Identification of Driver Mutations in MicroRNAs for Early Diagnosis of Lung Cancer
Article Information
Ramsha Nasir, Nisar A Shar*
Department of Biomedical Engineering, NED University of Engineering and Technology, Karachi, Pakistan
*Corresponding Authors: Nisar A Shar, Department of Biomedical Engineering, NED University of Engineering and Technology, Karachi, Pakistan
Received: 22 May 2020; Accepted: 29 July 2020; Published: 22 September 2020
Citation: Ramsha Nasir, Nisar A Shar. Identification of Driver Mutations in MicroRNAs for Early Diagnosis of Lung Cancer. Journal of Bioinformatics and Systems Biology 3 (2020): 074-080.
View / Download Pdf Share at FacebookAbstract
Over the past years, it has been discovered that microRNA possesses a critical role in lung cancer related deaths worldwide. Different research on molecular biology of lung cancer has improved our understanding that determines the behavior of malignant cells which has encouraged the discovery of driver mutations. The identification of driver mutations in lung cancer has led to a paradigm shift and provides us with new direction towards personalized therapies with improved survival rates. Therefore, it is of interest to explore microRNA driver mutations in this context. Hence, in this research different consistent point mutations are identified in candidate microRNAs at different sites and different numbers of transcription factors binding at these consistent mutations are examined. The highest consistency of mutation was 18 found in mir-23A at the genomic point (13880331) where total 5 transcription factors were binding. The second highest consistency was 12 found in different miRNAs at different genomic locals where 1-2 transcription factors were binding whereas; mir-25 is the only having consistency of 7 at genomic point (100089125) with 1 transcription factor attachment. The result of consistent mutation in regulatory regions indicates that these mutated sites may serve as potential biomarkers for lung cancer. Moreover, the comprehensive utilization of these microRNAs mutations, different drugs can be designed that mainly target these specified mutated loci and would be helpful in treating lung cancer.
Keywords
Lung cancer; MicroRNA; Transcription factors; Regulatory regions; Consistency
Lung cancer articles, MicroRNA articles, Transcription factors articles, Regulatory regions articles, Consistency articles
Lung cancer articles Lung cancer Research articles Lung cancer review articles Lung cancer PubMed articles Lung cancer PubMed Central articles Lung cancer 2023 articles Lung cancer 2024 articles Lung cancer Scopus articles Lung cancer impact factor journals Lung cancer Scopus journals Lung cancer PubMed journals Lung cancer medical journals Lung cancer free journals Lung cancer best journals Lung cancer top journals Lung cancer free medical journals Lung cancer famous journals Lung cancer Google Scholar indexed journals MicroRNA articles MicroRNA Research articles MicroRNA review articles MicroRNA PubMed articles MicroRNA PubMed Central articles MicroRNA 2023 articles MicroRNA 2024 articles MicroRNA Scopus articles MicroRNA impact factor journals MicroRNA Scopus journals MicroRNA PubMed journals MicroRNA medical journals MicroRNA free journals MicroRNA best journals MicroRNA top journals MicroRNA free medical journals MicroRNA famous journals MicroRNA Google Scholar indexed journals Transcription factors articles Transcription factors Research articles Transcription factors review articles Transcription factors PubMed articles Transcription factors PubMed Central articles Transcription factors 2023 articles Transcription factors 2024 articles Transcription factors Scopus articles Transcription factors impact factor journals Transcription factors Scopus journals Transcription factors PubMed journals Transcription factors medical journals Transcription factors free journals Transcription factors best journals Transcription factors top journals Transcription factors free medical journals Transcription factors famous journals Transcription factors Google Scholar indexed journals Regulatory regions articles Regulatory regions Research articles Regulatory regions review articles Regulatory regions PubMed articles Regulatory regions PubMed Central articles Regulatory regions 2023 articles Regulatory regions 2024 articles Regulatory regions Scopus articles Regulatory regions impact factor journals Regulatory regions Scopus journals Regulatory regions PubMed journals Regulatory regions medical journals Regulatory regions free journals Regulatory regions best journals Regulatory regions top journals Regulatory regions free medical journals Regulatory regions famous journals Regulatory regions Google Scholar indexed journals Consistency articles Consistency Research articles Consistency review articles Consistency PubMed articles Consistency PubMed Central articles Consistency 2023 articles Consistency 2024 articles Consistency Scopus articles Consistency impact factor journals Consistency Scopus journals Consistency PubMed journals Consistency medical journals Consistency free journals Consistency best journals Consistency top journals Consistency free medical journals Consistency famous journals Consistency Google Scholar indexed journals biomarkers articles biomarkers Research articles biomarkers review articles biomarkers PubMed articles biomarkers PubMed Central articles biomarkers 2023 articles biomarkers 2024 articles biomarkers Scopus articles biomarkers impact factor journals biomarkers Scopus journals biomarkers PubMed journals biomarkers medical journals biomarkers free journals biomarkers best journals biomarkers top journals biomarkers free medical journals biomarkers famous journals biomarkers Google Scholar indexed journals phenotype articles phenotype Research articles phenotype review articles phenotype PubMed articles phenotype PubMed Central articles phenotype 2023 articles phenotype 2024 articles phenotype Scopus articles phenotype impact factor journals phenotype Scopus journals phenotype PubMed journals phenotype medical journals phenotype free journals phenotype best journals phenotype top journals phenotype free medical journals phenotype famous journals phenotype Google Scholar indexed journals COSMIC database articles COSMIC database Research articles COSMIC database review articles COSMIC database PubMed articles COSMIC database PubMed Central articles COSMIC database 2023 articles COSMIC database 2024 articles COSMIC database Scopus articles COSMIC database impact factor journals COSMIC database Scopus journals COSMIC database PubMed journals COSMIC database medical journals COSMIC database free journals COSMIC database best journals COSMIC database top journals COSMIC database free medical journals COSMIC database famous journals COSMIC database Google Scholar indexed journals DNA articles DNA Research articles DNA review articles DNA PubMed articles DNA PubMed Central articles DNA 2023 articles DNA 2024 articles DNA Scopus articles DNA impact factor journals DNA Scopus journals DNA PubMed journals DNA medical journals DNA free journals DNA best journals DNA top journals DNA free medical journals DNA famous journals DNA Google Scholar indexed journals
Article Details
1. Introduction
Lung cancer is one of the leading cancer related mortality [1]. It is characterized by genetic diversity with relatively few persistent mutations, occurring at high frequency [2]. In this condition, the cells begin to divide in the lungs uncontrollably. These abnormal cells obstruct the functioning of lung that ultimately reduces the ability of a person to breath. Normally, our body has check and balance system by which our body programs cells to grow and die at certain stages of life cycle in order to avoid overgrowth of cells. Whereas in cancer, this instruction is overrides, causing cells to multiply and grow exponentially. This overgrowth of abnormal cells, leads to the development of lung cancer.
Moreover, many of these mutations are associated with several environment and heredity factors [3]. The changes in phenotype from normal lung cell to malignant occur in a series of genetic alterations and identification of these mutations has the potential to provide further therapeutic opportunities [4]. However, in recent years the vast improvements have been made in molecular biology that helps researchers to understand and underpins lung cancer. This has lead to a great improvement in prevention of diseases, treatments and early detection of tumors [5, 6]. The symptoms of lung cancer may comprise coughing with blood, stained mucus, persistent cough, wheezing sound, unexplained weight loss, breathlessness, pain when coughing or breathing [7]. The risk factors that are involved to take part in the development of lung cancer are smoking, asbestos fiber, pollutant air, exposure to radon, ceramics’, and fireworks [8]. These factors mutate the genome and some of these mutations may play aggressive role in driving lung cancer.
MicroRNAs also known as miRNAs or miRs, are short, non-coding, single stranded RNAs 20-25 nucleotide long [9]. The main function of MicroRNA is to regulate gene expressions post -transcriptionally [10]. MicroRNAs comprise of 1-2% of all genes [11]. The first microRNA was discovered as non-coding small RNA as growth regulator of Caenorhabditis elegans (lin-4 and let-7) [12]. MicroRNAs are unable to code proteins, but acquiring significant catalytic, regulatory and structural functions. Thus, microRNAs are capable of increasing and decreasing expressions of genes by binding to the untranslated region (3′-UTR) of their target messenger RNA (mRNA) and repress the production of protein by destabilizing the mRNA [13, 14], also known as gene silencing.
Different researches have proved that altered expression of miRNAs in tumors may also act as novel oncomiRs [15] and tumor suppressors [16]. These abnormal expression levels of microRNAs are characterized as a result of aberrant expressions of precursor microRNAs transcript. These aberrant expressions of genes have been observed to cause different abnormalities and cancers such as prostate, lung, breast, gastric, colorectal, lymphoma, leukemia, melanoma, and hepatocellular carcinoma [17]. Additionally to their role in physiological conditions, studies indicate that microRNAs is also playing significant role in the pathological mechanisms of neurodegenerative diseases [18].
2. Materials and Methods
2.1 Identification of Non-Coding Mutations in Non-coding variants
In this mutational analysis, the initial step is to identify mutations in non-coding variants. For this purpose, non-coding variant file is downloaded from COSMIC database (release v87) and overlapped with 100kb extended genomic locations of candidate miRNAs using bedtools intersect command. The non-coding variants file contains complete data of non-coding mutations. Genomic locations of microRNAs are identified from Ensemble genome browser GRch38. The genomic locations are extended, prior to overlapping because, it has been found in almost 70% of cases, that regulatory region of a gene lie within 100 kb [19]. The overlapped result determines vastly repetitive mutational regions in candidate miRNAs. Therefore; we developed python code to obtain non-repetitive mutations to identify unique sites of interest.
2.2 Identification of consistent mutations in Non-Coding variants
In the next step, we identified consistent mutations in non-coding variants by overlapping repetitive and non-repetitive file. It is also not necessary that all consistent point mutations possess the potential of driving lung cancer. It might be possible that these mutated sites are just repeating itself because of any interruption of any cellular process like DNA repair mechanism. Some of them can be passenger mutations because throughout the life of human beings many mutations can occur randomly, and many of the mutations occur due to several environmental factors, and few mutations are inherited from parents to their offspring’s. In any case, a portion of these consistent mutations can be driver mutations.
2.3 Identification of Consistent Non-Coding Mutations at IMR90 transcription factors Binding Sites
Non-coding region of DNA contain regulatory regions that are capable of increasing and decreasing the gene expressions. At these regulatory regions transcription factors bind which can either enhance or suppress the process of translation. Different studies indicate that mutations inside regulatory region are extensively significant in cancers and other pathogenic diseases. To further characterize and shortlist candidates, IMR90 Transcription factors (TFs) are mapped that are downloaded from UCSC ENCODE with identified consistent mutations to examine the point mutations in regulatory regions. IMR90 is the cancer cell line of human lung. There are total 10 Transcription Factors (TFs) for IMR90 cells out of which 5 (CTCF, MAFK, RAD21, POL, CEBPB) were downloaded from UCSC ENCODE (hg19). The files are then converted to GRCH38/hg38 genome coordinates using UCSC liftover tool. The overlapped result recognized significant TFs’ binding sites (TFBS) as well as the number of TFs binding at these consistent point mutations.
3. Results and Discussion
MIicroRNA are crucial in various biological processes such as regulatory functions, cellular differentiation, development and apoptosis. In cancers, mutations in non-coding regions can cause dysregulation of oncogenes and tumor suppressors. Measures of function in the non-coding genome usually reflect on binding of transcription factors. In this study, we identified potential driver mutations in candidate microRNAs. As shown in Figure 1, all non-coding mutations in microRNAs are first identified by intersecting non-coding variants with 100kb extended genomic locations of each microRNA using bedtools intersect command. This recognized repetitive mutations therefore; we used python code to extract non-repetitive mutations. In the next step, consistencies in mutations are identified by intersecting repetitive and non-repetitive files. Higher the consistencies of mutations have higher probabilities of driving lung cancer.
After the identification of consistencies, we map total five Transcription factors (CEBPB, CTCF, MAFK, POL2, RAD21) of IMR90 (lung cancer cell line) with consistent mutations of microRNA using bedtools intersect command. The result of intersecting consistent point mutations with Transcription factors of IMR90 cells examine number of Transcription Factors binding as well as significant Transcription Factors binding sites as shown in Table 1 (A, B). Highest consistency is found in MIR-23A as compared to other microRNA. At the highest consistency of 18 at a point mutation of (13880331) total 5 TFs are binding in MIR-23A. Recognition of this persistent mutated loci of non-coding variants, propose that it can play significant role in driving lung cancer and more likely to influence regulatory elements of genes that ultimately cause disruption in translation process because, this precise loci is having the highest probability of causing lung cancer.
|
MIR NAMES |
CHR: |
STAR-END |
MUTATED SITE |
CONSISTENCY |
NO. OF TFs BINDING |
TFs BINDING |
|
MIR-23A |
chr19 |
13836587-13836659 |
13880331 |
18 |
5 |
CEBPB, CTCF, MAFK, POL2, RAD21 |
|
MIR-21 |
chr17 |
59841266-59841337 |
59838122 |
12 |
1 |
POL2 |
|
59838178 |
12 |
1 |
POL2 |
|||
|
MIR-23A |
chr19 |
13836587-13836659 |
13829976 |
12 |
1 |
POL2 |
|
13830024 13830145 |
12 |
1 |
POL2 |
|||
|
12 |
1 |
POL2 |
||||
|
13831407 |
12 |
1 |
POL2 |
|||
|
13831773 |
12 |
1 |
POL2 |
|||
|
13836478 |
12 |
1 |
POL2 |
|||
|
13836558 |
12 |
1 |
POL2 |
|||
|
MIR-451A |
chr17 |
28861369-28861440 |
28854934 |
12 |
2 |
POL2, RAD21 |
|
28879226 |
12 |
2 |
RAD21 |
|||
|
MIR-372 |
chr19 |
53787890-53787956 |
53772987 |
12 |
2 |
CTCF |
|
53773139 |
12 |
2 |
CTCF |
|||
|
53773148 |
12 |
2 |
CTCF |
|||
|
MIR-16 |
chr3 |
160404745-160404825 |
160449652 |
12 |
2 |
POL2.RAD21 |
|
MIR-25 |
chr7 |
100093560-100093643 |
100101813 |
12 |
2 |
CEBPB, POL2 |
|
MIR-100 |
chr11 |
122152229-122158308 |
122115592 |
12 |
1 |
CEBPB |
Table 1A: Consistent non-coding mutations at IMR90 TF binding sites at specific positions.
|
MIR NAMES |
CHR: |
START-END |
MUTATED SITE |
CONSISTENCY |
NO. OF TFs BINDING |
TFs BINDING |
|
MIR-328 |
chr16 |
67202321-67202395 |
67180020 |
12 |
1 |
CTCF |
|
MIR-21 |
chr17 |
59841266-59841337 |
59841436 |
12 |
1 |
POL2 |
|
MIR-23A |
chr19 |
13836587-13836659 |
13789265 |
12 |
1 |
CEBPB |
|
13820939 |
12 |
1 |
CEBPB |
|||
|
MIR-155 |
chr21 |
25573980-25574044 |
25607308 |
12 |
1 |
POL2 |
|
25607316 |
12 |
1 |
POL2 |
|||
|
MIR-145 |
chr5 |
149430646-149430733 |
149517936 |
12 |
1 |
MAFK |
|
MIR-25 |
chr7 |
100093560-100093643 |
100089125 |
7 |
1 |
POL2 |
|
100091199 |
6 |
1 |
POL2 |
|||
|
100091597 |
6 |
1 |
POL2 |
|||
|
100091833 |
6 |
1 |
CTCF |
|||
|
100093937 |
6 |
1 |
MAFK |
|||
|
100094036 |
6 |
1 |
MAFK |
|||
|
100101469 |
6 |
1 |
MAFK |
|||
|
100148875 |
6 |
1 |
CEBPB |
|||
|
100148922 |
6 |
1 |
CEBPB |
|||
|
MIR-182 |
chr7 |
129770383-129770492 |
129833002 |
6 |
1 |
MAFK |
|
129865187 |
6 |
1 |
MAFK |
|||
|
MIR-126 |
chr9 |
136670602-136670686 |
136758603 |
6 |
1 |
CTCF |
|
MIR-370 |
chr14 |
100811139-101011213 |
100845392 |
6 |
1 |
POL2 |
|
100862420 |
6 |
1 |
MAFK |
Table 1B: Consistent non-coding mutations at IMR90 TF binding sites at specific positions.
At the consistency of 12 in different microRNAs, 1-2 TFs are binding. While, at least consistency of 6 in MIR-25, -182, -126, and MIR -370, only 1 TF is binding. The results of least consistency with least TFs are binding on mutated sites. It indicates that although, these point mutations are present in regulatory regions and possesses the potential of causing lung cancer, but having less probability as compared to the mutated sites having higher consistency in regulatory regions. The mutations at Transcription factors binding sites can be termed as driver mutations. The obtained results suggest, as these mutations are consistent and present at regulatory regions consequently; they can significantly affect genes expression level and might play a considerable role in driving lung cancer. It means either these genes are excessively expressed or unexpressed. Hence, these point mutations in regulatory regions can serve as potential biomarker for lung cancer. However, further evaluation is needed for the verification of acquired results to identify up to which extent these consistent mutations are involved in driving lung cancer.
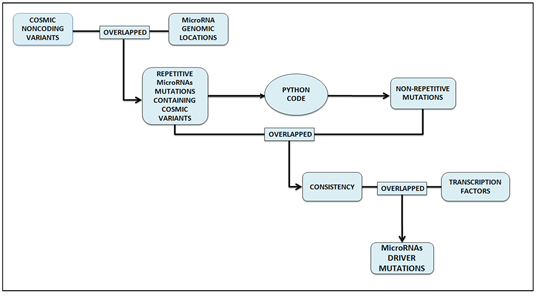
Figure 1: Methodology of finding driver mutations in microRNAs comprises of three steps mainly. In the initial step, non-coding mutations in non-coding variants are identified by overlapping cosmic non-coding variants with microRNA genomic locations. Overlapped result yields highly repetitive mutations. These repetitive mutations file is then given as input to the python code and the non-repetitive file is generated as output. The second step is the identification of consistency which is obtained by overlapping repetitive and non-repetitive mutations files. In the final step, consistent non-coding mutations in regulatory regions are identified.
4. Conclusion
The exploration for driver mutations in cancer genome is a crucial step in the progress towards personalized approaches to cancer treatments. The ability of miRNAs to regulate gene expression levels and their key role in progression of lung cancer endow with new direction for research on prognostic and predictive biomarkers [20]. By recognizing the molecular variations in microRNA responsible for driving lung cancers, drugs can be designed that particularly targets deregulated or mutated genes. Furthermore, new approaches may be designed that averts the damage or even restore these mutated regions of DNA due to which these drivers are formed. Hence, it is of importance to evaluate these driver microRNA mutations for further consideration.
Acknowledgement
NAS and RN are thankful to National Center in Big Data and Cloud computing (Genomics Lab) NED University of Engineering and Technology for their tremendous support in this study.
Conflict of Interest
The authors declare no conflict of interest.
References
- Dela Cruz CS, Tanoue LT, Matthay RA. Lung Cancer: Epidemiology, Etiology, and Prevention. Clinic in chest medicine 32 (2011): 605-644.
- Castellanos E, Feld E, Horn L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. Journal of thoracic oncology 12 (2017): 612-623.
- Kanwal M, Ding X, Cao Y. Familial Risk for Lung Cancer. Oncology letters 13 (2017): 535-542.
- Xiaohong Du, Jitai Zhang, Juping Wang, et al, Role of miRNA in Lung Cancer-Potential Biomarkers and Therapies. Current pharmaceutical design 23 (2018): 5997-6010.
- Vivien Wang, WeiWu. MicroRNA-based Therapeutics for Cancer. BioDrugs 23 (2009): 15-23.
- Barger JF, Nana-sinkam SP. MicroRNA as Tools and Therapeutics in Lung Cancer. Respiratory Medicine 109 (2015): 803.
- Kelly M Latimer, Timothy F Mott. Lung Cancer: Diagnosis, Treatment Principles, and Screening. American family physician 91 (2015): 250.
- Adrian Cassidy, Stephen W Duffy, Jonathan P Myles, et al. Lung Cancer Risk Prediction: A Tool for Early Detection. International journal of cancer 120 (2007): 1-6.
- Antony Rodriguez, Sam Griffiths-Jones, Jennifer L Ashurst, et al. Identification of Mammalian microRNA Host Genes and Transcription Units. Genome Research 14 (2004): 1902-1910.
- David P Bartel. MicroRNAs: Target Recognition and Regulatory Functions. Cell 136 (2009): 215-233.
- Joana A Vidigal, Andrea Ventura. The Biological Functions of miRNAs: Lessons From in Vivo Studies. Trends in cell biology 25 (2015): 137.
- Lee P Lim, Nelson C Lau, Earl G Weinstein, et al. The microRNAs of Caenorhabditis Elegans. Genes an development 17 (2003): 991.
- Lucy Barrett, Susan Fletcher, Steve Wilton. Regulation of Eukaryotic Gene Expression by the Untranslated Gene Regions and Other Non-Coding Elements. Cellular and molecular life sciences 69 (2012): 3613.
- Jin Wang Jinyun Chen Subrata Sen. MicroRNA as Biomarkers and Diagnostics. Journal of cellular physiology 231 (2016): 25-30.
- Aurora Esquela-Kerscher, Frank J Slack. Oncomirs-microRNAs with a role in cancer. Nature reviews 6 (2006): 259.
- Caiyan Zhang, Huimin Wang, Xiaomin Liu, et al. Oncogenic microRNA-411 Promotes Lung Carcinogenesis by Directly Targeting Suppressor Genes SPRY4 and TXNIP. Oncogene 38 (2019): 1892-1904.
- Weili Huang. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods in molecular biology 1617 (2017): 57-67.
- Sara Elramah, Marc Landry, Alexandre Favereaux, et al. MicroRNAs Regulate Neuronal Plasticity and Are Involved in Pain Mechanisms. Frontiers in cellular neuroscience 8 (2014).
- Mahesh Yaragatti, Claudio Basilico, Lisa Dailey. Identification of Active Transcriptional Regulatory Modules by the Functional Assay of DNA From Nucleosome-Free Regions. Genome research 18 (2008): 930-938.
- Huiyin Lan, Haiqi Lu, Xian Wang, et al. MicroRNAs as Potential Biomarkers in Cancer: Opportunities and Challenges. BioMed research international (2015).
