019): 75-83.
- Guo M, Bao EL, Wagner M, et al. SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res 45 (2017): e54.
- Feil G, Horres R, Schulte J, et al. Bacterial cellulose shifts transcriptome and proteome of cultured endothelial cells towards native differentiation. Molecular and Cellular Proteomics 16 (2017): 1563-1577.
- Kaida K, Honda Y, Hashimoto Y, et al. Application of green tea catechin for inducing the osteogenic differentiation of human dedifferentiated fat cells in vitro. International Journal of Molecular Sciences 16 (2015): 27988-28000.
- Zhang ZJ, Zhang SS, Niu BL, et al. (2019) A determining factor for insect feeding preference in the silkworm, Bombyx mori. PLoS Biology 17 (2019).
- Ogata N, Yokoyama T, Iwabuchi K (2012) Transcriptome responses of insect fat body cells to tissue culture environment. PloS One 7 (2012): e34940.
- Ogata N, Kozaki T, Yokoyama T, et al. Comparison between the Amount of Environmental Change and the Amount of Transcriptome Change. PloS One 10 (2015): e0144822.
- Kannan S, Farid M, Lin LB, et al. Transcriptomic entropy benchmarks stem cell-derived cardiomyocyte maturation against endogenous tissue at single cell level, PloS Comput Biol 17 (2021): e1009395.
- Liu B, Li C, Li Z, et al. An entropy-based metric for assessing the purity of single cell populations. Nat. Commun 11 (2020): 3155.
- Mitsuhashi J, Inoue H. Obtainment of a continuous cell line from the fat bodies of the mulberry tiger moth, Spilosoma imparilis (Lepidoptera: Arctiidae). Entomol. Zool 23 (1988): 488-490.
- Ogata N, Iwabuchi K. (2017) Relevant principal factors affecting the reproducibility of insect primary culture. In Vitro Cellular & Developmental Biology – Animal 53 (2017): 532-537.
- Xia Q, Zhou Z, Lu C, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306 (2004): 1937-1940.
- Kim D, Paggi JM, Park C, et al. (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 37 (2019): 907-915.
- R Core Team. R: A Language and Environment for Statistical Computiong (2011).
- Mujiono K, Witjaksono W, Putra NS. The Sex Pheromone Content of The Spodoptera Exigua (Hubner) Under Artificial and Natural Diets. International Journal of Science and Engineering 8 (2015): 146-150.
- Iizuka T, Tamura T, Sezutsu H, et al. Genetic Analysis of the Electrophysiological Response to Salicin, a Bitter Substance, in a Polyphagous Strain of the Silkworm Bombyx mori. PLoS ONE 7 (2012): e37549.
- Mase K, Iizuka T, Yamamoto T, et al. Genetic mapping of a food preference gene in the silkworm, Bombyx mori, using restriction fragment length polymorphisms (RFLPs). Genes & Genetic Systems 82 (2007): 249-256.
- Guo R, Xu X, Lu Y, et al. Physiological oxygen tension reduces hepatocyte dedifferentiation in in vitro culture. Scientific Reports 7 (2017): 5923.
- Teschendorff AE, Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome, Nat Commun 8 (2017): 15599.
- Wiesner K, Teles J, Hartnor M, et al. Haematopoietic stem cells: entropic landscapes of differentiation. Interface Focus 6 (2018): 20180040.
- Dongmei A, Ruocheng H, Jin W, et al. Integrated metagenomic data analysis demonstrates that a loss of diversity in oral microbiota is associated with periodontitis. BMC Genomics 18 (2017): 1041.
- Martinez O, Reyes-Valdes MH. Defining diversity specialization and gene specificity in transcriptomes through information theory, Natl. Acad. Sci. U.S.A 105 (2008): 9709-9714.
- Canguilhem G. La connaissance de la vie, Pari: Hachette (1952): 41.
- Ogata N, Hosaka A. Cellular liberality is measurable as Lempel-Ziv complexity of fastq files, 2022 IEEE 22nd International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan (2022): 321-326.
- Ogata N. Transcriptome Dedifferentiation Observed in Animal Primary Cultures is Essential to Plant Reprogramming. Bioinform. Syst. Biol 5 (2022):116-118.
- Ogata N. The Growing Liberality Observed in Primary Animal and Plant Cultures is Common to the Social Amoeba. J. Syst. Biol 6 (2023): 10-17.
