Identification of Hub Genes with Prognostic Values in Lung Cancer by Bioinformatics Analysis
Article Information
Meng Wang1, Tian Xie2, Jiubo Fan1*
1Department of Clinical Laboratory, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, 441021, P.R. China
2Department of Immunology, School of Basic Medical Sciences, Wuhan University, Wuhan, Hubei 430071, P.R. China
*Corresponding Author: Dr. Jiubo Fan, Department of Clinical Laboratory, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, 441021, P.R. China
Received: 17 July 2019, Accepted: 14 August 2019, Published: 28 August 2019
Citation: Meng Wang, Tian Xie, Jiubo Fan. Identification of Hub Genes with Prognostic Values in Lung Cancer by Bioinformatics Analysis. Journal of Cancer Science and Clinical Therapeutics 3 (2019): 131-136.
View / Download Pdf Share at FacebookAbstract
Lung cancer is the main cause of mortalities among all types of cancer. Lots of efforts have been made to elucidate the pathogenesis of lung cancer, but the molecular mechanisms are still not well understood. To identify the candidate genes in the carcinogenesis and progression of lung cancer, we acquired three datasets (GSE19188, GSE33532 and GSE30219) from Gene Expression Omnibus (GEO) database to identify differentially expressed genes (DEGs), then function enrichment analyses were performed. The protein-protein interaction network (PPI) was constructed and the module analysis was performed using STRING and Cytoscape. A total of 185 DEGs was identified, consisting of 32 upregulated genes and 153 down regulated genes. The enriched functions and pathways of the DEGs include angiogenesis, cell adhesion, G2/M transition of mitotic cell cycle, mitotic nuclear division, ECM-receptor interaction, PPAR signaling pathway, etc. we selected ten hub genes (UBE2C, RRM2, KIF11, CDKN3, KIAA0101, PRC1, ASPM, HMMR, TOP2A, and BIRC5) and further analyzed their biological functions. It was found that these genes play an active role in positive regulation of exit from mitosis, mitotic nuclear division, spindle organization, and cell division. Furthermore, Survival analysis showed that these 10 hub genes positively correlated with survival time of lung cancer patients. In conclusion, the discovery of the functions of DEGs and Hub genes can help us to understand the mechanism of the occurrence and development of lung cancer at the molecular level, and provide candidate targets for the diagnosis and treatment of lung cancer in the future.
Keywords
Lung cancer; Bioinformatics analysis; Differentially expressed genes; Hub genes; Prognosis
Lung cancer articles, Bioinformatics analysis articles, Differentially expressed genes articles, Hub genes articles, Prognosis articles
Lung cancer articles Lung cancer Research articles Lung cancer review articles Lung cancer PubMed articles Lung cancer PubMed Central articles Lung cancer 2023 articles Lung cancer 2024 articles Lung cancer Scopus articles Lung cancer impact factor journals Lung cancer Scopus journals Lung cancer PubMed journals Lung cancer medical journals Lung cancer free journals Lung cancer best journals Lung cancer top journals Lung cancer free medical journals Lung cancer famous journals Lung cancer Google Scholar indexed journals Bioinformatics analysis articles Bioinformatics analysis Research articles Bioinformatics analysis review articles Bioinformatics analysis PubMed articles Bioinformatics analysis PubMed Central articles Bioinformatics analysis 2023 articles Bioinformatics analysis 2024 articles Bioinformatics analysis Scopus articles Bioinformatics analysis impact factor journals Bioinformatics analysis Scopus journals Bioinformatics analysis PubMed journals Bioinformatics analysis medical journals Bioinformatics analysis free journals Bioinformatics analysis best journals Bioinformatics analysis top journals Bioinformatics analysis free medical journals Bioinformatics analysis famous journals Bioinformatics analysis Google Scholar indexed journals Differentially expressed genes articles Differentially expressed genes Research articles Differentially expressed genes review articles Differentially expressed genes PubMed articles Differentially expressed genes PubMed Central articles Differentially expressed genes 2023 articles Differentially expressed genes 2024 articles Differentially expressed genes Scopus articles Differentially expressed genes impact factor journals Differentially expressed genes Scopus journals Differentially expressed genes PubMed journals Differentially expressed genes medical journals Differentially expressed genes free journals Differentially expressed genes best journals Differentially expressed genes top journals Differentially expressed genes free medical journals Differentially expressed genes famous journals Differentially expressed genes Google Scholar indexed journals Hub genes articles Hub genes Research articles Hub genes review articles Hub genes PubMed articles Hub genes PubMed Central articles Hub genes 2023 articles Hub genes 2024 articles Hub genes Scopus articles Hub genes impact factor journals Hub genes Scopus journals Hub genes PubMed journals Hub genes medical journals Hub genes free journals Hub genes best journals Hub genes top journals Hub genes free medical journals Hub genes famous journals Hub genes Google Scholar indexed journals Prognosis articles Prognosis Research articles Prognosis review articles Prognosis PubMed articles Prognosis PubMed Central articles Prognosis 2023 articles Prognosis 2024 articles Prognosis Scopus articles Prognosis impact factor journals Prognosis Scopus journals Prognosis PubMed journals Prognosis medical journals Prognosis free journals Prognosis best journals Prognosis top journals Prognosis free medical journals Prognosis famous journals Prognosis Google Scholar indexed journals cancer mortalities articles cancer mortalities Research articles cancer mortalities review articles cancer mortalities PubMed articles cancer mortalities PubMed Central articles cancer mortalities 2023 articles cancer mortalities 2024 articles cancer mortalities Scopus articles cancer mortalities impact factor journals cancer mortalities Scopus journals cancer mortalities PubMed journals cancer mortalities medical journals cancer mortalities free journals cancer mortalities best journals cancer mortalities top journals cancer mortalities free medical journals cancer mortalities famous journals cancer mortalities Google Scholar indexed journals chemotherapy articles chemotherapy Research articles chemotherapy review articles chemotherapy PubMed articles chemotherapy PubMed Central articles chemotherapy 2023 articles chemotherapy 2024 articles chemotherapy Scopus articles chemotherapy impact factor journals chemotherapy Scopus journals chemotherapy PubMed journals chemotherapy medical journals chemotherapy free journals chemotherapy best journals chemotherapy top journals chemotherapy free medical journals chemotherapy famous journals chemotherapy Google Scholar indexed journals radiotherapy articles radiotherapy Research articles radiotherapy review articles radiotherapy PubMed articles radiotherapy PubMed Central articles radiotherapy 2023 articles radiotherapy 2024 articles radiotherapy Scopus articles radiotherapy impact factor journals radiotherapy Scopus journals radiotherapy PubMed journals radiotherapy medical journals radiotherapy free journals radiotherapy best journals radiotherapy top journals radiotherapy free medical journals radiotherapy famous journals radiotherapy Google Scholar indexed journals targeted therapy articles targeted therapy Research articles targeted therapy review articles targeted therapy PubMed articles targeted therapy PubMed Central articles targeted therapy 2023 articles targeted therapy 2024 articles targeted therapy Scopus articles targeted therapy impact factor journals targeted therapy Scopus journals targeted therapy PubMed journals targeted therapy medical journals targeted therapy free journals targeted therapy best journals targeted therapy top journals targeted therapy free medical journals targeted therapy famous journals targeted therapy Google Scholar indexed journals multimodal therapy articles multimodal therapy Research articles multimodal therapy review articles multimodal therapy PubMed articles multimodal therapy PubMed Central articles multimodal therapy 2023 articles multimodal therapy 2024 articles multimodal therapy Scopus articles multimodal therapy impact factor journals multimodal therapy Scopus journals multimodal therapy PubMed journals multimodal therapy medical journals multimodal therapy free journals multimodal therapy best journals multimodal therapy top journals multimodal therapy free medical journals multimodal therapy famous journals multimodal therapy Google Scholar indexed journals
Article Details
1. Introduction
Lung cancer is the leading cause of cancer?associated mortality [1]. It was estimated that 142,670 mortalities due to lung cancer occurred in the United States in 2019, which accounted for nearly a quarter of all cancer mortalities [2]. Despite recent improvements in multimodal therapy including surgery, chemotherapy, radiotherapy, and targeted therapy, its overall 5-year survival rate remains below 20% [3-4]. Therefore, there is an urgent need for the new therapeutic and diagnostic approaches, We usually rely on some bioinformatics tools when doing biomedical research. The most frequently used bioinformatics tools are microarray technology and bioinformatics analysis software. These bioinformatics tools are of great significance for us to study the relationship between genes and diseases. The information it provides enables us to have a comprehensive understanding of the functions of genes and their effects on diseases. As this research on lung cancer is, we used these tools to help us understand the differentially expressed genes (DEGs) related to the progression of lung cancer as well as their functions and pathways [5-7]. However, there are also some drawbacks, that is, when we conduct independent microarray analysis, there will be false positive rate. Therefore, in order to avoid the false positive rate affecting the experimental results, In this study, 3 GEO datasets was used to explore the DEGs in lung cancer tissues and non-cancer tissues. In addition, bioinformatics analysis is carried out through Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction (PPI) networks, so that we can understand the molecular mechanism of cancer more comprehensively. In conclusion, a total of 185 DEGs and 10 hub genes were identified, which may be candidate biomarkers for lung cancer.
2. Materials and Methods
2.1 Identification of DEGs
Three gene expression profiles (GSE19188, GSE33532 and GSE30219) were acquired from GEO database. The array data of GSE19188 consisted of 91 lung cancer tissues and 65 adjacent normal lung tissues [8]. GSE33532 contained 80 non?small cell lung cancer tissues and 20 normal lung tissues [9]. GSE30219 included 293 lung cancer tissues and 14 adjacent normal lung tissues [10]. DEG was obtained from GEO database by a way of GEO2R analysis (http://www.ncbi.nlm.nih.gov/geo/geo2r/). The adj. P<0.01 and |log2FC|>2 were set as DEGs cutoff criterion.
2.2 DEGs network construction and Hub genes identified
We used Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/) (version 11.0) to predict DEGs interactions [11]. Analyzing the functional interactions between proteins may provide insights into the mechanisms of generation or development of diseases. In this study, the combined score of interaction > 0.4 was a significantly difference. In addition, Cytoscape (version 3.6.0), a public bioinformatics platform, which makes it possible for visualizing DEGs interactions [12]. Molecular Complex Detection (MCODE) (version 1.5.1) serves as a plug-in of Cytoscape to conduct cluster analysis of the network system to obtain hub genes [13]. Ten genes with high degree of connectivity displayed on MCODE were got by using selection criteria (MCODE score > 5, degree cut-off=2, node score cut-off=0.2, k-score=2, Max depth=100).
2.3 KEGG and GO enrichment analyses of DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) contributed to the biological analysis of collected date. GO and KEGG analysis were performed using DAVID online tool [14, 15]. In this study, we only showed the results of biological process (BP) and KEGG. R package “ggplot2” was used to visualize GO terms and KEGG pathways.
2.4 Survival analysis of hub genes
Kaplan?Meier plotter (http://kmplot.com/analysis/) was used to assess the prognostic significance of hub genes in lung cancer. The database is capable to assess the effect of 54k genes on survival in 21 cancer types. It contains data of breast (n=6234), ovary (n=2190), lung (n=3452) and stomach (n=1440) cancer [16]. In order to better judge the prognostic value of hub gene accurately, we divided samples into two groups according to the median expression of the gene, namely, up-regulation group and down-regulation group. (high expression, top 50%; low expression, bottom 50%). The overall survival (OS) of lung cancer patients was evaluated using Kaplan?Meier survival plots. The P?values of the log?rank test and hazard ratio (HR) with 95% confidence intervals (CIs) were also calculated.
3. Results
3.1 Identification of DEGs
Three mRNA expression profiles (GSE19188, GSE33532 and GSE30219) were included in this study. Using adj. P < 0.01 and |log2FC| > 2 as cut-off criterion, 500 DEGs (144 upregulated and 356 downregulated) were identified in GSE19188, 610 DEGs (201 upregulated and 409 downregulated) were identified in GSE33532, and 303 DEGs (37 upregulated and 266 downregulated) were identified in GSE30219. a total of 185 overlapping DEGs in three datasets was obtained using Venn diagrams (Figure 1A), including 32 upregulated genes and 153 downregulated genes.
3.2 DEGs network construction and Hub genes identified
The network of DEGs was built with 176 nodes and 559 edges (Figure 1B). The hub genes which have a high degree of connectivity was identified by Cytoscape (Table 1 and Figure 1C).
3.3 KEGG and GO enrichment analyses of DEGs
DEGs were enriched in different BPs according to GO analysis. The top 10 terms were angiogenesis, cell adhesion, G2/M transition of mitotic cell cycle, response to wounding, single organismal cell-cell adhesion, mitotic nuclear division, BMP signaling pathway, activation of protein kinase activity, protein localization to cell surface, and cell division (Figure 2A). As for the pathways, the results of KEGG enrichment analysis showed that DEGs were enriched in ECM-receptor interaction, Malaria, PPAR signaling pathway, Adrenergic signaling in cardiomyocytes, Cell adhesion molecules (CAMs), and cGMP-PKG signaling pathway (Figure 2B). The functional analyses of genes in the most significant module were analyzed using DAVID. As shown in Table 2, we can find that the DEGs play a role in cell division, mitotic nuclear division, and G2/M transition of mitotic cell cycle. The biological process analysis of 10 hub genes revealed that these genes were mainly participated in positive regulation of exit from mitosis, mitotic nuclear division, spindle organization, and cell division (Table 3).
3.4 The prognostic value of hub genes
The prognostic value of 10 hub genes in PPI network was obtained from Kaplan?Meier plotter (http://kmplot.com/analysis/). As shown in Figure 3, a high expression of hub genes corresponded to a poorer OS for lung cancer patients. All the hub genes we identified had statistical significance.
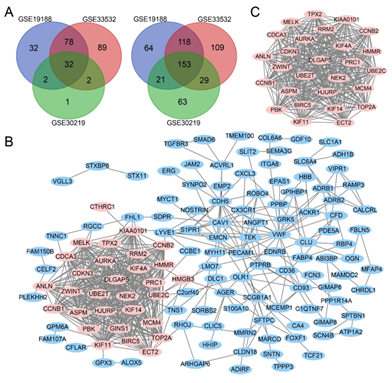
Figure 1: Venn diagram, PPI network and the most significant module of DEGs. (A) DEGs were selected with a |log2FC| >2 and adj. P-value <0.01 among the mRNA expression datasets GSE19188, GSE33532 and GSE30219. The 3 datasets showed an overlap of 32 upregulated DEGs (left) and 153 downregulated DEGs (right). (B) The PPI network of DEGs was constructed using Cytoscape. (C) The most significant module was obtained from PPI network. Upregulated genes are marked in light red; downregulated genes are marked in light blue.
|
Gene |
Full name |
Degree |
|
UBE2C |
ubiquitin conjugating enzyme E2 C |
30 |
|
RRM2 |
ribonucleotide reductase regulatory subunit M2 |
29 |
|
KIF11 |
kinesin family member 11 |
29 |
|
CDKN3 |
cyclin dependent kinase inhibitor 3 |
28 |
|
KIAA0101 |
KIAA0101 |
28 |
|
PRC1 |
protein regulator of cytokinesis 1 |
28 |
|
ASPM |
abnormal spindle microtubule assembly |
28 |
|
HMMR |
hyaluronan mediated motility receptor |
28 |
|
TOP2A |
topoisomerase (DNA) II alpha |
28 |
|
BIRC5 |
baculoviral IAP repeat containing 5 |
28 |
Table 1: The list of hub genes.
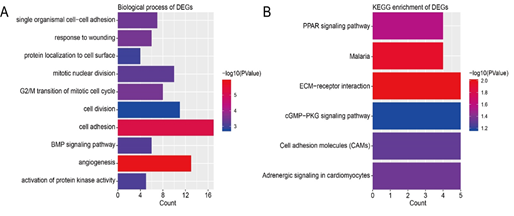
Figure 2: Functional and pathway enrichment analysis of DEGs. (A) GO analysis of DEGs. (B) KEGG pathway enrichment of DEGs.
|
Pathway ID |
Pathway description |
Count in gene set |
P-Value |
|
GO:0051301 |
cell division |
11 |
0.0000000000538 |
|
GO:0007067 |
mitotic nuclear division |
10 |
0.0000000000726 |
|
GO:0000086 |
G2/M transition of mitotic cell cycle |
8 |
0.00000000119 |
|
GO:0090307 |
mitotic spindle assembly |
4 |
0.0000227 |
|
GO:0007059 |
chromosome segregation |
4 |
0.000154 |
|
GO:0007051 |
spindle organization |
3 |
0.000273 |
|
GO:0007052 |
mitotic spindle organization |
3 |
0.000976 |
|
GO:0032147 |
activation of protein kinase activity |
3 |
0.002191 |
|
GO:0000910 |
cytokinesis |
3 |
0.002489 |
|
GO:0046602 |
regulation of mitotic centrosome separation |
2 |
0.00618 |
|
GO:0031145 |
anaphase-promoting complex-dependent catabolic process |
3 |
0.006601 |
|
GO:0007018 |
microtubule-based movement |
3 |
0.006929 |
|
GO:0007100 |
mitotic centrosome separation |
2 |
0.007719 |
|
GO:0031536 |
positive regulation of exit from mitosis |
2 |
0.009256 |
|
hsa04115 |
p53 signaling pathway |
3 |
0.00252 |
|
hsa04114 |
oocyte meiosis |
3 |
0.006781 |
|
hsa04110 |
cell cycle |
3 |
0.008406 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
Table 2: GO and KEGG pathway enrichment analysis of DEGs in the most significant module.
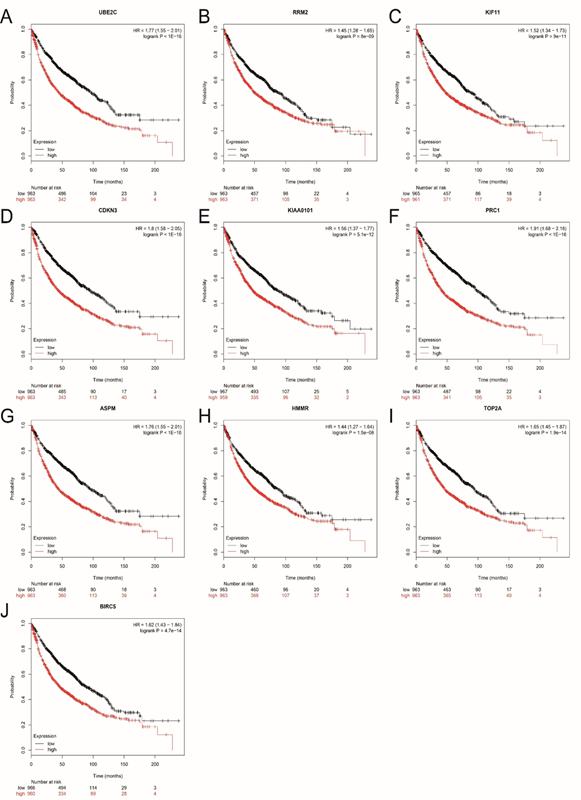
Figure 2: Functional and pathway enrichment analysis of DEGs. (A) GO analysis of DEGs. (B) KEGG pathway enrichment of DEGs.
Figure 3: Overall survival (OS) of the 10 hub genes in lung cancer based on Kaplan Meier-plotter. The patients were stratified into high-level and low-level groups according to median expression. (A) UBE2C. (B) RRM2. (C) KIF11. (D) CDKN3. (E) KIAA0101. (F) PRC1. (G) ASPM. (H) HMMR (I) TOP2A. (J) BIRC5.
|
Pathway ID |
Pathway description |
Count in gene set |
P-Value |
|
GO:0031536 |
positive regulation of exit from mitosis |
2 |
0.003212 |
|
GO:0007067 |
mitotic nuclear division |
3 |
0.007304 |
|
GO:0007051 |
spindle organization |
2 |
0.008545 |
|
GO:0051301 |
cell division |
3 |
0.014156 |
|
GO:0090307 |
mitotic spindle assembly |
2 |
0.019135 |
|
GO:0000910 |
cytokinesis |
2 |
0.02544 |
|
GO:0007059 |
chromosome segregation |
2 |
0.03587 |
GO, Gene Ontology.
Table 3: GO enrichment analysis of hub genes.
4. Discussion
To date, conventional surgery and chemotherapy remain the main treatment modalities for treating lung cancer. However, numerous lung cancer cases are diagnosed at an advanced stage. When treating patients with advanced lung cancer using conventional surgical resection and chemotherapy, the 5?year relative survival rate is only 4% [17]. Therefore, it is essential to explore the mechanisms of lung cancer progression to prevent its occurrence, guide pharmacy, indicate the prognosis, or improve survival rate.
The high-throughput platforms for detection of gene expression have been developing rapidly in diseases progression, which provides the basis of new target discovery for diagnosis, therapy, and prognosis of cancers [18,19]. Compared to the previous studies that only focused on several genes or a single cohort, this study selected 3 gene profile datasets from different research teams to integratedly explore the driven-genes and biological pathways in lung cancer. Finally, we identified 185 DEGs (32 upregulated and 153 downregulated). Biological pathway enrichment analysis revealed that cell adhesion, G2/M transition of mitotic cell cycle, response to wounding, single organismal cell-cell adhesion, mitotic nuclear division, BMP signaling pathway has certain influence on the development of lung cancer. The PPI network was constructed with 176 nodes and 559 edges. We then selected the most significant modules from the PPI network, and this module was mainly related to cell division, mitotic nuclear division, G2/M transition of mitotic cell cycle, mitotic spindle assembly, chromosome segregation, p53 signaling pathway, cell cycle, etc. The 10 hub genes in PPI network were selected by the degree of connectivity, including UBE2C, RRM2, KIF11, CDKN3, KIAA0101, PRC1, ASPM, HMMR, TOP2A, and BIRC5, which might play important roles in lung cancer. Several studies have proved that the expression of UBE2C was elevated abnormally in various cancers at both mRNA and protein levels. Increased UBE2C enhanced the ability of cell proliferation and predicted a poor prognosis [20]. RRM2 is a metabolic gene involved in nucleotide synthesis, which is abnormally expressed in many kinds of tumors and promotes the oncogenesis and development of tumors [21]. KIF11, a member of the actin family, is the motor protein needed to build bipolar spindles in cell division, could be used as a therapeutic target for tumors due to its function in tumorigenesis [22]. CDKN3 plays a crucial role in cell cycle regulation, Several studies have confirmed that overexpression of CDKN3 is closely related to tumor proliferation and metastasis [23]. KIAA0101 acts as a conserved PCNA-binding protein and plays an oncogene role in a variety of tumors [24]. PRC1 is essential for cytokinesis, Abnormal PRC1 expression will lead to chromosome instability and cancer evolution [25]. A large number of literatures have shown that ASPM is up-regulated in many cancers. Including prostate cancer, the proliferation and the invasive ability of cancer cells could be promoted by up-regulation of ASPM [26]. HMMR is involved in directectin the division of progenitor cells, supporting nerve growth in a PLK1-dependent pathway. Deregulation of HMMR was found in immortalized tumor cells [27]. TOP2A acts as a regulatory protein of topological state and involved in chromosome condensation and chromatid separation. It was proved to be a prognostic biomarker for bladder urothelial carcinoma [28]. BIRC5, also known as surviving, was an evolutionary conserved eukaryotic protein, played a vital role in the process of cell division [29]. Kaplan?Meier plotter was used to assess the prognostic value of hub genes, and the results indicated that high levels of these hub genes predicted poor OS in patients with lung cancer.
There were still several limitations in this study. First of all, In this topic, we have performed the function and prognostic value of hub genes, but only 10 hub genes have been analyzed, and the potential value of other genes that have not been studied, so the research in this field still needs to be further explored. Secondly, there are few biological information analysis tools used in this study, and TCGA and other databases can be used to verify this study in the future research.
5. Conclusion
In conclusion, the study identified 185 DEGs in lung cancer using bioinformatics analysis. The functions and pathways of DEGS were analyzed by using GO and KEGG enrichment analysis methods. In addition, 10 hub genes were identified and found to be reliable prognostic markers for lung cancer. And this study provided powerful basis for lung cancer studies, and in-depth experimental studies were needed.
Acknowledgments
This study was supported by Health and Family Planning Commission of Hubei Province project (WJ2015XB019 2015-2016) and Research and Development project of Xiangyang City (2014).
Authors' contributions
Jiubo Fan designed the study. Meng Wang and Tian Xie wrote the manuscript and performed the bioinformatics analysis. All authors read and approved the final manuscript.
Competing Interests
The authors declared no competing interests were existed.
References
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 389 (2017): 299-311.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 69 (2019): 7-34.
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 893 (2016): 1-19.
- Boolell V, Alamgeer M, Watkins DN, et al. The evolution of therapies in non-small cell lung cancer. Cancers 7 (2015): 1815-1846.
- Xu N, Chen S, Liu Y, et al. Profiles and Bioinformatics Analysis of Differentially Expressed Circrnas in Taxol-Resistant Non-Small Cell Lung Cancer Cells. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology 48 (2018): 2046-2060.
- Zhang N, Wang H, Xie Q, et al. Identification of potential diagnostic and therapeutic target genes for lung squamous cell carcinoma.Oncology Letters 18 (2019): 169-180.
- Wu Q, Zhang B, Sun Y, et al. Identification of novel biomarkers and candidate small molecule drugs in non-small-cell lung cancer by integrated microarray analysis.Onco Targets Ther 12 (2019): 3545-3563.
- Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 5 (2010): e10312.
- Meister M, Belousov A, Ec X, et al. Intra-tumor Heterogeneity of Gene Expression Profiles in Early Stage Non-Small Cell Lung Cancer 1 (2014).
- Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med 5 (2013): 186ra166.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47 (2019): D607-D613.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13 (2003): 2498-2504.
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4 (2003): 2.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4 (2009): 44-57.
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37 (2009): 1-13.
- Nagy A, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 8 (2018): 9227.
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet (London, England) 389 (2017): 299-311.
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Rev Clin Oncol 5 (2008): 588-599.
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 (2002): 207-210.
- Xie C, Powell C, Yao M, et al. Ubiquitin-conjugating enzyme E2C: a potential cancer biomarker. Int J Biochem Cell Biol 47 (2014): 113-117.
- Furuta E, Okuda H, Kobayashi A, et al. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta 1805 (2010): 141-152.
- Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer 12 (2012): 527-539.
- Cress WD, Yu P, Wu J. Expression and alternative splicing of the cyclin-dependent kinase inhibitor-3 gene in human cancer. Int J Biochem Cell Biol 91 (2017): 98-101.
- Yuan RH, Jeng YM, Pan HW, et al. Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research 13 (2007): 5368-5376.
- Li J, Dallmayer M, Kirchner T, et al. Linking Cytokinesis, Chromosomal Instability, and Cancer Evolution. Trends Cancer 4 (2018): 59-73.
- Pai VC, Hsu CC, Chan TS, et al. ASPM promotes prostate cancer stemness and progression by augmenting Wnt-Dvl-3-β-catenin signaling. Oncogene 38 (2019): 1340-1353.
- Connell M, Chen H, Jiang J, et al. HMMR acts in the PLK1-dependent spindle positioning pathway and supports neural development. Elife 10 (2017): 6.
- Zeng S, Liu A, Dai L, et al. Prognostic value of TOP2A in bladder urothelial carcinoma and potential molecular mechanisms. BMC Cancer 19 (2019): 604.
- Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci 132 (2019).
