In Silico Transcriptomic Analysis of Canadine Accumulation in Papaver Somniferum Cultivar
Article Information
Sai Batchu*
Department of Biology, The College of New Jersey, 2000 Pennington Rd. Pennington, NJ, USA
*Corresponding Author: Sai Batchu, Department of Biology, The College of New Jersey, 2000 Pennington Rd. Pennington, NJ, USA
Received: 20 February 2020; Accepted: 13 March 2020; Published: 20 March 2020
Citation:
Sai Batchu. In Silico Transcriptomic Analysis of Canadine Accumulation in Papaver Somniferum Cultivar. Journal of Biotechnology and Biomedicine 2 (2020): 024-028.
View / Download Pdf Share at FacebookAbstract
Papaver somniferum, colloquially known as opium poppy, currently remains as the only commercial source of pharmaceutically important benzylisoquinoline alkaloids, of which include canadine. Canadine is an important naturally occurring alkaloid in current demand for pharmaceutical studies. Understanding the biosynthesis in plantae will help in increasing production of this alkaloid. In the present study, a comparative transcriptomic analysis was performed between gene expression data procured from the normal cultivar and a high canadine cultivar. Results indicated differential expression of key enzymes in the phthalideisoquinoline pathway leading to canadine biosynthesis. Specifically, it was found that transcription of genes encoding berberine bridge enzyme canadine synthase, and scoulerine 9-O-methyltransferase were significantly increased in the high canadine cultivar compared to the normal cultivar. These findings provide basis for further elucidating the coordinated transcriptional processes underlying canadine accumulation in P. somniferum cultivars.
Keywords
Canadine, Papaver somniferum, Transcriptomic, In silico
Canadine articles Canadine Research articles Canadine review articles Canadine PubMed articles Canadine PubMed Central articles Canadine 2023 articles Canadine 2024 articles Canadine Scopus articles Canadine impact factor journals Canadine Scopus journals Canadine PubMed journals Canadine medical journals Canadine free journals Canadine best journals Canadine top journals Canadine free medical journals Canadine famous journals Canadine Google Scholar indexed journals Papaver somniferum articles Papaver somniferum Research articles Papaver somniferum review articles Papaver somniferum PubMed articles Papaver somniferum PubMed Central articles Papaver somniferum 2023 articles Papaver somniferum 2024 articles Papaver somniferum Scopus articles Papaver somniferum impact factor journals Papaver somniferum Scopus journals Papaver somniferum PubMed journals Papaver somniferum medical journals Papaver somniferum free journals Papaver somniferum best journals Papaver somniferum top journals Papaver somniferum free medical journals Papaver somniferum famous journals Papaver somniferum Google Scholar indexed journals Transcriptomic articles Transcriptomic Research articles Transcriptomic review articles Transcriptomic PubMed articles Transcriptomic PubMed Central articles Transcriptomic 2023 articles Transcriptomic 2024 articles Transcriptomic Scopus articles Transcriptomic impact factor journals Transcriptomic Scopus journals Transcriptomic PubMed journals Transcriptomic medical journals Transcriptomic free journals Transcriptomic best journals Transcriptomic top journals Transcriptomic free medical journals Transcriptomic famous journals Transcriptomic Google Scholar indexed journals In silico articles In silico Research articles In silico review articles In silico PubMed articles In silico PubMed Central articles In silico 2023 articles In silico 2024 articles In silico Scopus articles In silico impact factor journals In silico Scopus journals In silico PubMed journals In silico medical journals In silico free journals In silico best journals In silico top journals In silico free medical journals In silico famous journals In silico Google Scholar indexed journals myogenesis articles myogenesis Research articles myogenesis review articles myogenesis PubMed articles myogenesis PubMed Central articles myogenesis 2023 articles myogenesis 2024 articles myogenesis Scopus articles myogenesis impact factor journals myogenesis Scopus journals myogenesis PubMed journals myogenesis medical journals myogenesis free journals myogenesis best journals myogenesis top journals myogenesis free medical journals myogenesis famous journals myogenesis Google Scholar indexed journals muscle preservation articles muscle preservation Research articles muscle preservation review articles muscle preservation PubMed articles muscle preservation PubMed Central articles muscle preservation 2023 articles muscle preservation 2024 articles muscle preservation Scopus articles muscle preservation impact factor journals muscle preservation Scopus journals muscle preservation PubMed journals muscle preservation medical journals muscle preservation free journals muscle preservation best journals muscle preservation top journals muscle preservation free medical journals muscle preservation famous journals muscle preservation Google Scholar indexed journals antioxidant activity articles antioxidant activity Research articles antioxidant activity review articles antioxidant activity PubMed articles antioxidant activity PubMed Central articles antioxidant activity 2023 articles antioxidant activity 2024 articles antioxidant activity Scopus articles antioxidant activity impact factor journals antioxidant activity Scopus journals antioxidant activity PubMed journals antioxidant activity medical journals antioxidant activity free journals antioxidant activity best journals antioxidant activity top journals antioxidant activity free medical journals antioxidant activity famous journals antioxidant activity Google Scholar indexed journals biosynthesis articles biosynthesis Research articles biosynthesis review articles biosynthesis PubMed articles biosynthesis PubMed Central articles biosynthesis 2023 articles biosynthesis 2024 articles biosynthesis Scopus articles biosynthesis impact factor journals biosynthesis Scopus journals biosynthesis PubMed journals biosynthesis medical journals biosynthesis free journals biosynthesis best journals biosynthesis top journals biosynthesis free medical journals biosynthesis famous journals biosynthesis Google Scholar indexed journals Benzylisoquinoline alkaloids articles Benzylisoquinoline alkaloids Research articles Benzylisoquinoline alkaloids review articles Benzylisoquinoline alkaloids PubMed articles Benzylisoquinoline alkaloids PubMed Central articles Benzylisoquinoline alkaloids 2023 articles Benzylisoquinoline alkaloids 2024 articles Benzylisoquinoline alkaloids Scopus articles Benzylisoquinoline alkaloids impact factor journals Benzylisoquinoline alkaloids Scopus journals Benzylisoquinoline alkaloids PubMed journals Benzylisoquinoline alkaloids medical journals Benzylisoquinoline alkaloids free journals Benzylisoquinoline alkaloids best journals Benzylisoquinoline alkaloids top journals Benzylisoquinoline alkaloids free medical journals Benzylisoquinoline alkaloids famous journals Benzylisoquinoline alkaloids Google Scholar indexed journals pharmaceutically articles pharmaceutically Research articles pharmaceutically review articles pharmaceutically PubMed articles pharmaceutically PubMed Central articles pharmaceutically 2023 articles pharmaceutically 2024 articles pharmaceutically Scopus articles pharmaceutically impact factor journals pharmaceutically Scopus journals pharmaceutically PubMed journals pharmaceutically medical journals pharmaceutically free journals pharmaceutically best journals pharmaceutically top journals pharmaceutically free medical journals pharmaceutically famous journals pharmaceutically Google Scholar indexed journals
Article Details
1. Introduction
Opium poppy (Papaver somniferum) is an important source of benzylisoquinoline alkaloids, a group of plant secondary metabolites that exhibits a multitude of pharmacological activities including antimalarial [1], antispasmodic [2], analgesic [3], and antitussive [4]. Canadine is a benzylisoquinoline alkaloid implicated in myogenesis, muscle preservation, and antioxidant activity [5, 6]. Given that canadine is an important naturally occurring alkaloid in current demand for pharmaceutical studies and procurement is limited to natural producers, it is imperative to explore novel methods for producing this compound. Understanding the biosynthesis of canadine in its natural producers, such as opium poppy, will provide insight for engineering metabolons, selective breeding, and prompting exploration for new sources of canadine. Analyzing gene expression profiles can allow for the elucidation of molecular pathways leading to synthesis of metabolites, especially for characterizing the transcriptional regulation of crucial enzymes [7, 8]. Previous studies have used transcriptome analysis to characterize the biosynthesis of benzylisoquinoline alkaloids in P. somniferum [9, 10]. However, no previous study has utilized comparative transcriptomics to focus on canadine biosynthesis in plantae between different cultivars of opium poppy.
2. Materials and Methods
Publically available transcriptome datasets of opium poppy leaves [11], were retrieved from the NCBI Sequence Read Archive under accession numbers SRR8327180, SRR8327186, SRR8327187 for three replicates of the high canadine cultivar T and SRR8325825, SRR8325826, SRR8325833 for three replicates of the normal B1 cultivar. Sequences were filtered with Trim Galore [12] to remove low quality reads and adaptor sequences. FastQC [13] was used to determine final quality of reads. Cleaned reads were mapped to indexed Papaver somniferum genome assembly version 1 [14] with HISAT2 aligner run with default parameters [15]. Alignment output was assembled and quantified using StringTie [16]. DESeq2 [17], an R software package [18], was used for data normalization. Genes with adjusted p value <0.05 and log2fold change <-1 or log2fold change >1 were deemed as differentially expressed. Additional figures were generated with R package ggplot2 [19].
3. Results
The specific cultivar, T, analyzed in this study has been shown to accumulate canadine in greater amount compared to normal cultivar B1 (Table1). Further analysis revealed an increased expression of key enzymes in the canadine biosynthesis pathway such as canadine synthase, berberine bridge enzyme, and scoulerine 9-O-methyltransferase (Figure 1A, 1B). Genes for other enzymes in the canadine biosynthesis, such as pathway did not display significant differential regulation compared to B1 cultivar.
|
BIA |
B1 |
T |
|
Norcoclaurine |
46.2 ± 13.25 |
55.1 ± 9.56 |
|
Scoulerine |
632.3 ± 116.43 |
107.8 ± 4.53 |
|
Papaverine |
175.2 ± 44.88 |
99.9 ± 12.25 |
|
Canadine |
1150.9 ± 257.82 |
1641.6 ± 372.12 |
Table 1: Benzylisoquinoline alkaloids (BIA) detected in opium poppy cultivars. All data are represented as mean ± SD from three independent plants (n = 3). The unit is mg/kg dry leaves [7].
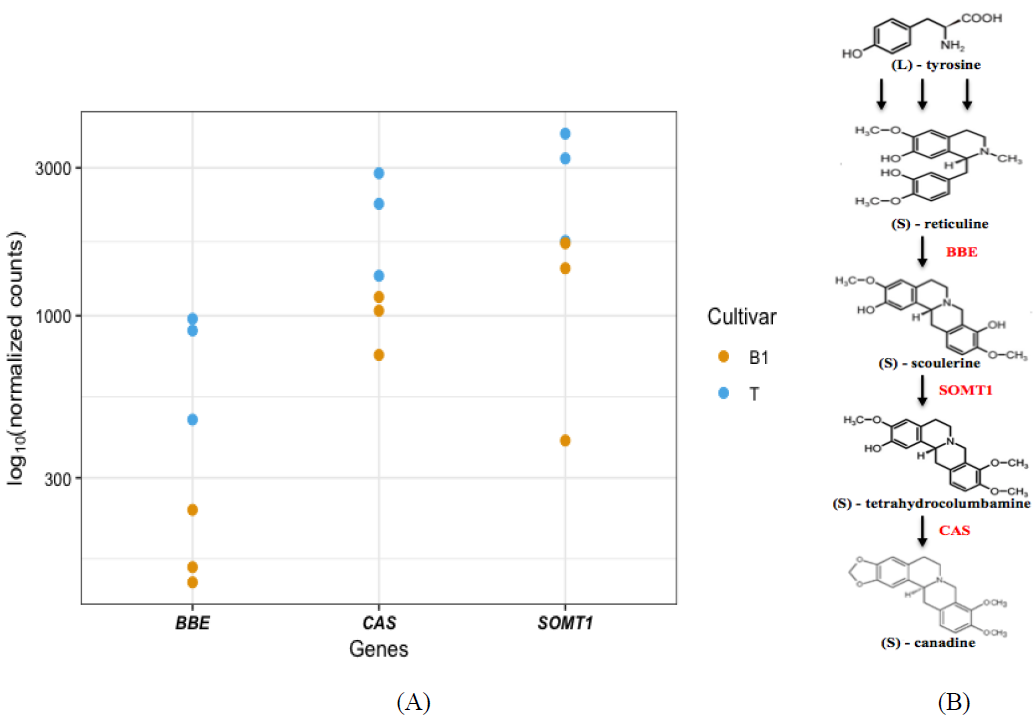
Figure 1: (A) Normalized counts of berberine bridge enzyme (BBE), canadine synthase (CAS) and scoulerine 9-O-methyltransferase (SOMT1) in the two opium poppy cultivars; (B) Biosynthesis of canadine in opium poppy. (S)-Scoulerine is formed from (S)-reticuline by berberine bridge enzyme (BBE). (S)-Canadine is then formed sequentially from (S)-scoulerine by scoulerine 9-O-methyltransferase (SOMT1) yielding tetrahydrocolumbamine, which then acts as a substrate for the methylenedioxy bridge forming enzyme canadine synthase (CAS).
4. Discussion
The results indicate transcriptional upregulation for certain genes in the phthalideisoquinoline pathway leading to the synthesis of canadine in high canadine accumulating cultivars. The three enzymes in Figure 2a act sequentially and may provide further evidence for sequential enzyme expression [20]. The increased expression of scoulerine 9-O-methyltransferase may prevent scoulerine from accumulating in appreciable amounts compared to B1 cultivar. It is also important to note that not all of the genes encoding enzymes in the pathway exhibited increased expression. Further genome annotation of the P. somniferum genome will allow us to uncover possible alterations in transcriptional regulator expression specific to enzymes involved in canadine biosynthesis. Briefly herein, a novel characterization of the canadine biosynthetic pathway from a transcriptomic perspective in a high-canadine cultivar was reported. The putative findings establish a basis for additional investigations and validation experiments to clarify the transcriptomic regulation underlying the biosynthesis of this important benzylisoquinoline alkaloid.
Acknowledgments
Special thanks to Electronic Laboratory for Science and Analysis (ELSA) High Performance Computing cluster provided by The College of New Jersey for enabling the analysis of this high-throughput data.
References
- Allen R, Millgate A, Chitty J, et al. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nature Biotechnology 22 (2004): 1559-1566.
- Hocking KM, Putumbaka G, Wise ES, et al. Papaverine Prevents Vasospasm by Regulation of Myosin Light Chain Phosphorylation and Actin Polymerization in Human Saphenous Vein. PloS one 11 (2016): e0154460.
- Vree TB, vanDongen RT, Koopman-Kimenai PM. Codeine analgesia is due to codeine-6-glucuronide, not morphine. International Journal of Clinical Practice 54 (2000): 395-398.
- Rida PC, LiVecche D, Ogden A, et al. The Noscapine Chronicle: A Pharmaco-Historic Biography of the Opiate Alkaloid Family and its Clinical Applications. Medicinal research reviews 35 (2015): 1072-1096.
- Lee H, Lee SJ, Bae GU, et al. Canadine from Corydalis turtschaninovii Stimulates Myoblast Differentiation and Protects against Myotube Atrophym. International journal of molecular sciences 18 (2017): 2748.
- Correché ER, Andujar SA, Kurdelas RR, et al. Antioxidant and cytotoxic activities of canadine: biological effects and structural aspects. Bioorg Med Chem 16 (2008): 3641-3651.
- Liu Y, Chen X, Wang J, et al. Transcriptomic analysis reveals flavonoid biosynthesis of Syringa oblata Lindl. in response to different light intensity. BMC Plant Biology 19 (2019): 487.
- Harðardóttir S, Wohlrab S, Hjort DM, et al. Transcriptomic responses to grazing reveal the metabolic pathway leading to the biosynthesis of domoic acid and highlight different defense strategies in diatoms. BMC Molecular Biology 20 (2019): 7.
- Hagel JM, Morris JS, Lee EJ, et al. Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biology 15 (2015): 227.
- Pathak S, Lakhwani D, Gupta P, et al. Comparative transcriptome analysis using high papaverine mutant of Papaver somniferum reveals pathway and uncharacterized steps of papaverine biosynthesis. PloS one 8 (2013): e65622.
- Zhao Y, Zhang X, Li M, et al.Transcriptomic profiles of 33 opium poppy samples in different tissues, growth phases, and cultivars. Scientific Data 6 (2019): 66.
- Trim Galore. GitHub.
- Andrews S. FastQC: a quality control tool for high throughput sequence data (2010).
- Guo L, Winzer T, Yang X, et al. The opium poppy genome and morphinan production. Science 362 (2018): 343-347.
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12 (2015): 357-360.
- Pertea M, Pertea GM, Antonescu MA, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology 33 (2015): 290-295.
- Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15 (2014): 550.
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2013).
- Wickham H. Elegant Graphics for Data Analysis. Springer-Verlag New York (2016).
- Oyarzun D, Ingalls BP, Middleton RH, et al. Sequential activation of metabolic pathways: a dynamic optimization approach. Bulletin of Mathematical Biology 8 (2009): 1851-1872.
