In Silico and In Vitro Study of Rice Bran Peptides for the Treatment of Oxidative Stress Diabetes and Hypertension
Article Information
Md. Ruhul Amin¥1, Md Nijamuddin Mojumder¥2, Md Ratul Rahman2, Nilufa Ferdous3, Md. Omar Faruque1, Muhammad Ali Siddiquee4, Zakir Hossain Howlader2 and Md Alauddin1,5*
1Department of Nutrition and Food Technology, Jashore University of Science and Technology, Jashore-7408,
Jashore, Bangladesh.
2Department of Biochemistry and Molecular Biology, University of Dhaka, Dkaka-1000, Dhaka, Bangladesh.
3Grain Quality and Nutrition Division, Bangladesh Rice Research Institute (BRRI), Gazipur, Bangladesh
4The Aristocrat Agro Ltd. Gulshan-1, Dhaka-1212, Bangladesh.
5Department of Biochemistry and Molecular Biology, Jashore University of Science and Technology, Jashore-7408,
Jashore, Bangladesh.
¥Md. Ruhul Amin and Md Nijamuddin Mojumder both are the first authors of this article.
*Corresponding author: Md. Alauddin, Department of Nutrition and Food Technology Jashore, University of Science and Technology, Jashore-7408, Jashore, Bangladesh. and Department of Biochemistry and Molecular Biology, Jashore University of Science and Technology, Jashore-7408, Jashore, Bangladesh.
Received: 26 March 2024; Accepted: 01 April 2024; Published: 29 April 2024
Citation: Md. Ruhul Amin, Md Nijamuddin Mojumder, Md Ratul Rahman, Nilufa Ferdous, Md. Omar Faruque, Muhammad Ali Siddiquee and Zakir Hossain Howlader and Md Alauddin. In Silico and In Vitro Study of Rice Bran Peptides for the Treatment of Oxidative Stress Diabetes and Hypertension. Cardiology and Cardiovascular Medicine. 8 (2024): 177-194.
View / Download Pdf Share at FacebookAbstract
The study explores the health benefits of rice bran protein hydrolysates and bioactive peptides, focusing on their anti-oxidative, anti-diabetic, and anti-hypertensive properties through in-silico and in-vitro analyses. Rice bran proteins were isolated and in vitro enzymatically digested to assess soluble peptide concentration, degree of hydrolysis (DH), anti-oxidative properties, and inhibitory activity against α-amylase and angiotensin- I-converting enzyme (ACE). This study indicates a higher degree of protein hydrolysis (84.0-99.1%) in various rice bran protein fractions, demonstrating increased hydrolysis with both single and multiple enzyme digestion. The alcalase enzyme was notably efficient for the DH of all protein hydrolysates, and the combination of enzymes (alcalase-trypsin) exhibited the highest DH in the prolamin fraction. Moreover, alcalasetrypsin (4h digested) demonstrated significant inhibitory activity against α-amylase and ACE, respectively. Additionally, in-silico studies were implemented to investigate bioactive peptides binding affinity to the target protein compared to reference drugs. Our study discovered that YY and IP peptides exhibit the highest binding affinity to ACE and α-amylase target proteins, respectively. Moreover, these peptides demonstrated favorable oral bioavailability and non-toxic behavior compared to reference drugs in molecular dynamics (MD) simulations. This encourages the development of nutraceuticals and dietary supplements based on rice bran protein hydrolysates, supported by additional in-vivo research.
Keywords
Rice Bran Proteins; Peptides; In-silico and In-vitro study; Anti-oxidative; Anti-hypertensive; Anti-hyperglycemic
Rice Bran Proteins articles; Peptides articles; In-silico and In-vitro study articles; Anti-oxidative articles; Anti-hypertensive articles; Anti-hyperglycemic articles
Article Details
Abbreviations
DH - degree of hydrolysis
ACE - angiotensin-I-converting enzyme
MD - molecular dynamics
YY - Tyr-Tyr
IP - Ile-Pro
NCDs - Non-communicable diseases
HTN - hypertension
AAI - α-amylase inhibitors
RAS - renin-angiotensin systems
RBP - rice bran protein
RBPHs - bran protein hydrolysates
BSA - bovine serum albumin
DPPH - 2,2-diphenyl-1-picrylhydrazyl
ABTS+ - 2,2´-azino-bis (ethylbenzothiazoline-6-sulfonic acid)
DNS - dinitrosalicylic
FAPGG - N-[3-(2-Furyl) acryloyl]-Phe-Gly-Gly
RMSD - Root Mean Square Deviation
RMSF - Root Mean Square Fluctuation
Rg - Radius of Gyration
SASA - Solvent Accessible Surface Area
SMILES - Simplified Molecular Input Line Entry System
RO5 - Lipinsinks rule of 5
DRB - defatted rice bran
ADMET - Absorption, distribution, metabolism, excretion, and toxicological
Highlights:
- Isolation of rice bran proteins (total protein, albumin, globulin, glutelin and prolamin).
- Rice bran proteins were hydrolyzed with alcalase (4 h) have a greater degree of hydrolysis compared to single and multiple enzyme treatment.
- Enzyme (alcalase and trypsin) treated protein hydrolysates showed prodigious inhibitory activity against the α-amylase and ACE.
- Insilico study showed that YY and IP peptides have the highest binding affinity to the ACE and α-amylase target proteins, respectively.
- Absorption-distribution-metabolism-excretion-toxicological (ADMET) and molecular dynamic (MD) simulation studies of peptides have showed good oral bioavailability and no toxicity behavior compared to reference drugs.
1. Introduction
Non-communicable diseases (NCDs), including diabetes and cardiovascular disease, are caused by various factors such as genetic, environmental, physiological, and behavioral [1]. Approximately 41 million people are estimated to
die from non-communicable diseases, accounting for 74% of all-cause deaths worldwide. Diabetes and hypertension (HTN) have emerged as the most severe and chronic metabolic disorders [1-4]. The primary goal of numerous therapies developed for the prevention and development restriction of diabetes and hypertension is the control of postprandial hyperglycemia and blood pressure. Reducing glucose levels by blocking α-amylase and α-glucosidase activity to prevent carbohydrate cleavage is the one common treatment strategy for managing type 2 diabetes [5-10]. α-amylase and glucosidase inhibitors are substances primarily active in the gastrointestinal system, where they suppress the enzymatic breakdown of complex carbohydrates by α-amylase and α-glucosidase, thereby retarding and reducing the absorption of monosaccharides from the intestines in the context of diabetes treatment. Acarbose has been widely used as an oral α-amylase inhibitors (AAI) for managing hyperglycemia for over 20 years [11,12]. Nevertheless, prolonged acarbose use causes an accumulation of undigested carbohydrates in the colon that triggers gastrointestinal complications like diarrhoea, flatulence, and abdominal distention as a result of chronic inhibition of starch hydrolysis and excessive pancreatic α-amylase inhibition [13,14]. The management of hypertension in humans is governed by the renin-angiotensin systems (RAS), which consist of predominantly three enzymes: renin, angiotensin-I-converting enzyme (ACE), and chymase [15]. ACE plays a pivotal role within the RAS system, contributing to elevated blood pressure by transforming the inactive decapeptide angiotensin-I into the potent vasoconstrictor octapeptide angiotensin-II, while also deactivating the vasodilatory nonapeptide bradykinin [16]. Consequently, inhibiting ACE activity is a promising strategy for managing hypertension [17]. Numerous chemical ACE inhibitors have been synthesized after identifying ACE inhibitors within snake venom [18], including captopril, alacepril, enalapril, and lisinopril [19]. Nevertheless, these synthetic ACE inhibitors are associated with diverse adverse effects, such as skin rashes, coughing, and angioedema [20,21]. Hence, numerous research endeavors have been undertaken to formulate novel and safe ACE inhibitors and α-amylase inhibitors (AAI) inhibitors, derived from natural origins. Until now, a diverse range of protease-digested plant hydrolysates originating from walnuts, beans, hemp seed protein, ocimum tenuiflorum seeds, rice, soybean, and dark tea has been recognized as significant contributors to the suppression of α-amylase and α-glucosidase activity [22-29]. Similarly, over the past few years, a variety of ACE inhibitory peptides have been discovered from a diverse selection of food sources, including casein, corn, gelatin, soybeans, egg white, tuna, and dried bonito[30-36]. Rice, a staple food for over fifty percent of the global populace, is cultivated by over 100 countries, with Asia accounting for 90% of worldwide production. Rice bran is produced as a by-product of rice milling, is estimated that the world's annual production of rice bran amounts to 76 million tons. At present, rice bran is mainly used as animal and poultry feed, with a tiny proportion also being extracted for cooking oil and used as a dietary supplement or component of the microbiological medium. Additionally, rice bran contains 11.0-17.0% protein, 12.0–22.0% fat, 6.0– 14.0% fiber, 8.0-17.0% ash, and 10.0–15.0% moisture, as well as anthocyanins in red and black rice bran[37,38]. However, even though rice bran is widely available, little research has focused on creating products from rice bran protein. As a result, this valuable protein source isn't being used to its full potential. Lately, notable research has focused on exploring the health-promoting bioactive properties of peptides generated through the enzymatic hydrolysis of cereal proteins [39]. The unrefined hydrolysates of cereal proteins and the peptides extracted from them have demonstrated a range of physiological roles, encompassing attributes such as anti-hypertensive, anti-diabetic, anti-oxidant, antimicrobial, and opioid activities[39]. An in vitro study has demonstrated the anti-diabetic properties of rice bran bioactive peptides, including Leu-Pro (LP) and Ile-Pro (IP) [40]. Moreover, another study has indicated that rice bran contains additional bioactive peptides, such as Leu-Arg-Ala (LRA), Tyr-Tyr (YY), and Tyr-Ser-Lys (YSK), which exhibit anti-hypertensive effects [41]. The ability of cereal protein hydrolysates to potentially prevent diseases suggests they could become valuable constituents for the formulation of functional foods and nutraceuticals. Our research focuses on clarifying the inhibition of α-amylase and ACE enzymes using rice bran protein (RBP) hydrolysates. Through in vitro analysis, we assess their effects on single and multiple enzyme digestion, as well as their anti-oxidant properties. Additionally, we identify the most potent anti-diabetic and anti-hypertensive bioactive peptide via in silico study.
2. Materials and Methods
2.1 Samples and reagents
In this study, the rice bran variety BRRI-dhan-74 was collected from the Bangladesh Rice Research Institute, BRRI. Enzymes, DPPH, ABTS, and all other reagents were of analytical grade.
2.2 Sequential extraction of proteins from rice bran
Raw organic rice bran (BRRI-dhan-74) was defatted with n-Hexane (1:6, w/v) by stirring at room temperature for 1.0h, followed by the slurry centrifuged at 1200g for 10 minutes. The precipitate was collected and re-extracted twice. Then the defatted rice bran was air-dried. Both the raw rice bran and defatted rice bran were kept in a plastic zip lock bag at -20 °C before use in further experiments. The sequential extraction of proteins such as albumin, globulin, glutelin, and prolamin was done according to the Osborne method [42]. Nevertheless, the extraction process was adjusted to eliminate potential phenolic impurities found in rice bran, which could disrupt antioxidant and other bioactivity assessments [43]. The first step in sequential extraction was to combine samples of the bran (50 g) with 5 volumes of 70% (v/v) ethanol for 60 minutes. The prolamin fraction was then obtained by centrifuging the mixture at 10,000 g for 15 minutes at 4°C. This process also removed free phenolic compounds from the rice bran in addition to prolamin. The extract's prolamin was purified by precipitating it with double volumes of acetone for 24 hours at -18°C, followed by centrifuging it at 10,000 g for 15 minutes at 4°C and twice washing it with acetone. The phenolic-containing supernatant was thrown away. To isolate the water-soluble albumin portion from the remnants of the initial 70% ethanol extraction-prolamin was extracted using 5 volumes of distilled water for a duration of 1.0 hour. Following this procedure, the remaining residue was subjected to an additional extraction with 5 volumes of 5% (w/v) NaCl for 1.0h and the supernatant was filtered to get the globulin fraction. Finally, the remnant was extracted with 5 volumes of 0.1M NaOH for 1h and the supernatant was filtered to yield the glutelin fraction. Each extraction step was repeated twice to improve the recovery of protein. The protein components were lyophilized and stored at -20°C until use.
2.3 Enzymatic hydrolysis of rice bran protein
The four types of rice bran protein (RBP) fractions were hydrolyzed with two commercial proteases, Alcalase and Trypsin at their respective optimum conditions. Both single-enzyme digestion and multiple-enzyme digestion methods were used.
2.4 Single enzyme digestion
For digestion with alcalase and trypsin separately, albumin (0.7g), globulin (0.5g), glutelin (1.0g), and prolamin (0.1g) were dissolved in 20 mL Tris-HCL buffer separately. Adding 0.1M NaOH or 0.1M HCl maintained the pH at 8.0, and 7.4 for Alcalase and Trypsin, respectively. The ratio of enzymes to substrate (E/S), calculated in terms of protein content, was set at 1:50 (w/w) for both enzymes. After adjusting the pH, the mixture was poured into several Eppendorf tubes and kept in a water bath at 60 °C (Alcalase) and 37 °C (Trypsin), and samples of hydrolysates were taken at 2.0 and 4.0 hours. Subsequently, enzyme inactivation was done by keeping all the tubes at -20 °C for 15 minutes and then centrifuging at 10,000 g for 15 minutes. The supernatant was collected which contained protein hydrolysates and stored at 4 °C for further experiments.
2.5 Multiple enzyme digestion
In the context of multiple enzyme digestion techniques, enzymes can be simultaneously introduced only when they
exhibit their best catalytic performance under comparable pH and temperature conditions. As the optimum pH and temperature for Alcalase and Trypsin catalytic activities are different, consecutive enzyme addition was performed. Firstly, trypsin digestion was performed at optimum conditions (pH 7.4, 37 °C for 4.0h) as discussed above single enzyme digestion. Then, pH was adjusted at 8.0 by adding 0.1M NaOH for alcalase digestion. Subsequently, Alcalase solution was added with the same tubes and digestion was performed at 60 °C for 4.0h. Then, enzyme inactivation was done by keeping all the tubes at -20 °C for 15 minutes. After that, the tubes were centrifuged at 10,000 g for 15 minutes. The supernatant was collected which contained protein hydrolysates and stored at 4 °C for further experiments.
2.6 Determination of soluble peptide in protein hydrolysates
The concentration of soluble peptides in rice bran protein hydrolysates (RBPHs) was estimated by the Biuret method where the bovine serum albumin (BSA) was used as the standard. Protein hydrolysates, obtained as dissolved in sodium phosphate buffer, were used as a sample for the determination of soluble peptide concentration. RBPHs or BSA samples were mixed with biuret reagent thoroughly by vertexing and incubated for 30 minutes at 25 °C. Next, the solutions were transferred to appropriate cuvettes, and the absorbance values were measured at a wavelength of 540 nm, subsequently compared to the reference BSA standard curve.
2.7 Determination of degree of hydrolysis (DH)
The degree of hydrolysis (DH) of rice bran protein following enzymatic digestion was quantified in the sample. This measurement was conducted using the TNBS colorimetric technique, as outlined by Adler-Nissen with minor adjustments [44]. 0.25 mL of hydrolysate sample was mixed with 2 mL of 0.2 M sodium phosphate buffer, pH 8.2, and 2.0 mL of 0.1% (w/v) TNBS solution. The mixture was mixed uniformly and incubated at 50 °C in a water bath for 1.0h in the dark. The reaction was then terminated by the addition of 4 mL of 0.1 M HCl and allowed to cool at room temperature for 30 min. Absorbance was then measured at 340 nm and compared with an L-leucine standard curve (0.0–1.0 mM) to determine the level of unbound amino groups. The overall count of amino groups within the sample (htot) was determined through the complete hydrolysis of the peptide bonds present in the sample (0.25 mL) first with 3 mL of 6 M HCl at 110 °C for 24 h, followed by the same colorimetric procedure described above. The degree of hydrolysis (DH), defined as the percentage of peptide bonds broken (h) relative to the total bonds per unit weight (htot), expressed by the formula,
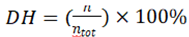
2.8 Determination of antioxidant activities
2.8.1 Determination of DPPH radical scavenging activity
DPPH (2,2-diphenyl-1-picrylhydrazyl) radical scavenging activity indicates the activity of anti-oxidants. The stable DPPH radical-scavenging activity was measured using the modified method described by Chang et al., 2001 [45]. Three mL of 0.1 mM methanol DPPH solutions was added to 1 mL of extract solution at different concentrations and the contents were stirred vigorously for a few seconds. Then the solutions were allowed to stand in a dark place at room temperature for 10 minutes for the reaction to occur. After 10 minutes, absorbance was measured against a blank at 517 nm with a double-beam Thermo Scientific UV-VIS spectrophotometer (Model, Evolution 300). The percentage of DPPH radical-scavenging activity of each sample was calculated as,
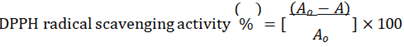
Where Ao = absorbance of the control solution (containing all reagents except plant extracts); A = absorbance of the DPPH solution containing plant extract and ascorbic acid was used as a positive control standard.
2.8.2 Determination of ABTS+ radical scavenging activity
This assay was based on the ability of different substances to scavenge 2,2´-azino-bis (ethylbenzothiazoline-6 sulfonic acid) (ABTS+) radical cation [46]. The radical cation was prepared by mixing 7mM ABTS stock solution with 2.45mM potassium persulfate (1/1, v/v) and leaving the mixture for 4-16 h until the reaction was complete and the absorbance was stable. The ABTS+ solution was diluted with ethanol to an absorbance of 0.700 ± 0.05 at 734 nm for measurements. The photometric assay was conducted on 3.0 mL of ABTS+ solution was added to 1.0 mL of extract solution at different concentrations and the contents were stirred vigorously for a few seconds. Then the solutions were allowed to stand at room temperature for 15 minutes for the reaction to occur. After 15 minutes, absorbance was measured against a blank at 734 nm with a double-beam Thermo Scientific UV-VIS spectrophotometer (Model, Evolution 300). The percentage of ABTS+ radical-scavenging activity of each plant extract was calculated as,
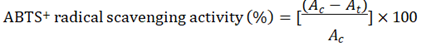
Where Ac = absorbance of the control solution (containing all reagents except plant extracts); At = absorbance of the ABTS solution containing plant extract and ascorbic acid was used as a positive control standard.
2.10 Determination of α-amylase inhibitory activity
The α-Amylase inhibitory capability was assessed using the procedure outlined by Yu and colleagues, with minor adjustments [47].Taking 50μL of α-amylase (50 U/mL) was pre-mixed with 100μL of protein hydrolysates sample and 400μL 20 mM sodium phosphate buffer, pH 6.9. After a 10-minute incubation period, we initiated the reaction by introducing 500 µL of 1% starch solution (in 20 mM sodium phosphate buffer, pH 6.9). The reaction continued for 5.0 min at 25 °C and was halted by the addition of 500μL of dinitrosalicylic (DNS) acid color reagent (12 g of sodium potassium tartrate tetrahydrate in 8.0 mL of 2 M NaOH and 20 mL of 96 mM of 3,5-dinitrosalicylic acid solution) and incubation at 90 °C for 10 min. After the samples had cooled to room temperature, 200µL was removed from each tube and diluted by adding 2 mL of water, and the absorbance was measured at 540 nm. A control with 100% enzyme activity was prepared by replacing the protein hydrolysates with 100μl of the buffer. The pharmacological α-amylase inhibitor acarbose was subjected to the same assay methodology and served as a reference standard. The percentage of α-amylase inhibition was determined using the following formula,
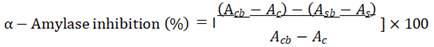
where Acb represents the absorbance of the control blank, Ac stands for the absorbance of the control, Asb corresponds to the absorbance of the sample blank, and As represents the absorbance of the sample, and Acarbose was used as a positive control standard.
2.11 Determination of ACE-inhibitory activity
The ACE-inhibitory activity was assayed according to the published method [48] with slight modification. The 20μL of ACE solution (0.25 U/mL in distilled water) and 60μL of protein hydrolysates were mixed in the Eppendorf tube and incubated at 37°C for 5 min. 300μL of 0.88 mM N-[3-(2-Furyl) acryloyl]-Phe-Gly-Gly (FAPGG) (in 50mM Tris–HCl buffer containing 300mM NaCl, pH 7.5) was added and the decay in absorbance at 340nm due to the degradation of FAPGG by ACE was monitored for 50 min. A control with 100% enzyme activity was prepared by replacing the protein hydrolysates with 60μL of the buffer. The pharmacological ACE inhibitor captopril was assayed the same way and used as a positive control. The ACE-inhibitory effectiveness was determined through the subsequent formula,
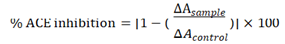
where ΔAsample and ΔAcontrol represent the slopes of the samples and the control, respectively and captopril was used as a positive control standard.
2.12 Molecular docking studies
According to a recent study, rice bran protein (RBP) hydrolysates include both anti-hypertensive peptides like Leu Arg-Ala (LRA), Tyr-Tyr (YY) and Tyr-Ser-Lys (YSK) as well as anti-diabetic peptides like Leu-Pro (LP) and Ile-Pro (IP) [40,41]. These bioactive peptides’ 3D structure construction and energy minimization were done by using ChemOffice 2020. Positive control, Captopril, and Acarbose drug structures were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). The crystal structure of human ACE complexing with Lisinopril (1O86.pdb) and human α-amylase complexing with Acarbose (3BAI.pdb) obtained from the protein data bank (https://www.rcsb.org) was used as the target protein. The energy-minimized peptides (LRA, YY, and YSK) and reference drug (Captopril) were used as ligands against the ACE target protein. Moreover, energy-minimized peptides (LP and IP) and reference drugs (Acarbose) were used as ligands against the α-amylase target protein. The molecular docking experiment was performed by Autodock Vina integrated PyRx software [49,50]. Before docking, water molecules, native ligands, and other hetero atoms were removed and polar hydrogen was added to the target proteins by DS Studio 2021. Subsequently, the receptors (ACE and α-amylase) underwent an energy minimization process using Swiss-Pdb Viewer v4.1. In PyRx software, during ACE docking, the grid box was set with coordinates 40.59, 37.24, and 43.6 (x, y, and z), and the dimensions of the grid box were 62.78, 73, and 67.46 (x, y, and z). For α-amylase docking, the grid box was set with coordinates 8.38, 28.61, and 50.18 (x, y, and z), and the dimensions of the grid box were 58.61, 74.97, and 59.08 (x, y, and z). The molecular docking was evaluated according to the scores and binding-energy value to obtain the best poses for peptides. DS 2.5 software was also used to identify the hydrogen bonds as well as the hydrophobic, hydrophilic, electrostatic, and coordination interactions between residues located at the ACE and renin active sites. The analysis and creation of illustrations for protein-ligand complexes were performed using Discovery Studio Visualizer 4.0. This software was also employed to assess and define the interactions within the docked protein-ligand complex, with a specific focus on identifying the polar and hydrophobic interactions between the ligand and its target.
2.13 Molecular dynamics studies
Molecular dynamics (MD) simulations were conducted using the CHARMM36 force field in GROMACS 2023[51]. After the docking study, the receptor-ligand complex with the best binding energy was used as the initial structure for molecular dynamics simulation. MD simulation was performed on the ACE-YY complex and α-Amylase-IP complex. Similarly, MD of ACE-Captopril complex and α-Amylase-Acarbose complex was also performed as a control. In the MD, each complex was solvated in a TIP3P cubic box water model with a distance of 1.2 nm between the complex and each side of the solvated box, and Na+ and Cl- ions were added to neutralize the total charge of the system. Periodic boundary conditions (PBC) were used. Then the systems were energy-minimized using 5000 steps from the steepest descent algorithm. The subsequent step involved two-staged equilibration where the initial equilibration phase advanced in a consistent manner, maintaining a constant Number of particles, Volume, and Temperature (NVT) ensemble using the Berendsen temperature coupling technique for maintaining the temperature within the 3D box [52]. Next, the second equilibration phase was conducted within an NPT (Number of particles, Pressure, and Temperature) ensemble at 1 atm and 303.15 K, following the guidance of the Parrinello-Rahman barostat [53]. Furthermore, 10 ns of MD production of each complex was performed with a 2 fs timestep. After simulation, the results were analyzed using common GROMACS functions Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Hydrogen bond, Radius of Gyration (Rg), and Solvent Accessible Surface Area (SASA) by using gmx_rmsd, gmx_rmsf, gmx_hbond, gmx_gyrate, and gmx_sasa commands respectively.
2.14 Prediction of physiochemical and pharmacokinetics properties and toxicity analysis
The selected peptides and reference drugs were evaluated for their physicochemical and pharmacokinetic characteristics using SwissADME (http://www.swissadme.ch/)[54]. The transformation of peptide sequences into the Simplified Molecular Input Line Entry System (SMILES) format was executed using ChemDraw; Reference drugs’ canonical SMILES codes were copied from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Subsequently, these SMILES strings were then submitted to SwissADME as input for analysis. The DataWarrior software was used to calculate the drugscore of peptides and standard drugs [55]. Peptides and drugs sdf structure format was obtained from ChemDraw and PubChem, respectively. Then these sdf files were used as input files for DataWarrior software. Lipinsinks rule of 5 (RO5) was followed to carry out drug-likeness prediction of selected peptides and compare them with standard drugs. Bioavailability radar obtained from SwissADME to predict oral bioavailability.
3. Results
3.1 Isolation of rice bran proteins and concentration of soluble peptides in protein hydrolysates
Rice bran proteins (total protein, albumin, globulin, glutelin, and prolamin) were extracted in lyophilized form. The amount of total protein was found 3.096 ± 0.006 g per 100 g defatted rice bran (DRB) whereas the amount of albumin, globulin, glutelin, and prolamin was found 0.410 ± 0.006, 0.239 ± 0.003, 3.736 ± 0.004, and 0.185 ± 0.005 g per 100g (DRB) respectively. Percentage of the protein content of extracted total protein, albumin, globulin, glutelin, and prolamin was found 98.4%, 99.1%, 97.7%, 99.1%, and 84.2% respectively. The soluble protein content of total protein, albumin, globulin, glutelin, and prolamin was found 3.046 ± 0.010, 0.406 ± 0.001, 0.233 ± 0.001, 3.703 ± 0.008 and 0.156 ± 0.002 g per 100 g (DRB) respectively. All data has been presented in Table 1. The extracted proteins were hydrolyzed by applying both single enzyme digestion and multiple (trypsin + alcalase) enzyme digestion methods. The peptides were extracted as a solution of sodium phosphate buffer and the concentration of peptides was measured using the Biuret method. Table 2 shows the concentration (mg/mL) of soluble peptides in different rice bran protein hydrolysates (RBPHs) such as albumin, globulin, glutelin, and prolamin. Among these protein hydrolysates, the concentration (mg/mL) of soluble peptide in albumin hydrolysates decreases more with multiple enzyme digestion (5.648 ± 0.021) than single enzyme (4h of alcalase; 5.925 ± 0.021) digestion compared with undigested protein hydrolysate (6.949 ± 0.021). In our study, we found that peptide concentration decreased in the case of trypsin, alcalase, and (trypsin + alcalase) digestion compared with before digestion, indicating that protein hydrolysis is occurred. In the case of single enzyme digestion, significant differences in the peptide concentrations were observed between 2 h and 4 h of enzymatic digestion.
Table 1: Rice bran protein extraction from defatted rice bran sample. Data is presented as mean value ± standard error of the mean (SEM)
|
Type of Protein |
Protein Yield (g/100g defatted rice bran)a |
Soluble protein content defatted rice bran)b |
(g/100g Percent of Peptides content |
|
Total protein |
3.096 ± 0.006 |
3.046 ± 0.010 |
98.4 |
|
Albumin |
0.410 ± 0.006 |
0.406 ± 0.001 |
99.1 |
|
Globulin |
0.239 ± 0.003 |
0.233 ± 0.001 |
97.7 |
|
Glutelin |
3.736 ± 0.004 |
3.703 ± 0.008 |
99.1 |
|
Prolamin |
0.185 ± 0.005 |
0.156 ± 0.002 |
84.2 |
|
a Calculated as dry weight of extract from 100 g of defatted rice bran. b Calculated as the dry weight of water-soluble rice bran protein in the extract from 100 g of defatted rice bran |
|||
Table 2: Concentration (mg/mL) of soluble peptides in protein hydrolysates
3.2 Degree of hydrolysis (DH) of rice bran protein hydrolysates
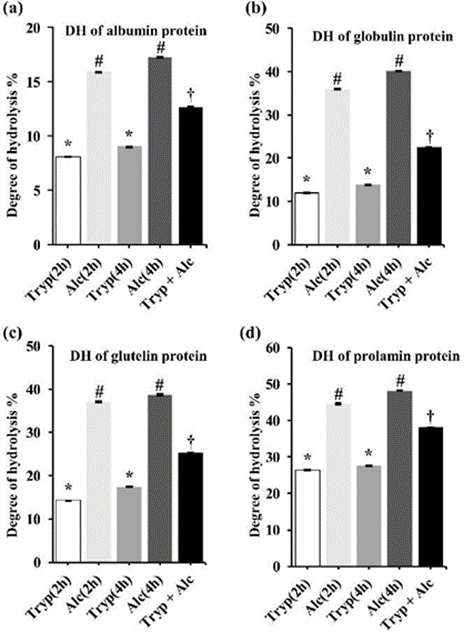
The degree of hydrolysis is the proportion of cleaved peptide bonds in the protein hydrolysates. Significant differences in the degree of hydrolysis were observed between trypsin (2 h) and trypsin (4 h), alcalase (2 h) and alcalase (4 h), trypsin (2 h) and alcalase (2 h), and trypsin (4 h) and alcalase (4 h) of digestion for albumin, globulin, glutelin, and prolamin, adjusted p-value was <0.05. Figure 1 shows that the degree of hydrolysis is greater with single alcalase digestion, and with multiple (trypsin + alcalase) digestion, and lower with trypsin digestion. Overall, the degree of hydrolysis of all the proteins was efficient with alcalase 2h and alcalase 4h digestion. In our study, we found that albumin, globulin, glutelin, and prolamin hydrolyzed with alcalase (4 h) have a greater degree of hydrolysis compared to trypsin (2 h), trypsin (4 h), alcalase (2 h), and (trypsin + alcalase).
Figure 1: The degree of hydrolysis is the proportion of cleaved peptide bonds in the protein hydrolysates. This figure shows a) Albumin; b) Globulin; c) Glutelin and d) Prolamin hydrolysis with trypsin (2h), alcalase (2h), trypsin (4h), alcalase (4h), and (trypsin + alcalase; (2h). Data are presented as mean value ± standard error of the mean (SEM). To analyze the significance among the mean values, one-way ANOVA and Tukey test were performed as a post hoc test. Different symbols (*, #, †) indicates (p <0.05) significant differently
3.3 Antioxidant properties of rice bran protein hydrolysates
DPPH (2,2-diphenyl-1-picrylhydrazyl) and 2,2´-azino-bis (ethylbenzothiazoline-6-sulfonic acid) (ABTS+) radical cation scavenging activity indicates the activity of antioxidants to scavenge free radicals by using DPPH and ABTS+. DPPH and ABTS+ radical serves as free radicals to be reduced by the antioxidant and as the indicator for the reaction. In our study, significant differences in the % of DPPH inhibition were observed between untreated and trypsin (2 h), untreated and alcalase (2 h), untreated and trypsin (4 h), untreated and alcalase (4 h), untreated and (trypsin + alcalase) for albumin, globulin, glutelin, and prolamin. Table 3 states the DPPH and ABTS+ radical scavenging activity of RBP hydrolysates. Our study found that trypsin hydrolyzed (4 h) prolamin has the highest DPPH radical scavenging activity (74.832 ± 0.067%) compared to untreated proteins and other protein hydrolysates. Moreover, we found that glutelin (digested with Trypsin + Alcalase) and prolamin digested with alcalase (4 h) have the highest ABTS+ radical scavenging activity (98.636 ± 0.152%) compared to untreated proteins and other protein hydrolysates.
Table 3: Antioxidant (DPPH and ABTS+ radical scavenging activity), antidiabetic (α-Amylase inhibitory activity), and antihypertensive (ACE inhibitory activity) properties of RBP hydrolysates
|
DPPH radical scavenging activity % |
||||||
|
Protein hydrolysates |
Untreated |
2h of digestion |
4h of digestion |
Tryp + Alc |
||
|
Trypsin |
Alcalase |
Trypsin |
Alcalase |
|||
|
Albumin |
16.913 ± 0.134 |
18.993 ± 0.067 |
11.611 ± 0.201* |
20.537 ± 0.268 |
15.570 ± 0.134 |
14.161 ± 0.201 |
|
Globulin |
7.852 ± 0.201 |
13.154 ± 0.134* |
12.617 ± 0.134* |
14.027 ± 0.201* |
13.423 ± 0.134* |
14.899 ± 0.134* |
|
Glutelin |
23.960 ± 0.201 |
27.114 ± 0.134* |
23.557 ± 0.067 |
34.094 ± 0.268* |
26.711 ± 0.134 |
23.423 ± 0.067 |
|
Prolamin |
71.007 ± 0.134 |
73.826 ± 0.134 |
51.275 ± 0.134* |
74.832 ± 0.067 |
54.497 ± 0.134* |
50.134 ± 0.201* |
|
ABTS+ radical scavenging activity % |
||||||
|
Albumin |
85.152 ± 0.429 |
88.636 ± 0.214 |
88.333 ± 0.214 |
90.455 ± 0.643* |
89.545 ± 0.214 |
90.758 ± 0.214* |
|
Globulin |
82.273 ± 0.455 |
90.758 ± 0.152* |
79.242 ± 0.152 |
92.121 ± 0.303* |
80.758 ± 0.152 |
80.758 ± 0.152 |
|
Glutelin |
89.697 ± 0.303 |
97.273 ± 0.303* |
94.697 ± 0.152 |
98.333 ± 0.152* |
95.606 ± 0.152* |
98.636 ± 0.152* |
|
Prolamin |
94.848 ± 0.303 |
97.121 ± 0.152 |
97.424 ± 0.152 |
98.485 ± 0.303* |
98.636 ± 0.152* |
96.212 ± 0.152 |
|
α-Amylase inhibitory activity (%) |
||||||
|
Protein hydrolysates |
Untreated |
2h of digestion |
4h of digestion |
Tryp + Alc |
||
|
Trypsin |
Alcalase |
Trypsin |
Alcalase |
|||
|
Albumin |
72.480 ± 0.302 |
73.690 ± 0.101 |
79.234 ± 0.202* |
77.218 ± 0.202* |
81.653 ± 0.202* |
76.815 ± 0.202* |
|
Globulin |
71.069 ± 0.302 |
75.000 ± 0.202 |
78.427 ± 0.202* |
76.008 ± 0.202* |
79.637 ± 0.202* |
76.411 ± 0.202* |
|
Glutelin |
74.597 ± 0.202 |
78.125 ± 0.101 |
81.754 ± 0.101* |
81.552 ± 0.302* |
83.165 ± 0.302* |
75.504 ± 0.101 |
|
Prolamin |
59.173 ± 0.504 |
73.387 ± 0.403* |
78.528 ± 0.101* |
75.504 ± 0.302* |
80.847 ± 0.202* |
77.016 ± 0.202* |
|
ACE inhibitory activity (%) |
||||||
|
Albumin |
17.118 ± 0.530 |
29.882 ± 0.471* |
18.018 ± 0.370 |
44.738 ± 0.621* |
23.970 ± 0.441* |
32.104 ± 0.351* |
|
Globulin |
31.962 ± 0.391 |
49.580 ± 0.421* |
34.824 ± 0.471 |
59.265 ± 0.441* |
44.616 ± 0.498* |
61.438 ± 0.326* |
|
Glutelin |
31.838 ± 0.514 |
58.353 ± 0.471* |
37.906 ± 0.329* |
67.934 ± 0.286* |
44.501 ± 0.384* |
52.528 ± 0.414* |
|
Prolamin |
23.059 ± 0.471 |
37.685 ± 0.550* |
14.915 ± 0.210* |
44.721 ± 0.603* |
26.941 ± 0.471 |
32.623 ± 0.271* |
Data are presented as mean value ± standard error of the mean (SEM). To analyze the significance of the mean values, one-way ANOVA and Tukey test were performed as a post hoc test, and *p-value <0.05 is considered significant compared to untreated.
3.4 α – Amylase and ACE inhibitory activity of rice bran protein hydrolysates
The α-Amylase and the ACE (Angiotensin-I-Converting Enzyme) inhibitory activity of rice bran protein hydrolysates indicate anti-diabetic and anti-hypertensive activity, respectively. Table 3 also states that α-Amylase and ACE inhibitory activity of RBP hydrolysates (albumin, globulin, glutelin, and prolamin) digested with trypsin (2 h), trypsin (4 h), alcalase (2 h), alcalase (4 h), and (trypsin + alcalase) as well as untreated conditions. In our study, significant differences in the percent of α-amylase and ACE inhibition were observed between untreated and trypsin (2 h), untreated and alcalase (2 h), untreated and trypsin (4 h), untreated and alcalase (4 h), untreated and (trypsin + alcalase) for albumin, globulin, glutelin, and prolamin. We found that untreated albumin, globulin, glutelin, and prolamin inhibited 72.480 ± 0.302, 71.069 ± 0.302, 74.597 ± 0.202, and 59.173 ± 0.504% α-amylase respectively. Moreover, the untreated albumin, globulin, glutelin, and prolamin inhibited 17.118 ± 0.530, 31.962 ± 0.391, 31.838 ± 0.514, and 23.059 ± 0.471% ACE respectively. In contrast, single enzyme digestion with different times and multiple enzyme digestion of RBPHs showed a higher amount of inhibitory activity than undigested protein hydrolysates. Our study found that glutelin digested with trypsin (4 hr) has significant ACE inhibitory activity (67.934 ± 0.286%) compared to untreated proteins and other protein hydrolysates. Similarly, alcalase hydrolyzed (4 h) glutelin has the highest α- Amylase (83.165 ± 0.302%) inhibitory activity compared to untreated proteins and other protein hydrolysates. From the evidence in Table 3, enzyme-digested protein hydrolysates exert more inhibitory effects on the α-Amylase and ACE than untreated protein hydrolysates.
3.5 Molecular docking results
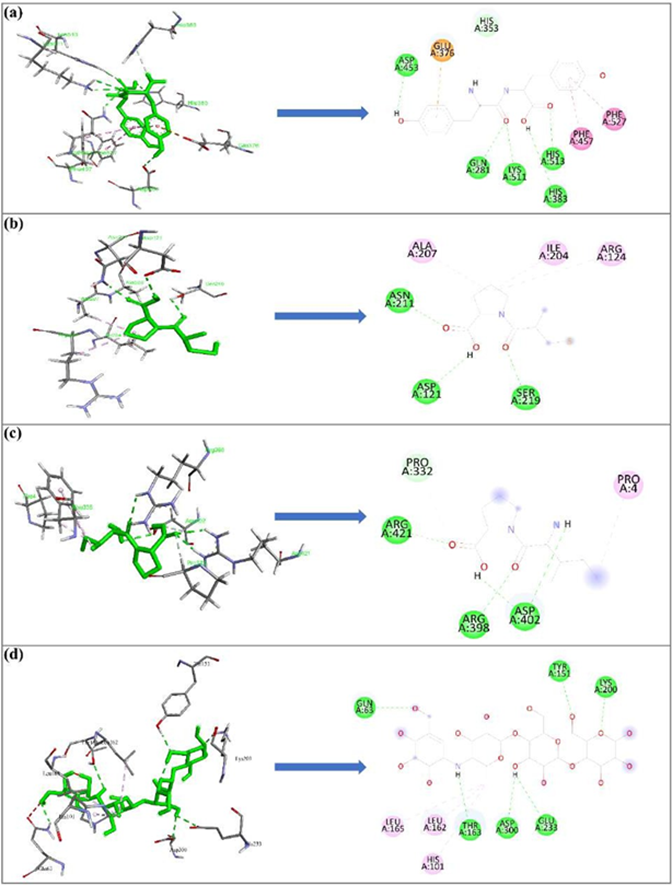
Molecular docking was conducted to predict the potential molecular interactions of the previously identified RBPHs- derived peptides with the binding site of human ACE and α-Amylase enzyme. Table 4 displays the binding affinity of the peptides, and reference drugs with both target proteins of interest. Table 1 also states different types of interactions between ligands and receptors such as hydrogen bonds, hydrophobic bonds, and electrostatic bonds with their corresponding bond length in angstrom units. From Table 4, it can be concluded that the YY peptide among other anti-hypertensive peptides, and the IP peptide among other anti-diabetic peptides have a significantly strong binding affinity with -7.8Kcal/mol and -6.2Kcal/mol, respectively. The 2D and 3D interaction of standard drugs and the best two ligands (YY and IP) with their target receptors are illustrated in Figure 2. Figure 2a shows the ACE – YY complex which is stabilized by a total of 10 bonds including 7 hydrogen bonds (GLN281, LYS511, LYS511, HIS513, HIS383, ASP453, HIS353), 2 hydrophobic bonds (PHE457, PHE527), and single electrostatic bonds (GLU376). On the other hand, Figure 2b shows the ACE – Captopril (standard drug) complex which is stabilized by a total of 6 bonds including 3 hydrogen bonds (ASN211, SER219, ASP121) and 3 hydrophobic bonds (ARG124, ALA207, ILE204). Moreover, Figure 2c shows the α-Amylase – IP complex which is stabilized by a total of 6 bonds including 5 hydrogen bonds (ARG398, ARG421, ASP402, ASP402, PRO332) and a single hydrophobic bond (PRO4). Additionally, Figure 2d shows the α-Amylase – Acarbose (standard drug) complex which is stabilized by a total of 9 bonds including 6 hydrogen bonds (GLN63, TYR151, LYS200, THR163, GLU233, ASP300) and 3 hydrophobic bonds (LEU162, LEU165, HIS101).
Table 4: Docking interactions of rice bran bioactive peptides and reference drugs with target proteins
Figure 2: The 2D and 3D interaction of standard drugs and ligands (YY and IP) is represented as a green bold line with their target receptors obtained by Autodock Vina and DS Studio 2021. Figure 2(a) ACE – YY, 2(b) ACE – Captopril, 2(c) α-Amylase – IP, 2(d) α-Amylase – Acarbose. The green and pink broken lines indicate the hydrogen bond and Pi bond between the target protein residue and ligand, respectively
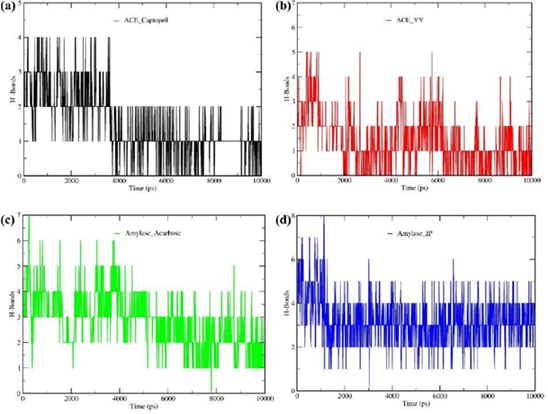
3.6 Molecular dynamics simulation result
A molecular dynamics (MD) simulation study was conducted for 10 ns to confirm the stability of each ligand-receptor complex. The best two ligand-receptor docked complexes (ACE_YY and α-Amylase – IP) among experimental peptides and standard drug complexes were subjected to MD simulation. To evaluate the protein-ligand complex stability, the geometric parameters such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), the radius of gyration (Rg), hydrogen bond, and solvent accessible surface area (SASA) were calculated.
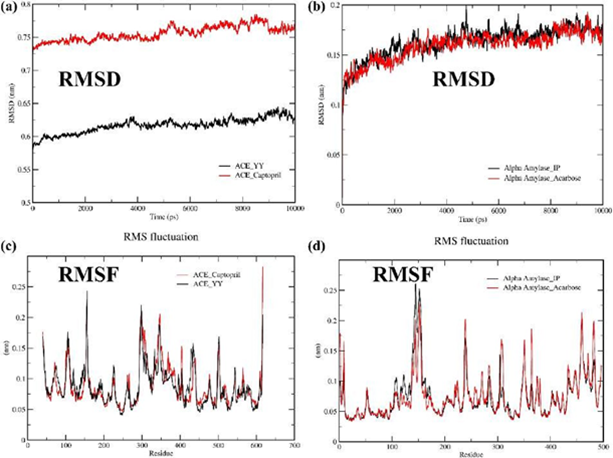
3.7 Root Mean Square Deviation (RMSD)
Root mean square deviation (RMSD) measures the deviation of ligand-protein complex structure from its primary reference geometry. Figure 3a shows the RMSD of the ACE_YY complex and ACE_Captopril complex with respect to their protein backbone. The average RMSD value for the ACE_YY is 0.59 while ACE_Captopril is 0.73. Both complexes' RMSD values gradually increased from their primary structure throughout the simulation period. It indicates that a lower RMSD value of ACE_YY complex is more stable than ACE_Captopril. Additionally, Figure 3b displays the RMSD of the α-Amylase_IP complex and α-Amylase_Acarbose complex with respect to their protein backbone. The mean RMSD values for the α-Amylase_IP and α-Amylase_Acarbose were the same (0.16). It reveals the α-Amylase_IP RMSD value coincides with the α-Amylase_Acarbose RMSD throughout the simulation.
Figure 3: Molecular dynamics (MD) simulation study was conducted for 10 ns to confirm the stability of each ligand-receptor complex. The best two ligand-receptor docked complexes (ACE_YY and α-Amylase_IP) among experimental peptides and standard drug complexes were subjected to MD simulation. Figures 3a and 3b displayed the geometric parameters root mean square deviation (RMSD) of complexes (ACE_YY and α-Amylase_IP) among experimental peptides and standard drugs, Figures 3c and 3d displayed the root mean square fluctuation (RMSF) of complexes (ACE_YY and α-Amylase_IP) among experimental peptides and standard drug
3.8 Root Mean Square Fluctuation (RMSF)
The residual fluctuations in the ligand complexes were observed by Root mean square fluctuation (RMSF). Figure 3c represents the RMSF graph of the ACE_YY and ACE Captopril. In addition, the RMSF graph of the α-Amylase_IP and α-Amylase_Acarbose is illustrated in Figure 3d. From the RMSF graph, it is evident that multiple peaks are shown throughout the entire simulation, and these peaks are a consequence of the fluctuations that occurred during the simulation period. The average RMSF value for the YY complex and Captopril complex was 0.08 while the IP complex and Acarbose complex was 0.07. All the ligands and reference drug docking complexes fluctuated around 0.05 – 0.25 nm in the simulation period.
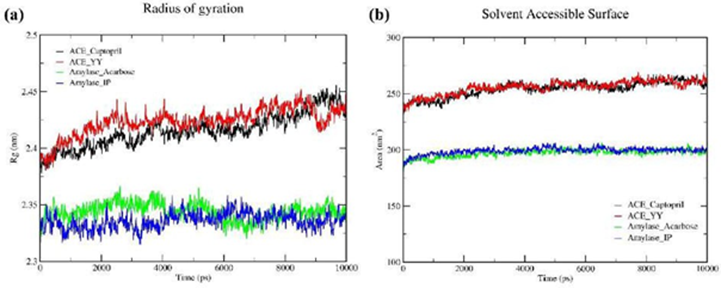
3.9 Radius of gyration (Rg)
Figure 4a displays the radius of gyration (Rg) of all the ligand-receptor complexes and this parameter measures the compactness of protein throughout the simulation. The average Rg for ACE_YY complex and ACE_Captopril complex were 2.42 and 2.41, respectively. Additionally, the mean Rg for α-Amylase_IP complex and α- Amylase_Acarbose complex were 2.33 and 2.34, respectively. From Figure 4a, as we can see, experimental peptides’ impact on protein compactness was the same as standard drugs.
Figure 4: A molecular dynamics (MD) simulation study was conducted for 10 ns to confirm the stability of each ligand-receptor complex. The best two ligand-receptor docked complexes (ACE_YY and α-Amylase_IP) among experimental peptides and standard drug complexes were subjected to MD simulation. To evaluate the protein-ligand complex stability, Figure 4a displays the radius of gyration (Rg) and Figure 4b displays the solvent accessible surface area (SASA) throughout the simulation
3.10 Solvent Accessible Surface Area (SASA)
SASA analysis was conducted to forecast the degree of protein conformation alterations throughout a 10-nanosecond MD trajectory simulation, as illustrated in Figure 4b. This analysis allowed us to assess the influence of water molecules on the protein's structural changes. The SASA analysis states the value of the ACE_YY complex was 255.69 nm2 whereas the ACE_Captopril was 254.30 nm2. The SASA values of the α-Amylase_IP complex and α- Amylase_Acarbose were 199 nm2 and 197 nm2, respectively. From the evidence of Figure 4b, the experimental peptides SASA showed very similar results concerning their standard drugs SASA.
3.11 Hydrogen bond
We calculated the number of hydrogen bonds formed during the complete run of MD simulations for selected complexes, as presented in Figure 5. As shown in Figure 5b, the ACE_YY complex formed 5 hydrogen bonds during 10 ns simulation whereas the standard ACE inhibitor drug complex (ACE_Captopril, Figure 5a) formed a maximum of 4 hydrogen bonds. Additionally, Figure 5d displays the α-Amylase_IP complex formed a maximum of 8 hydrogen bonds throughout the simulation while the α-Amylase_Acarbose complex (Figure 5c) formed a maximum of 7 hydrogen bonds. We can infer from the evidence in Figure 5 that, in the context of hydrogen bonding, our experimental peptides (YY and IP) exhibited behavior with their target proteins that were very comparable to how standard medications react with target proteins.
Figure 5: A molecular dynamics (MD) simulation study was conducted for 10 ns to confirm the stability of each ligand-receptor complex. The best two ligand-receptor docked complexes (ACE_YY and α-Amylase_IP) among experimental peptides and standard drug complexes were subjected to MD simulation. Figure 5a displayed the ACE_Captopril hydrogen bonds during 10 ns simulation, Figure 5b displayed the ACE_YY complex hydrogen bonds during 10 ns simulation; Figure 5c the α-Amylase_Acarbose complex hydrogen bonds throughout the simulation, and Figure 5d displays the α-Amylase_IP complex hydrogen bonds during 10 ns simulation
3.12 Prediction of physiochemical, toxicity risk, and pharmacokinetics properties of rice bran peptides.
SwissADME and DataWarrior are freely available software applications employed in certain in-silico investigations to evaluate the potential toxicity, pharmacokinetics, physicochemical properties, and drug-likeness of bioactive peptides. Table 5 shows that both experimental peptides (YY and IP) were non-mutagenic, non-irritant, non-tumorigenic, and without any adverse effects on reproductive health that are very comparable with reference drugs. Lipinski’s rule of five (RO5) and Veber’s rule were applied to estimate the drug likeliness of both selected peptides. According to Lipinski’s rule, a small drug molecule should maintain 4 criteria to be morally acceptable, including, (i) molecular weight (MW) ≤ 500 (ii) cLogP (partition coefficient between n-octanol and water) ≤ 5, (iii) the number of hydrogen bond donors (HBD) ≤ 5, and (iv) the number of hydrogen bond acceptors (HBA) ≤ 10. Additionally, as reported by the Veber rule, the small molecules should maintain two criteria to be orally bioactive, including, i) rotatable hydrogen bonds (RB) < 10 and topological polar surface area (TPSA) ≤ 140 Å2. Table 5 states that both peptides obey all the RO5 and Veber rules criteria. Moreover, YY and IP peptides drug scores were 0.45 and 0.55, respectively, which was relatable with their standard drugs. Table 5 also shows the pharmacokinetic properties of the selected peptides. Both peptides were labeled as highly gastrointestinal (GI) absorbable and not able to pass Blood Brain Barrier (BBB) which is desirable.
Furthermore, both peptides were predicted not to act as P-Glycoprotein (P-gp) substrates. Consequently, the oral bioavailability of these peptides would remain unaffected by P-glycoprotein, and there would be no potential for these peptides to cause drug-drug interactions. Moreover, both peptides were projected to have no inhibitory effects on any of the five cytochrome P450 (CYP) isozymes, namely CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4, which indicate that there are less drug-drug interactions in the course of metabolism. Moreover, both peptide’s log Kp (skin permeation) values were highly negative which means they are less skin permeation.
Table 5: Physiochemical, Toxicity risk, and Pharmacokinetics properties of selected bioactive peptides and reference drugs
4. Discussion
From the evidence of the result section, Table 1 suggests that obtained protein fraction such as albumin, globulin, glutelin, and prolamin from rice bran by sequential extraction contains soluble peptide. Additionally, in Table 2 throughout the digestion period, soluble peptide concentration decreased in all protein hydrolysates at different extents with different combinations of digestion time and enzyme indicating protein hydrolysis. Furthermore, the degree of hydrolysis of protein is directly associated with anti-oxidant activity. The data in Figure 1 suggests that Alcalase is the most efficient enzyme for hydrolysis of four protein fractions. The combined enzyme (Alcalase + Trypsin) is the second most efficient way for the digestion of all four protein fractions while trypsin is the least efficient for all protein fractions. A previous study also demonstrated that the Alcalase enzyme is more efficient for hydrolyzing wheat gluten than acid proteases such as trypsin and pancreatin[56]. These values of the degree of hydrolysis in this study showed a higher extent compared with results reported in the literature. Zhao et al. (Zhao et al., 2012), for example, reported a DH of 16.9% and 3.0% for rice dreg protein hydrolyzed by Protamax and Flavourzyme, respectively while Hamad [58] reported a DH of 8.8% for rice bran protein hydrolyzed by Flavourzyme. It is essential to highlight that the methodology employed by these authors involved the utilization of crude proteins, subjected to digestion with Flavourzyme and Protamax enzymes during their studies. In contrast, our current investigation employs a more refined approach, involving the digestion of purified protein fractions using single enzymes such as alcalase and trypsin, as well as multiple enzyme treatments.
Table 3 suggests that the anti-oxidant properties of RBPHs increase with the increase of the extent of hydrolysis of protein fraction. It indicates that a more extended digestion period changes the amino acid and peptide concentration in protein hydrolysates. It also reflects the enzyme used in the hydrolysis process and the period of digestion influences the anti-oxidant activity of hydrolysates. The results align with prior research findings that reported the anti-oxidant effectiveness of protein hydrolysates from both walnut and pumpkin meal showed an enhancement as the degree of hydrolysis (DH) increased[59,60]. The anti-oxidant properties of peptides are attributed to their capacity to effectively neutralize free radicals. Peptides function as potent radical inhibitors by providing electrons while maintaining structural stability through the resonance within their molecular configuration[61,62]. Studies showed that peptides with a molecular weight (MW) below 3 kDa exhibited the highest DPPH radical-scavenging activity[63]. Compared with the other hydrolysates, the highest DPPH inhibition performed by prolamin fractions digested with trypsin (4h) could be related to the major presence of peptides below 3kDa. Peptide characterization is conducted through the protein hydrolysis fractionation method, with molecules weighing less than 3 kDa, demonstrating the potential for both anti-oxidant and anti-hypercholesterolemic properties[64]. Based on the data presented in Table 3, it is evident that enzyme-digested protein hydrolysates exhibit a greater degree of inhibition on both α-amylase and ACE compared to untreated protein hydrolysates. The α -amylase is a pivotal enzyme in the breakdown of dietary starch, liberating oligosaccharides that can subsequently be converted into glucose, and rapidly assimilated by the body[65]. Consequently, the inhibition of α-amylase is considered a highly efficacious strategy in the management of diabetes. All of the four protein fractions digested with trypsin (4h) and alcalase (4h) showed significant inhibitory activity, followed by combined enzyme (tryp + alc) treatment. These trends align closely with the degree of protein hydrolysis, indicating that the peptides within the hydrolysates are the primary agents likely responsible for inhibiting α-Amylase activity. There is a scarcity of literature addressing the α-Amylase inhibitory potential of cereal or plant protein hydrolysates. However, a recent investigation has unveiled the α-Amylase inhibitory properties of hydrolysates derived from barley proteins [66]. Additionally, ACE (angiotensin-I-converting enzyme) plays a pivotal role in the Renin-Angiotensin System (RAS), regulating blood pressure by preventing the conversion of angiotensin I to angiotensin II[67]. Consequently, ACE inhibition represents an effective strategy for blood pressure reduction. Among the various rice bran protein fractions, it is noteworthy that trypsin-digested glutelin (for 4 hours) demonstrates a more pronounced inhibitory effect on ACE. This observation suggests a higher concentration of inhibitory bioactive peptides in glutelin hydrolysates compared to both untreated proteins and other protein hydrolysates. It's worth noting that a prior study also documented the ACE- inhibitory potential of tripeptides derived from rice protein hydrolysates [68]. The current in silico study was performed to elucidate the therapeutic potential of bioactive anti-hypertensive and anti-diabetic peptides derived from rice bran hydrolysates (RBP) protein which is previously reported. In association with in-vitro enzyme inhibition activity, it is useful to carry out in-silico molecular docking studies to predict the orientation and binding affinity of the bioactive peptides at the receptor’s active site. We used AutoDock Vina for the current analysis, which applies an automated protocol for predicting receptor-ligand binding and has thereby been widely applied in computer-aided drug design (CADD). Binding affinity is crucial to assay the docking interaction between ligand and target receptor[69]. The best-docked complex was considered based on the binding affinity of ligands to the receptor. The higher negative docking score indicates better binding. From the given result, anti-hypertensive peptides YY, YSK, LRA, and Captopril (anti-hypertensive drug) have binding affinity to target protein (ACE) -7.8 Kcal/mol, -7 Kcal/mol, -7.2 Kcal/mol and -5.9 Kcal/mol, respectively. On the other hand, anti-diabetic peptides IP, LP, and Acarbose (anti-diabetic drug) have binding affinity to target protein (α-Amylase) -6.2 Kcal/mol, -5.9 Kcal/mol, and - 7.7 Kcal/mol, respectively. Due to more negative binding scores, the YY and IP peptide has more potential to inhibit ACE and α-Amylase enzymes, respectively, compared with other peptides. Hydrogen bond interactions and bond length were demonstrated to play a crucial role in stabilizing the docked ligand complexes[70]. Pina and Roque have reported that ACE contained three active pockets, S1, S2, and S1’. S1 included Ala354, Glu384 and Tyr523 residues, S2 included Gln281, His353, Lys511, His513 and Tyr520 residues, while S1’ contained Glu162 residue[71]. Furthermore, previous studies stated that the ACE-inhibitory drugs Lisinopril, and Enalapril interact with ACE amino acid residues Gln281, His353, Glu384, Lys511, His 513, and Tyr520 [72]. Our experimental peptide YY also binds with the active site residues of the ACE such as Gln281, Lys511, His513, His383, Asp453, and His353. The distance of hydrogen bond interactions between the RBP-derived peptides and both ACE α-Amylase amino acid residues typically were short (< 3.0Å; Table 3) indicating that the peptides’ binding affinity to ACE and α-Amylase was strong. The docking results herein corroborate these assumptions and suggest that most probably these are potent ACE inhibitors that will contribute to the ACE inhibitory and antihypertensive activity in vivo. Further work will be needed to confirm ACE inhibition and activity in vivo using pure synthesized peptides. In association with the molecular docking study, performing molecular dynamics (MD) simulation is crucial to view the insight into the best-docked complex’s structural stability. We used GROMACS in this present study which applies CHARMM36 force fields to the simulation under specified conditions. We analyzed different structural properties of the protein-ligand complexes including RMSD, RMSF, Rg, SASA, and Hydrogen bonds. In the context of these parameters, both experimental peptides YY and IP showed very similar behavior to their target proteins as control drugs (Figure 2-5). Though the IP peptide has a slightly lower binding affinity than the control drug acarbose, molecular dynamics studies reveal that the IP peptide effect on α-Amylase coincides with the standard drug acarbose.
Using computational methods to test the potential drug metabolites helps reduce the number of experimental studies and improve the success rate in pharmacokinetics studies. The interaction of inhibitors with a target receptor cannot guarantee the suitability of bioactive compounds as drugs for the target pathology. Therefore, ADMET screening of compounds is critical in drug discovery. Unfavorable characteristics of ADMET in the biological system are the main reasons for the failure of drug molecules during clinical experiments. Absorption, distribution, metabolism, excretion, and toxicological (ADMET) studies were also conducted by using DataWarrior v5.5.0 and SwissPDB to validate this work for therapeutic agent discovery. From the evidence in Table 5, both bioactive peptides will be safe for consumption which is predicted. To be a compound orally bioactive, it should follow Lipinski’s rule of five (RO5) and Veber’s rules of two, any compound violating more than one of these rules could have problems related to its bioavailability[73,74]. Table 5 depicts that all the experimental peptides can easily pass through the cell membrane since their molecular weight is less than 500g/mol. If the size increases, it will create barriers such as the prevention of passive diffusion through the tight aliphatic side chains of the bilayer membrane.
According to the Lipinski rule, cLogP should be less than or equal to 5, Both peptides have negative cLogP (affinity for lipids), so they have good permeability through a biological membrane. A drug molecule is expected to be in an aqueous solubility, range of −1 to −5, and the cLogS values of YY peptide and standard drug (Captopril) fall within the range, indicating that the compounds have good absorption and distribution potential IP peptide with cLogS value of 0.2 which is slight deviate but not far from the standard value. Both peptides fit with Lipinski’s rule of five and Veber’s rule of two.
The overall theoretical ADMET properties justify that YY and IP peptides showed potential lead-like features and can be used for further assessment for hypertension and diabetes treatment, respectively.
5. Conclusion
This study comprehensively analyzed the rice bran proteins and their hydrolysates to evaluate the inhibitory activity of α-Amylase and ACE as well as anti-oxidant properties through an in vitro approach. Among all protein fractions, glutelin digested with trypsin (4h) and alcalase (4h) mostly inhibit ACE and α-Amylase, respectively. Furthermore, in silico molecular docking predicted that bioactive peptides of rice bran protein hydrolysate have a significant role in the inhibition of ACE and α-amylase enzyme. Additionally, ADMET and MD simulation studies revealed that rice bran peptides are safe, do not have any toxicity, and behave like control drugs. These findings demonstrate that rice bran protein hydrolysate has a significant medicinal or pharmaceutical potential that could be utilized in food, food products, and dietary or nutraceutical supplements to combat diabetes and hypertension with further proven in vivo data.
Author Contribution
Md. Ruhul Amin, Md Nijamuddin Mojumder, and Md Ratul Rahman played pivotal roles in the conception, design, analysis, and interpretation of data, as well as in the initial drafting of the manuscript. Nilufa Ferdous and Zakir Hossain Howlader provided crucial analytical support throughout the study. Md. Omar Faruque and Muhammad Ali Siddiquee reviewed and edited the manuscript. Md Alauddin's supervision played a crucial role in guiding the entire research project.
Conflict of Interest
All other authors declare no conflict of interest.
Declarations
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Ethical approval is not applicable. This study received funding from the Bangladesh Bureau of Educational Information and Statistics (BANBEIS), Ministry of Education, Project ID-LS2019858. All data were generated by our laboratory. Any data set can be accessed on request by our laboratory.
References
- Riaz BK, Islam MZ, Islam ANMS, Zaman MM, Hossain MA, et al. Risk factors for non-communicable diseases in Bangladesh: findings of the population-based cross-sectional national survey 2018. BMJ Open 10 (2020): e041334.
- World Health Organization. Noncommunicable diseases (2023).
- Chan JCN, Lim LL, Wareham NJ, Shaw JE, Orchard TJ, et al. The Lancet Commission on diabetes: using data to transform diabetes care and patient lives. The Lancet 296 (2020): 2019-2082.
- Kearney PM, Whelton MB, Reynolds K, Muntner P, Whelton PK, et al. Global burden of hypertension: analysis of worldwide data Lancet 365 (2005): 217-223.
- Dowarah J, Singh VP. Anti-diabetic drugs recent approaches and advancements. Bioorg Med Chem 28 (2020).
- Brandt SJ, Götz A, Tschöp MH, Müller TD. Gut hormone polyagonists for the treatment of type 2 diabetes. Peptides 100 (2018): 190-201.
- Yan J, Zhao R, Yang R, Zhao W. Bioactive peptides with antidiabetic properties: a review. Int J Food Sci Technol (2019).
- Cheung BMY, Li C. Diabetes and Hypertension: Is There a Common Metabolic Pathway?. Curr Atheroscler Rep 14 (2012): 160-166.
- Wang J, Wu T, Fang L, Liu C, Liu X, et al. Anti-diabetic effect by walnut (Juglans mandshurica Maxim.)-derived peptide LPLLR through inhibiting α-glucosidase and α-amylase, and alleviating insulin resistance of hepatic HepG2 cells. J Funct Foods 69 (2020).
- Zhang Y, Wang N, Wang W, Wang J, Zhu Z, et al. Molecular mechanisms of novel peptides from silkworm pupae that inhibit α-glucosidase. Peptides 76 (2016): 45-50.
- Weng J, Soegondo S, Schnell O, Sheu WHH, Grzeszczak W, et al. Efficacy of acarbose in different geographical regions of the world: analysis of a real-life database. Diabetes Metab Res Rev (2015)
- Balfour JA, McTavish D, Lebovitz H, Lefebvre P. Nutrition and Disease Prevention. Drugs (1993)
- Kim G-N, Kwon Y-I, Jang H-D. Mulberry leaf extract reduces postprandial hyperglycemia with few side effects by inhibiting α-glucosidase in normal rats. J Med Food 14 (2011): 712-717.
- Lee KH, Ha KS, Jo SH, Lee CM, Kim YC, et al. Effect of Long-Term Dietary Arginyl-Fructose (AF) on Hyperglycemia and HbA1c in Diabetic db/db Mice. Int J Mol Sci 15 (2014): 8352-8359.
- Crowley SD, Coffman TM. Recent advances involving the renin–angiotensin system. Exp Cell Res 318 (2012): 1049-1056.
- Peach MJ. Renin-angiotensin system: biochemistry and mechanisms of action. Physiol Rev 57 (1977).
- Du L, Fang M, Wu H, Xie J, Wu Y, et al. A novel angiotensin I-converting enzyme inhibitory peptide from Phascolosoma esculenta water-soluble protein hydrolysate. J Funct Foods 5 (2013): 475-483.
- Ferreira SH, Bartelt DC, Greene LJ. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry 13 (1970): 2583-2593.
- Vercruysse L, Van Camp J, Smagghe G. ACE Inhibitory Peptides Derived from Enzymatic Hydrolysates of Animal Muscle Protein: A Review. J Agric Food Chem 53 (2005): 8106-8115.
- Antonios TFT, MacGregor GA. Angiotensin converting enzyme inhibitors in hypertension: potential problems. J Hypertens 13 (1995): 11-16.
- Vermeirssen V, Van Camp J, Verstraete W. Optimisation and validation of an angiotensin-converting enzyme inhibition assay for the screening of bioactive peptides. J Biochem Biophys Methods 51 (2002): 71-82.
- Wang J, Wu T, Fang L, Liu C, Liu X, et al. Anti-diabetic effect by walnut (Juglans mandshurica Maxim.)-derived peptide LPLLR through inhibiting α-glucosidase and α-amylase, and alleviating insulin resistance of hepatic HepG2 cells. J Funct Foods 69 (2020).
- Zhao B, Su K, Mao X, Zhang X. Separation and identification of enzyme inhibition peptides from dark tea protein. Bioorg Chem 99 (2020).
- Valencia-MejíaE, Batista KA, Fernández JJA, Fernandes KF. Antihyperglycemic and hypoglycemic activity of naturally occurring peptides and protein hydrolysates from easy-to-cook and hard-to-cook beans (Phaseolus vulgaris L.). Food Res Int 121 (2019): 238-246.
- Karthiraj T, Harish Babu B, Senthil Kumar R. Task-specific deep eutectic solvent based extraction coupled cascade chromatography quantification of α-glucosidase inhibitory peptide from Ocimum tenuriflorum seeds. Microchemical J 157 (2020).
- González-Montoya M, Hernández-Ledesma B, Mora-Escobedo R, Martínez-Villaluenga C. Bioactive Peptides from Germinated Soybean with Anti-Diabetic Potential by Inhibition of Dipeptidyl Peptidase-IV, α-Amylase, and α-Glucosidase Enzyme. Int J Mol Sci 19 (2018).
- Uraipong C, Zhao J. In vitro digestion of rice bran proteins produces peptides with potent inhibitory effects on α-glucosidase and angiotensin I converting enzyme. J Sci Food Agric (2018).
- Ren Y, Liang K, Jin Y, Zhang M, Chen Y, et al. Identification and characterization of two novel α-glucosidase inhibitory oligopeptides from hemp (Cannabis sativa L.) seed protein. J Funct Foods 26 (2016): 439-450.
- Wang R, Zhao H, Pan X, Orfila C, Lu W, et al. Preparation of bioactive peptides with antidiabetic, antihypertensive, and antioxidant activities and identification of α-glucosidase inhibitory peptides from soy protein. Food Sci Nutr (2019).
- Oshima G, Shimabukuro H, Nagasawa K. Peptide inhibitors of angiotensin I-converting enzyme in digests of gelatin by bacterial collagenase. Biochim Biophys Acta 566 (1979): 128-137.
- Maruyamaand S, Suzuki H. A Peptide Inhibitor of Angiotensin I Converting Enzyme in the Tryptic Hydrolysate of Casein. Agri and Biol Chem 6 (1982): 1393-1394.
- Kohama Y, Matsumoto S, Oka h, Teramoto T, Okabe M, et al. Isolation of angiotensin-converting enzyme inhibitor from tuna muscle. Biochem Biophys Res Commun 155 (1988): 332-337.
- Miyoshi S, Ishikawa H, Kaneko T, Fukui F, Tanaka H, et al. Structures and Activity of Angiotensin-converting Enzyme Inhibitors in an α-Zein Hydrolysate. Agric Biol Chem 32 (1991): 1313-1318.
- Kuba M, Tana C, Tawata S, Yasuda M. Production of angiotensin I-converting enzyme inhibitory peptides from soybean protein with Monascus purpureus acid proteinase. Process Biochemistry 40 (2005): 2191-2196.
- Mine Y, Kovacs-Nolan J. New insights in biologically active proteins and peptides derived from hen egg. Worlds Poult Sci J 2 (2006): 87-96.
- Yokoyama K, Chiba H, Yoshikawa M. Peptide Inhibitors for Angiotensin I-Converting Enzyme from Thermolysin Digest of Dried Bonitot. Biosci Biotechnol Biochem 56 (1992): 1541-1545.
- Chen XQ, Nagao N, Itani T, Irifune K. Anti-oxidative analysis, and identification and quantification of anthocyanin pigments in different coloured rice. Food Chem 135 (2012): 2783-2788.
- Pengkumsri N, Chaiyasut C, Sivamaruthi BS, Saenjum C, Sirilun S, et al. The influence of extraction methods on composition and antioxidant properties of rice bran oil. Food Sci Technol 35 (2015).
- Gong X, An Q, Le L, Geng F, Jiang L, et al. Prospects of cereal protein-derived bioactive peptides: Sources, bioactivities diversity, and production. Crit Rev Food Sci Nutr 62 (2022): 2855-2871.
- Liu YQ, Strappe P, Shang WT, Zhou ZK. Functional peptides derived from rice bran proteins. Crit Rev Food Sci Nutr 10 (2019): 349-356.
- Shobako N, Ohinata K. Anti-Hypertensive Effects of Peptides Derived from Rice Bran Protein. Nutrients 12 (2020): 3060.
- Agboola S, Ng D, Mills D. Characterisation and functional properties of Australian rice protein isolates. J Cereal Sci 41 (2005): 283-290.
- Uraipong C, Zhao J. Rice bran protein hydrolysates exhibit strong in vitro α-amylase, β-glucosidase and ACE-inhibition activities. J Sci Food Agric 96 (2016): 1101-1110.
- Adler-Nissen J. Enzymic Hydrolysis of Food Proteins (1986).
- Chang ST, Wu JH, Wang SY, Kang PL, Yang NS, et al. Antioxidant activity of extracts from Acacia confusa bark and heartwood. J Agric Food Chem 49 (2001): 3420-3424.
- Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med 26 (1999): 1231-1237.
- Yu Z, Yin Y, Zhao W, Liu J. Anti-diabetic activity peptides from albumin against α-glucosidase and α-amylase. Chen, Food Chem 135 (2012): 2078-2085.
- Adjonu R, Doran G, Torley P, Agboola S. Screening of whey protein isolate hydrolysates for their dual functionality: Influence of heat pre-treatment and enzyme specificity. Food Chem 136 (2013): 1435-1443.
- Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J Chem Inf Model 61 (2021): 3891-3898.
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263 (2015): 243–250.
- Lemkul J. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package. Living J Comput Mol Sci 1 (2019).
- Golo VL, Shaitan KV. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. Biofizika (2002)
- Tuble SC, Anwar J, Gale JD. An approach to developing a force field for molecular simulation of martensitic phase transitions between phases with subtle differences in energy and structure. J Am Chem Soc 126 (2004): 396-405.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7 (2017).
- Sander T, Freyss J, Von Korff M, Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55 (2015): 460-473.
- Kong X, Zhou H, Qian H. Enzymatic hydrolysis of wheat gluten by proteases and properties of the resulting hydrolysates. Food Chem 102 (2007): 759-763.
- Zhao Q, Xiong H, Selomulya C, Chen XD, Zhong H, et al. Zhou. Enzymatic hydrolysis of rice dreg protein: effects of enzyme type on the functional properties and antioxidant activities of recovered proteins. Food Chem 134 (2012): 1360-1367.
- Hamada JS. Characterization and Functional Properties of Rice Bran Proteins Modified by Commercial Exoproteases and Endoproteases. J Food Sci 28 (2000).
- Chen N, Yang H, Sun Y, Niu J, Liu S. Purification and identification of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Peptides 38 (2012): 344-349.
- Venuste M, Zhang X, Shoemaker CF, Karangwa E, Abbas S, et al. Influence of enzymatic hydrolysis and enzyme type on the nutritional and antioxidant properties of pumpkin meal hydrolysates. Food Funct 5 (2013).
- Manzoor M, Singh J, Gani A. Exploration of bioactive peptides from various origin as promising nutraceutical treasures: In vitro, in silico and in vivo studies. Food Chem 373 (2022).
- López-garcía G, Dublan-garcía O, Arizmendi-cotero D, Oliván LMG. Antioxidant and Antimicrobial Peptides Derived from Food Proteins. Molecules 27 (2022): 1343.
- Bougatef A, Hajji M, Balti R, Lassoued I, Triki-Ellouz Y, et al. Antioxidant and free radical-scavenging activities of smooth hound (Mustelus mustelus) muscle protein hydrolysates obtained by gastrointestinal proteases. Food Chem 114 (2009): 1198-1205.
- Mahdi C, Untari H, Padaga MC. IOP Conference Series. Mater Sci Eng (2018).
- Gropper SS, Smith JL, Carr TP. Advanced Nutrition and Human Metabolism (2017).
- Connolly A, Piggott CO, FitzGerald RJ. In vitro α-glucosidase, angiotensin converting enzyme and dipeptidyl peptidase-IV inhibitory properties of brewers' spent grain protein hydrolysates. Food Research International 56 (2014): 100-107.
- Erdcs EG. Conversion of angiotensin I to angiotensin II. Am J Med 60 (1976): 749-759.
- Chen J, Liu S, Ye R, Cai G, Ji B, et al. Angiotensin-I converting enzyme (ACE) inhibitory tripeptides from rice protein hydrolysate: Purification and characterization. J Funct Foods 4 (2013): 1684-1692.
- Gu Y, Zhang X, Xu A, Chen W, Liu K, et al. Protein-ligand binding affinity prediction with edge awareness and supervised attention. IScience 26 (2023).
- Varma AK, Patil R, Das S, Stanley A, Yadav L. Sudhakar. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One 5 (2010): e12029.
- Pina AS, Roque ACA. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J Mole Recog 22 (2009): 162-168.