High Resistance Mutation to cART in HIV-1 Exposed Infected Children and Recent Emergence of CRF02_AG Variant in Bouar, A Rural Environment of Central African Republic
Article Information
Ulrich Vickos1, 2*, Giovanni Gaiera3, Nicola Cotugno4, Christelle Luce Bobossi Gadia5, Ornella Anne Sibiro Demi6, Angelo Sala7, Michela Sampaolo8, Alain Le Faou9, Enzo Boeri8
1Infectious and tropical diseases Unit, Amitie Hospital, Bangui CAR
2Laboratory of Immunity, Department of systemics medicine, Faculty of medicine and surgery, Tor Vergata University, Rome, Italy
3Clinic of Infectious Diseases, IRCCS San Raffaele Scientific Institute, Milan, Italy
4Academic Department of Pediatrics, Research Unit of Congenital and Perinatal Infection, Children's Hospital Bambino Gesù, Rome, Italy
5Molecular biology Unity, Clinical biology and public health national Laboratory, Bangui, CAR
6Laboratory of viral hepatitis, Institut Pasteur of Bangui, CAR
7Saint Michel Center, Society of Priests of the Sacred Heart of Betharram, Health Pastoral Diocese of Bouar, Bouar, CAR.
8Laboratory of Microbiology, San Raffaele Scientific Institute, Milan, Italy
9Faculty of Pharmacy Brabois Santé, Vandoeuvre-lès-Nancy, France
*Corresponding Author: Ulrich Vickos, Infectious and tropical diseases Unit, Amitie Hospital, 2, Ouango-Bangui Mango Street, Bangui, 94045, Central African Republic
Received: 17 February 2022; Accepted: 25 February 2022; Published: 22 March 2022
Citation:
Ulrich Vickos, Giovanni Gaiera, Nicola Cotugno, Christelle Luce Bobossi Gadia, Ornella Anne Sibiro Demi, Angelo Sala, Michela Sampaolo, Alain Le Faou, Enzo Boeri. High Resistance Mutation to cART in HIV-1 Exposed Infected Children and Recent Emergence of CRF02_AG Variant in Bouar, A Rural Environment of Central African Republic. Journal of Pediatrics, Perinatology and Child Health 6 (2022): 155-176.
View / Download Pdf Share at FacebookAbstract
Introduction: The emergence of HIV-1 recombinant forms and drug combined antiretroviral therapy (cART) resistance are frequent in the therapeutic course of HIV-infected children in Low and Middle-Income Countries (LMIC) precisely in Central African Republic (CAR) as evidenced by studies carried out in the Bangui capital. Vertical transmission rate including breastfeeding is 12.4. The aim is to analyze retrospectively the molecular characterization of sequencing results and mutation detected in HIV infected children under cART since infancy.
Methods: The 2019 retrospective review of the clinical, therapeutical, and immunological-molecular records of six children who were performed the genome sequencing, followed in Bouar, at the St Michel IST/HIV Center, in the north-west of the CAR. These perinatal HIV-infected children were not tested early and started late cART regimens, used for vertical transmission prevention and treatment initiation of HIV infection in CAR.
Results: We analyzed results from viral RNA extracted amplification and sequencing of 6 children plasma samples collected under first line ART. Sequencing of viral genomes revealed high level resistance mutations to NRTIs (ABC/FTC/3TC) and to NNRTIs (EFV/NVP used locally and DOR/ETR/ RPV unused) with ambiguous positions in amino-acids comparison and deletion. Three CRF02_AG strains formed a cluster by strongly detaching from other CAR and worldwide strains with robust boostrap at 91.
Conclusions: The genomes sequencing showed that resistance mutations made the treatment inefficient confirming the clinical, immunological, and virol-ogical failure and an emerging CRF02_AG genotype variant, probably of foreign origin. This discovery clearly highlights the importance of ART genetic resistance testing and personalized medicine.
Keywords
Children, cART Resistance Mutations, HIV-1 Subtype, Central African Republic
Article Details
Abbreviations:
1st line sub: 1st line substitution; 3TC: Lamivudine; /r: ritonavir; ABC: Abacavir; ARI: Acute respiratory Infections; CAR: Central African Republic, cART: combined ART; CTM: Cotrimoxa-zole; DRI: Drug Resistance Interpretation; HAART: highly active ART; IR: Intermediate resistance; LLR: Low-level resistance; MoH: Ministry of Health; MS: Mutation Scoring; PI: Protease Inhibitor; NA: Not applicable; NNRTI: Non-nucleoside Reverse Trans-criptase Inhibitors; NRTI: Nucleoside Reverse Transcriptase Inhibitors; HLR: High-Level Resis-tance; PLLR: Potential low-level resistance; PLWH: people living with HIV; S: Sensible/Susceptible; ATV/r: Atazanavir/r; DRV/r: Darunavir/r; LPV/r: Lopinavir/r; AZT: Zidovudine; FTC: Emtricitabine; TDF: Tenofovir; TBC: Tuberculosis; DOR: Doravirine; EFV: Efavirenz; ETR: Etravirine; NVP: Nevirapine; RPV: Rilpivirine
1. Introduction
HIV-1 prevalence is lightly decreasing in Central African Republic (CAR) with, in 2019, 4,900 new HIV infections, 3,800 AIDS-related deaths, and 3.5% people living with HIV (PLWH) as compared to 4.9% in 2010 [1, 2]. In 2016, an estimated 100,000 PLWH, among whom only 47% had access to antiretroviral therapy (ART). For the children, about 6,900 were infected and 3,156 were on ART. Treatment or prophylaxis of mother-to-child HIV transmission were available for 81% of pregnant women. Thus, it is estimated that less than 1000 newborns are infected each year [1]. The mana-gement of pediatric infections and the prevention of mother-to-child transmission (PMTCT) of HIV in sub-Saharan Africa has dramatically improved. This has been possible by a widespread use of combined ART (cART) and highly active ART (HAART). However, this has been accompanied by the emergence of strains of HIV highly resistant to antiretroviral drugs (ARV). Numerous studies have evaluated the impact of their use on the emergence of drug resistance mutations (DRM) in infected children, mainly infants born to mother presenting either a failure of PMTCT or an absence of prophy-laxis [3, 4]. However, less studies have been conducted in Central African rural areas [5-7]. It is easier to carry out studies in major cities given the accessibility and availability of biological and thera-peutic means for the monitoring of HIV patients.
In Bangui, this is related to high rates of resistance to nucleotide reverse transcriptase inhibitor (NRTI), non-nucleotide reverse transcriptase inhibitor (NNRTI) DRM (45%) and protease inhibitors (PI) (24%) [8]. Overall, 55% of children receiving first-line treatment are eligible for a second-line regimen of whom 64% need a third-line treatment including an integrase strand transfer inhibitor (INSTI). Most HIV-infected African children fail to respond to the first- or second-line regimens recommended by WHO which is problematic as the available ARV drugs are in limited numbers [9, 10]. For pediatric HIV infections they are: Azidothymidine (AZT), Abacavir (ABC), Lamivudine (3TC), Truvada (Emtricitabine/Tenofovir (FTC/TDF)), Efavirenz (EFV), Nevirapine (NVP), Duovir-N (Lamivudine/ Zidovudine-Nevirapine (3TC/ZDV-NVP)), Atazana-vir/Ritonavir (ATV/r) and Kaletra (Lopinavir/ Ritonavir (LPV/r) junior). NRTI and NNRTI are favored for first line treatments. Besides no other ARV drug classes are available (Supplementary Table 2).
Two types of resistance to the available drugs are known. First two competitive mechanisms: one by preventing the insertion of the NRTI in the DNA chain, the other specific of thymidine analogues which once incorporated are released by the activity of pyrophosphorylase. The mutations associated with this process are known as thymidine analogue mutations (TAM). Second, non-competitive mecha-nisms are specific of the NNRTI. These molecules which impair the activity of the transcriptase by fixation in the vicinity of the active site of the enzyme, can no longer adhere to the enzyme [8]. The mutations associated with these resistances are known as primary mutations. Secondary mutations, alone, have in most cases a limited effect, but they increase the resistance level due to primary mutations when both present.
Each mutation is associated with resistance mech-anism of its own and the corresponding resistance level varies widely from one to the other [11]. We have investigated the profile of resistance mutations to cARTs in 6 HIV+ children followed in the rural area of ??Bouar for whom, complete clinical, immune-ovirological and phylogenetic data have been obtained despite the limited number of patients.
2. Methods
2.1 Study design
This is a retrospective observational study of 6 HIV-infected children who belong to a cohort of more than 1,639 people living with HIV (PLWH) attending the “Saint Michel HIV/STI Center of Bouar for the Antiretroviral Therapy”, a non-profit structure with limited resources. The mean time of ARV therapy initiation for the 6 children was 30 months after birth. These children have been exposed-infected by their HIV-infected mothers. They were prescribed a first-line treatment at inclusion which was not modified thereafter. For the 6 children complete socio-demo-graphic, clinical, immunovirological and therapeutic data were collected from their follow-up medical file, at the time of obtaining blood samples for genotyping analysis. All information were gathered from the patient' files. The Center has only limited possibilities of diagnostic test thus genomic analyses were performed in Italy for the sequencing of integrase, polymerase, and protease genes.
2.2 Genomic analysis
Plasma samples from the 6 selected children, sent to the virology laboratory of the San Raffaele Hospital, Milan, Italy in ice pack. GenExpert HIV (Cepheid, Sunnyvale, CA), were used for viral load deter-mination primarily in the Center. The viral integrase (IN), reverse transcriptase (RT) and protease (PR) genes were sequenced in Italy with the ViroSeq HIV-1 genotyping system (Stanford University, Stanford, CA), according to the manufacturer’s instructions, as described previously [12].
2.3 Drug resistance associated mutations and genomic analysis
Mutations of RT and PR genes associated with resistance to NRTIs, NNRTIs and PIs were identified and their consequences on resistance to ARV drugs interpreted according to the mutations scoring of the Stanford University genotypic resistance inter-pretation algorithm and the HIV Drug Resistance Database (https://hivdb.stanford.edu/page/release-notes/). The scores are the sum of each mutation penalty score for a given drug. Scores less than 10 indicate susceptibility; between 10 and 14 a potential low-level resistance; between 15 and 29 low-level resistance; between 30 and 59 intermediate resistance. Scores of 60 or greater indicate high-level resistance.
For genomic analysis, first, HIV-1 subtypes were determined using the IN gene sequences according to the genotyping tool of the NIH available on line (https://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi). From INSDC database, 1,227,800 coding sequences (CDS) belonging to taxonomic IDs 11,676 (HIV-1 M), 11,709 (HIV-2) and 12,721 (others HIV-1 O et N) were retrieved. CDS were dereplicated (884,488 unique CDS) and clustered (636 groups) with Biomanda Data to group similar CDS based on the sequence similarity and the gene annotation. CDS cluster composed of 578 229 CDS for 454,944 unique ones was selected.
Second, for strains comparisons, after the raw sequences from the 6 patients have been curated to remove sequencing errors, the nucleotide positions 2096 to 2 551 of the genome of the reference HIV strain AF033819 was retained. It corresponds to the 195 first amino acids of the A chain of the reverse transcriptase. Eighty-four unique sequences of the target region and from the gag-pol gene CDS cluster were collected from GenBank. They correspond to the most frequent sequences available from African Countries and other main countries (USA, France, Canada, United Kingdom, China, etc.). Moreover, 300 sequences from CAR were included. CDS were aligned to the reference genome sequence using Clustal Omega algorithm. A phylogenetic reconstr-uction by maximum likelihood from PhyML algorithm (Model GTR) with a bootstrap analyzes of 1 000 replicates were performed on nucleic acid sequences. Phylogenetic tree was annotated with the available metadata on sequences. HIV-1 IN, RT and PR sequences were deposited at the GenBank Nucleotide Archive database with the accession numbers MW373071 to MW373076.
2.4 Statistical analyses
The studied children characteristics and the analyzes results were entered into a Microsoft Excel data sheet.
3. Results
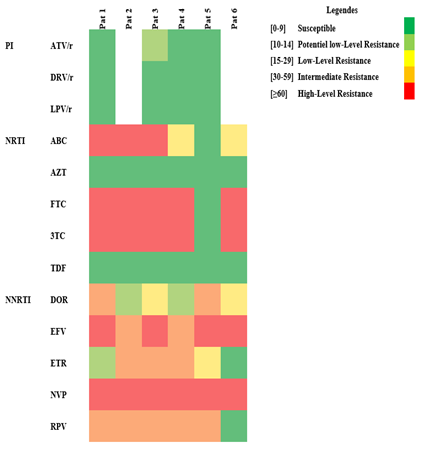
The available and complete 6 patients' data are displayed in Table 1. The mutation scorings and drug used are detailed (Supplementary Table 1 and Figure 1a/1b).
- Patient 1: In the PR gene an accessory mutation of resistance to PI (V32S) was found. Several major mutations of the RT gene and were associated with resistance to NRTI (L74LI, M184V) and NNRTI (K103N, E138Q, Y188F). The mutation scoring is from -10 to 130. The patient was an immunological and virological partial responder.
- Patient 2: The PR gene sequence was not obtained. The resistance mutations to NRTI (L74V, Y115F, M184V) and NNRTI (Y181C) give a mutation scoring varying from -10 to 120, high-level resistance for ABC (Supplementary Table 1). A high viral load was associated with a low CD4 count.
- Patient 3: Accessory resistance mutations in PR gene was found (G73A along with several others) and comprising the highly uncommon L76 deletion. In the RT gene, resistance mutations to NRTIs (L174V, Y115F) and NNRTIs (K103N, Y181C) were noted. The mutations scoring was from -10 to 120 (Supplementary Table 1), high-level resistance for TDF, ABC and NVP. Despite a high CD4 count, the viral load was elevated. Patient 2 and patient 3 were from the same mother.
- Patient 4: No major resistance mutations were observed in the PI gene. The presence in the RT gene of low-level resistance mutations to NRTI (M184I) and intermediate one to NNRTI (Y181C) were found. The mutation scoring varied from -10 to 60 (Supplementary Table 1). This child has been lost to follow-up for 2 years. The last analyzes were made 1 year and a half after his return to follow-up.
- Patient 5: No major resistance mutation for PIs has been noted. However, resistance mutations were found solely to NNRTI (K103N, V106I, H221Y, F227L, Y181C) and none to NRTI. In this patient, mutation scoring varied from 25 to 115 (Supple-mentary Table 1), resistance exclusively to NNRTI. The viral load was remarkably high and the CD4 count low.
- Patient 6: The sequencing of the PI gene was unsuccessful. Resistance mutations to NRTI (M184V) and to NNRTI (K103N, V108I) were observed with a scoring of -10 to 75. The viral load was moderately elevated while the CD4 count result was satisfactory.
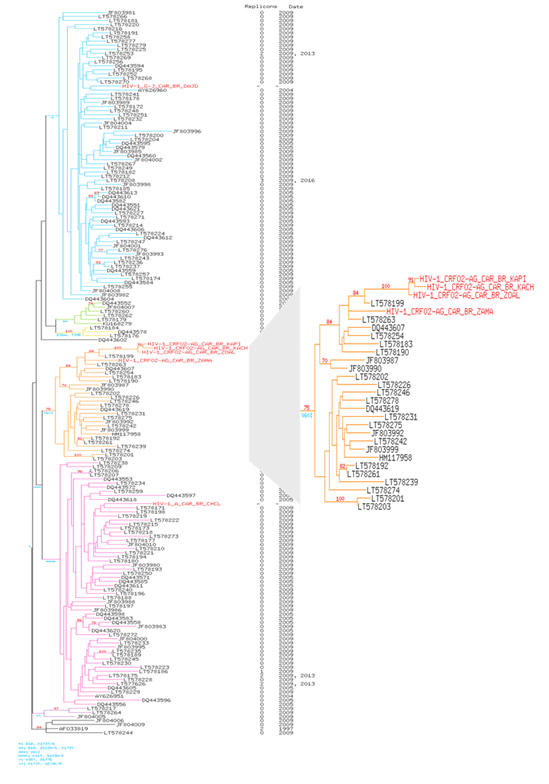
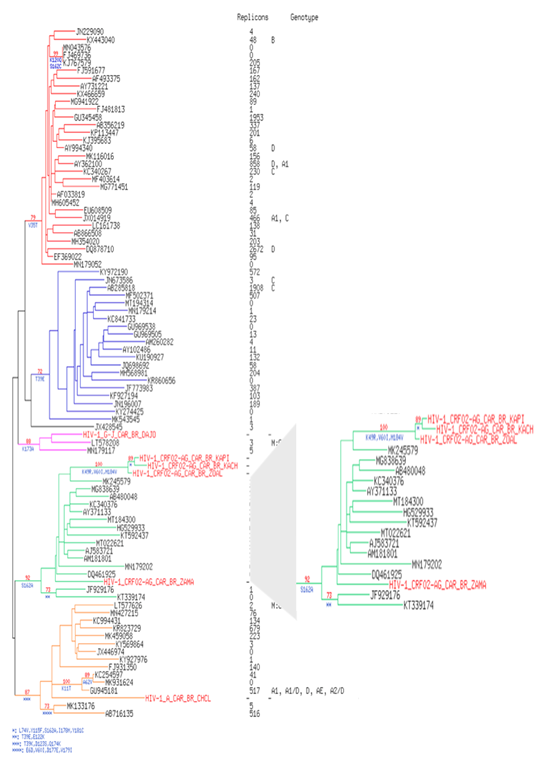
The 6 patients' strains harbored the M184V mutation (corresponding to a hyper susceptibility to Zidovudine/Azidothymidine (ZDV or AZT). This mutation has been shown to make HIV susceptible to AZT(ZDV) by reduction of HIV-1 replication (13,14) which is the case of our patients (Figure 1a/b). Two accessory RT resistance mutations are also present in the 6 strains (Q174E/K, V35I/K/T). The phylogenic comparison of RT sequences shows that HIV-1 A (patient 1), G-J (patient 5) and 1 of the CRF02_AG strains (patient 4) were found into their respective genotype. On the contrary, 3 CRF02_AG strains (patients 2, 3 and 6), are isolated in the corresponding genotype with a branch longer than the others and a boostrap at 90, testifying the robustness of the relationship between these three strains when compared to Central African strains (Figure 2). The results were identical when compared to strains of different origin (Supplementary Figure 1).
NA: not applicable; Immunization/Vaccination program: BCG, VPO, DTC-HepB-Hib1, VPO/VPI1, PCV13, Rota, VAM/MM, VAA; 1st line sub: 1st line substitution (cf. supplementary Table 2), CTM : Cotrimoxazole ; ARI : Acute respiratory Infections ; TBC: Tuberculosis ; MoH : Ministry of Health.
Table 1: Demographic, clinical, immunovirological, and Drug cART characteristics of HIV-1 Exposed Infected (HEI) children.
Figure 1a: Heatmap of mutations analyzis to pharmacoresistance sequencing. PI: Protease Inhibitor, NRTI: Nucleoside Reverse Transcriptase Inhibitors, NNRTI: Non-nucleoside Reverse Transcriptase Inhibitors, ritonavir: /r, Atazanavir/r (ATV/r), Darunavir/r (DRV/r), Lopinavir/r (LPV/r), Abacavir (ABC), Zidovudine (AZT), Emtricitabine (FTC), Lamivudine (3TC), Tenofovir (TDF), Doravirine (DOR), Efavirenz (EFV), Etravirine (ETR), Nevirapine (NVP), Rilpivirine (RPV).
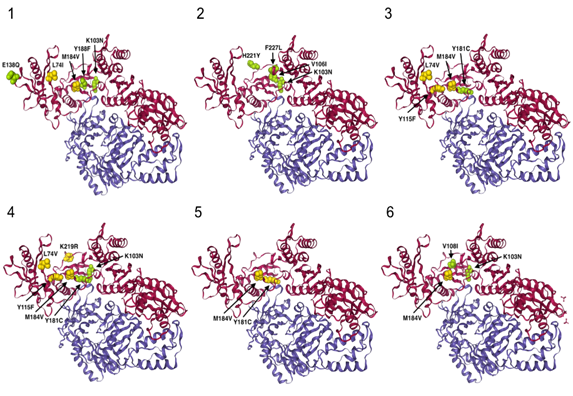
Figure 1b: Crystal structure (cartoon style 3D representation) of the HIV reverse transcriptase (PDB id: 3LAK) for the 6 patients. A (Patient 1), B (Patient 5), C (Patient 2), D (Patient 3), E (Patient 4) and F (Patient 6). Chain A and B are colored in magenta and blue, respectively. Only NRTI resistance and NNRTI resistance mutations are shown, in yellow and green, respectively. Here we show the location of the main resistance mutations for each patient on their HIV-1 RT 3D representation.
Figure 2: Phylogenetic tree of HIV-1 RT pol gene of our 6 HIV-1 isolates of present study in 300 Central African HIV-1 reference strains of clade A, B, C, D and CRF submitting in GenBank representing the various genetic subtypes. The viral strains from our study are named in Red Color. 585 conserved sites were used to perform the phylogenetic reconstruction by maximum likelihood and with 1000 bootstrap replicates. For each leaf, accession number of the sequence, number of replicons and collection date were indicated. Bootstrap score over 70% are shown in red with the characteristic amino acid mutations in blue. Six clusters are highlighted in color: in blue for the cluster I, in green for the cluster II, in yellow for the cluster III, in orange for the cluster IV, in pink for the cluster V and in purple for cluster VI. Patients are highlighted in red. Tree is rooted with the reference HIV sequence AF033819’s cluster.
4. Discussion
This is the first study performed in a rural area of CAR. Its interest resides in the fact that it concerns people suffering hardship in their everyday life because of the recurrent troubles which plague the country. No children were administered PI and two of them (patients 4 and 5) had no major PI resistance mutations while strains of patients 1 and 3 harbors resistance mutation: V32S is highly unusual while G73S/T/C/A are non-polymorphic accessory PI-selected mutations. Both are associated primarily with reduced susceptibility to ATV [11, 15]. L76V, a non-polymorphic mutation selected by LPV and DRV, reduces susceptibility to the three PIs. The highly unusual L76 deletion was found in patient 3 but his strain remains susceptible to LPV [16]. However, the strains of the two former patients remained susceptible or hypersusceptible to ATV, and LPV (Figure 1a). Similar results have been reported in CAR [17-20] and other African countries such as Cameroon, Democratic Republic of Congo (DRC) and Congo Republic [21, 22]. The mutations described in these countries are identical to those found in Bouar and 96% of these strains were susceptible to PI [21].
The treatment combining NRTI and NNRTI failed to render undetectable the viral genome but ensured a satisfactory CD4 count in patients 1, 3 and 6. Patient 3 despite a high CD4 count, had absolutely no control of viral replication. The three remaining patients have both high viral load and low CD4 counts. Considering the NRTI, five patients harbored the M184V/I resistance mutation of the RT gene (Figure 3a/b). The 19 months infant (patient 5) alone had no major resistance to NRTI which may be due to his young age and a treatment for only one year. However, the virus remained resistant to NNRTI and the administration of two NRTI (ABC/3TC) did not prevent an extremely high viral load. This may be related to an observance problem which favored resistance outcome. The mutations L74V/I (patients 1, 2 and 3) and Y115F (patients 2 and 3) were associated with NRTI resistance. The patient 5 was susceptible to the 6 NRTI (Figure 1a). All the six viral strains have retained susceptibility to TDF which may explain the partial efficacy of the treatments of patient 4 and 6. HIV of five patients have developed high level resistance to ABC, FTC, 3TC varying from 60 to 130 on the scoring mutation. The Y181C mutation causes intermediate resistance to ABC and low-level resistance to TDF. The number of resistance mutations to NNRTIs is elevated. Accessory mutations are mainly compensatory which have alone little effect on drug resistance. However, their accumulation can increase the level of pheno-typic resistance [23]. Major mutations associated with resistance are K103N (patients 1, 3, 5, 6) and Y181C (patients 2, 3, 4) the latter being the only one present in patients 2 and 4. Patient 6 have the K103N, associated to the V106I. Y181C is a non-polymor-phic mutation associated with a reduced response to an EFV-containing regimen in NNRTI-experienced patients [15, 24]. K103N is a non-polymorphic mutation that causes high-level resistance to NVP and EFV. Mutation V106I (patient 5) occurs in 1% to 2% of viruses from untreated persons [25], (https://hi vdb.stanford.edu/dr-summary/resistance-notes/ NNRTI/). It is associated with a reduced NNRTI susceptibility in association with other mutations. H221Y is a non-polymorphic accessory mutation selected primarily by NVP. It frequently occurs in combination with Y181C (but it is found isolated in patients 5). In the same patient, F227L is a non-polymorphic mutation which in most cases is associated to V106A. The former is selected in vivo and in vitro by NVP and is associated with high-level resistance to NVP and intermediate activity of EFV. The major V106I mutation was present in patient 5 and 6. NVP has been and remains the frequent first molecule administered even if it is no longer used for prophylaxis of vertical maternal-fetal transmission. It is commonly part of first line treatment and as in patients 1, 2, 3 and 5. Resistance to this drug was observed in patient 6 and an intermediate activity in patient 4. There is structural difference between EFV (given to patients 4 and 6) and the other NNRTIs which explains the differences of susceptibility between this drug and the other NNRTI. The previous studies on HIV and resistance to ARVs in children which were held in CAR were limited to Bangui. They confirm the resistance to the different classes of ARVs which has grown over time since in different social groups [18, 26-28]. HIV-1 infected children born to HIV-infected mothers living in rural areas have an underestimated prevalence of clinical, virologic and immunologic failures. It is interesting to note that some mutations correspond to resistance to drugs unavailable in CAR. The IP Darunavir (DRV) remains susceptible which is not surprising as IP are not prescribed to the 6 children. On the contrary, intermediate resistances are present for Doravirine (DOR) in patients 1 and 5, Rilpivirine (RPV) in patients 1 to 5) and Etravirine (ETR) in patients 2 and 4, three NNRTI not available yet in the country [29]. For example, the rare mutation F227I/V which is said to be selected in vitro by DOR is present in patient 5 with a corresponding low-level resistance (figure 1a). The presence of numerous resistance mutations of the PI (mean 8) et RT genes (mean of 2 for NRTI, and NNRTI but 16 for accessory mutations) is surprising considering the young age of patients and sometimes the short time since they were treated. Some of these mutations are likely due to the transmission of virus by mother at birth but for some an occurrence by chance is also a possibility. In the absence of maternal results, one cannot conclude in favor of one or the other hypothesis. For example, patients 2 and 3 are from the same mother and were administered the same cART. They both harboured the L74F, Y115F, M184V, Y181C mutations which may come from their mother. However, the patient 3 has an additional one (K103N) who may have been selected during his treatment despite his younger age. Nonetheless, these two children respond differently to treatment. While the viral load is high for both, patient 3 have a far higher CD4 count maybe, as being younger, its treatment duration was shorter, and he has retained a good immunity. Considering the patient 5, the viral strain was susceptible to all NRTI. Because he is still an infant, the mutations responsible for resistance to NNRTI are likely of maternal origin. The lowest number of mutations of the RT in this patient may be related to his young age. Considering that an undetectability of the viral load was never achieved, all these patients are prone to develop mutations of the RT genes (15 to 23 were observed). Studies carried out in Bangui in HIV+ children at the Bangui Pediatric University Hospital Center found 60% of non-responders among the cohort of pediatric patients. The main mutations of the RT found were M184V and K103N [20, 30, 31]. The 6 patients have presented opportunistic infections which is related to the absence of control of the viral replication (Table 1/Figure 3a/b). Besides the maternal transmission, and a limited efficacy of the administered ART, local factors may contribute to the poor evolution of the virological and immunological results of these young patients. The prescription was adapted to age, weight, and body surface. Parents were given treatment for one month with the recommendations for their administration. However, the drug supply, particularly of pediatric formulations may be interrupted and solely a limited number of ART is available. Moreover, they are not always available, the limited quantity of syrup formulation renders the administration of ART difficult in young infant. The viral load is not systematically performed at each visit because of the limited resources. The follow-up consists of quarterly visits with a clinical exami-nation, hematological, chemical, and immunological tests and finally the distribution of ARTs. Once the treatment is initiated and if the evolution is satisfactory, visits are programmed every three months. The households are visited weekly by community staff. Self-report is not a good predictor of adherence, and the actual level of non-adherence is certainly higher than the one reported. Adherence to therapy plays a central role in the development of resistance mutations. Compliance must be greater than 95% to obtain an adequate virological response [21, 32]. Moreover, in places lacking electricity, a good conservation of the drug is not ascertained and in a hot climate they may lose their efficacy with time [32, 33]. The recurring politico-military crises which have repercussions on the supply of ART. Nonetheless the Center can continuously deliver these molecules to its planned reserve stock if deliveries are insured. The insecurity may prevent the attendance to visits, with, consequently, an interruption of treatment. All these factors are in favor of a discontinuity of drug administration and thus, of the emergence of resistances. The frequency of drug administration may also be a factor in the development of resistance mutations [34]. Some studies have shown the development of mutations at different times, without correlation with frequency 60 to 80%. Additionally, the health center cannot afford the sequencing of the viral genome. Thus, the actual resistance of the virus to ART drugs is not determined and the treatment cannot be adapted adequately, and a mere switch of cART does not warrant an efficacy. The insecurities also favor the migration of populations seeking stability on the other side of the borders. This may explain the presence in three patients of the CRFO2_AG genotype, constituting a single cluster, totally separate from all the CRF02_AG strains in CAR and the world and probably of foreign origin or the emergence of sub-CRFs with specific mutations. In Bangui, strains consist mainly of CRF (especially CRF11) and BA and rarely CRF02_AG [26, 28].
5. Conclusions
Children in Bouar, a rural area, show high level resistance to RT and susceptibility to PI and emergence of CRF_AG variant. It is striking that all these young patients were in treatment failure although at different levels. In this difficult environment, the success of cART is not warranted. More complete survey must be done to evaluate the rate of success of ART administration in this population to make a good comparison with what occurs in Bangui urban environment. Thus, some of the difficulties and low success of cART described in Bouar must exist in Bangui [28]. Making the accessibility to viral load determination for all patients would ameliorate their follow-up with better budget. The viral RNA sequencing would permit a better prescription of ARV. Additionally, the introduction of more recent ART would permit to overcome the observed dramatic emergence of resistances. The more recent PI, NRTI and NNRTI may have already a reduced activity (e.g. DOR, ETR, RPV), due to the accumulation of mutations, thus drugs of other classes (e.g. integrase inhibitors) would be helpful for controlling the infection.
Ethics Considerations and Patient Consent Statement
The expert committee of the HIV/AIDS national control program of the CAR Ministry of Health of the approved this program of resistance surveillance to cART (Arrêté n°0277/MSPP/CAB/DGSPP/DMPM/ SMEE du 05 août 2002) as part of the surveillance of communicable diseases requiring compulsory cover-age. Our retrospective study does not require patient consent to participate.
Consent for Publication
Not applicable
Availability of Data and Materials
The data and materials used are available on request. The sequences of the strains are already in GenBank.
Competing Interests
Any conflicts of interests for all authors should be declared
Funding
This work was supported by personal authors contributions.
No funding.
Not applicable.
Authors’ Contributions
UV acquired the data and wrote the draft paper. GG, EB AS and UV conceived and designed the experiments. SM and EB performed the experiments and analyzed the data with AS, GG and UV. NC helped to draft. GG, EB, ALF, NC and UV reviewed the paper. All authors have read and approved the final manuscript.
Acknowledgments
The authors would like to express their sincere thanks to all the Participants; Healthcare workers of HIV/STI Centre St Michel of Bouar (CAR) who helped of data collection with patient recruitment and performing health assessments. The authors thank Julien Gardès (Biomanda, Nice in France) for the expert biotechnical assistance in phylogeny.
Author Information
1Infectious and tropical diseases Unit, Amitie Hospital, Bangui CAR.
2Laboratory of Immunity, Department of systemics medicine, Faculty of medicine and surgery, Tor Vergata University, Rome, Italy.
3Clinic of Infectious Diseases, IRCCS San Raffaele Scientific Institute, Milan, Italy.
4Academic Department of Pediatrics, Research Unit of Congenital and Perinatal Infection, Children's Hospital Bambino Gesù, Rome, Italy.
5Molecular biology Unity, Clinical biology and Public health national Laboratory, Bangui, CAR,
6Laboratory of viral hepatitis, Institut Pasteur of Bangui, CAR.
7Saint Michel Center, Society of Priests of the Sacred Heart of Betharram, Health Pastoral Diocese of Bouar, Bouar, CAR.
8Laboratory of Microbiology, San Raffaele Scientific Institute, Milan, Italy.
9EA 3452 CITHEFOR, Faculty of Pharmacy Brabois Santé, Vandoeuvre-lès-Nancy, France.
ALF is Emeritus Professor in faculty of pharmacy Brabois Santé and faculty of maieutical medicine and health professions of Nancy university in France.
References
- AIDS Info. Country factsheets Central African Republic. UNAIDS (2019).
- Diemer SCH, Ngbale RN, Longo JDD, et al. Risk factors for transmission of HIV from mother to child in Bangui. Med Sante Trop 27 (2017): 195-199.
- Diemer S-CH, Longo J de D, Tekpa G, et al. Factors associated with non-disclosure of HIV status among women attending prenatal clinics in Bangui. Sante Publique 30 (2018): 397-403.
- Gamba EP, Nambei WS, Kamandji L. Integrated screening for HIV, syphilis, and toxoplasmosis among pregnant women in the Central African Republic. Med Sante Trop 23 (2013): 421-426.
- Bélec L, Gresenguet G, Georges-Courbot MC, et al. Sero-epidemiologic study of several sexually transmitted diseases (including HIV infection) in a rural zone of the Central African Republic. Bull Soc Pathol Exot Filiales 81 (1988): 692-698.
- Matsika-Claquin, Massanga M, Ménard D, et al. HIV epidemic in Central African Republic: high prevalence rates in both rural and urban areas. J Med Virol 72 (2004): 358-362.
- Punzi G, Saracino A, Brindicci G, et al. HIV infection and protease genetic diversity in a rural area of the Southern Central African Republic. J Med Virol 77 (2005): 457-459.
- Mossoro-Kpinde CD, Gody J-C, Mboumba Bouassa R-S, et al. High levels of virological failure with major genotypic resistance mutations in HIV-1-infected children after 5 years of care according to WHO-recomm-ended 1st-line and 2nd-line antiretroviral regimens in the Central African Republic: A cross-sectional study. Medicine (Baltimore) 96 (2017): e6282.
- Edessa D, Sisay M, Asefa F. Second-line HIV treatment failure in sub-Saharan Africa: A systematic review and meta-analysis. PLoS One 14 (2019): e0220159.
- Fokam J, Santoro MM, Takou D, et al. Evaluation of treatment response, drug resis-tance and HIV-1 variability among adoles-cents on first- and second-line antiretroviral therapy: a study protocol for a prospective observational study in the centre region of Cameroon (EDCTP READY-study). BMC Pediatr 19 (2019): 226.
- Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs 72 (2012): e1-e25.
- Nijhuis M, Deeks S, Boucher C. Implications of antiretroviral resistance on viral fitness. Curr Opin Infect Dis 14 (2001): 23-28.
- Zhang YM, Imamichi H, Imamichi T, et al. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol 71 (1997): 6662-6670.
- Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4 (2009): e4724.
- Wensing AM, Calvez V, Ceccherini-Silberstein F, et al. 2019 update of the drug resistance mutations in HIV-1. Top Antivir Med 27 (2019): 111-121.
- Wong-Sam A, Wang Y-F, Zhang Y, et al. Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease. ACS Omega 3 (2018): 12132-12140.
- Moussa S, Pinson P, Pelembi P, et al. First data on HIV-1 resistance mutations to anti-retroviral drugs in Central African Republic. AIDS Res Hum Retroviruses 26 (2010): 1247-1248.
- Charpentier C, Gody J-C, Tisserand P, et al. Surveillance of antiretroviral drug resistance mutations in untreated young children living in the Central African Republic. Antivir Ther 16 (2011): 1347-1350.
- Charpentier C, Gody J-C, Mbitikon O, et al. Virological response and resistance profiles after 18 to 30 months of first- or second-/third-line antiretroviral treatment: a cross-sectional evaluation in HIV type 1-infected children living in the Central African Republic. AIDS Res Hum Retroviruses 28 (2012): 87-94.
- Mboumba Bouassa R-S, Mossoro-Kpinde CD, Gody J-C, et al. High predictive efficacy of integrase strand transfer inhibitors in perinatally HIV-1-infected African children in therapeutic failure of first- and second-line antiretroviral drug regimens recommended by the WHO. J Antimicrob Chemother 74 (2019): 2030-2038.
- Aghokeng AF, Monleau M, Eymard-Duvernay S, et al. Extraordinary hetero-geneity of virological outcomes in patients receiving highly antiretroviral therapy and monitored with the World Health Organi-zation public health approach in sub-saharan Africa and southeast Asia. Clin Infect Dis 58 (2014): 99-109.
- Fokam J, Sosso SM, Yagai B, et al. Viral suppression in adults, adolescents and children receiving antiretroviral therapy in Cameroon: adolescents at high risk of virological failure in the era of ‘test and treat’. AIDS Res Ther 16 (2019): 36.
- Rhee S-Y, Varghese V, Holmes SP, et al. Mutational Correlates of Virological Failure in Individuals Receiving a WHO-Recomm-ended Tenofovir-Containing First-Line Regimen: An International Collaboration. EBioMedicine 18 (2017): 225-235.
- Gatanaga H, Ode H, Hachiya A, et al. Combination of V106I and V179D poly-morphic mutations in human immuno-deficiency virus type 1 reverse transcriptase confers resistance to efavirenz and nevirapine but not etravirine. Antimicrob Agents Chemother 54 (2010): 1596-1602.
- F R, D S, Jl L, et al. HIV-I and HIV-II double infection in Central African Republic. Lancet (London, England) (1986).
- Rs MB, H P, Cd M-K, et al. Purifying Selection in Human Immunodeficiency Virus-1 pol Gene in Perinatally Human Immunodeficiency Virus-1-Infected Children Harboring Discordant Immunological Response and Virological Nonresponse to Long-Term Antiretroviral Therapy. Journal of clinical medicine research (2020).
- Charpentier C, Gody J-C, Tisserand P, et al. Usefulness of a genotypic resistance test using dried blood spot specimens in African HIV-infected children with virological failure according to the 2010-revised WHO criteria. Arch Virol 156 (2011): 1603-1606.
- Mossoro-Kpinde CD, Gody J-C, Mboumba Bouassa R-S, et al. Escalating and sustained immunovirological dissociation among antiretroviral drug-experienced perinatally human immunodeficiency virus-1-infected children and adolescents living in the Central African Republic: A STROBE-compliant study. Medicine (Baltimore) 99 (2020): e19978.
- Diouara AAM, Ndiaye HD, Guindo I, et al. Antiretroviral treatment outcome in HIV-1-infected patients routinely followed up in capital cities and remote areas of Senegal, Mali and Guinea-Conakry. J Int AIDS Soc 17 (2014): 19315.
- Rajesh L, Ramesh K, Hanna LE, et al. Emergence of drug resistant mutations after single dose nevirapine exposure in HIV-1 infected pregnant women in south India. Indian J Med Res 132 (2010): 509-512.
- Asahchop EL, Wainberg MA, Oliveira M, et al. Distinct resistance patterns to etravirine and rilpivirine in viruses containing nonnucleoside reverse transcriptase inhibitor mutations at baseline. AIDS 27 (2013): 879-887.
- Tebit DM, Ganame J, Sathiandee K, et al. Diversity of HIV in rural Burkina Faso. J Acquir Immune Defic Syndr 43 (2006): 144-152.
- Hirsch MS, Günthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Top HIV Med 16 (2008): 266-285.
- Boullé C, Guichet E, Kouanfack C, et al. Virologic Failure and Human Immunodeficiency Virus Drug Resistance in Rural Cameroon With Regard to the UNAIDS 90-90-90 Treatment Targets. Open Forum Infect Dis 3 (2016): ofw233.
Supplementary
|
Drugs Resistance |
HIV-Exposed Infected Children ID (DRI, MS) |
||||||
|
Class |
Generic name |
Patient 1 |
Patient 2 |
Patient 3 |
Patient 4 |
Patient 5 |
Patient 6 |
|
PI |
Atazanavir/r (ATV/r) |
S, 0 |
NA |
PLLR, 10 |
S, 0 |
S, 0 |
NA |
|
Darunavir/r (DRV/r) |
S, 0 |
NA |
S, 0 |
S, 0 |
S, 0 |
NA |
|
|
Lopinavir/r (LPV/r) |
S, 0 |
NA |
S, 5 |
S, 0 |
S, 0 |
NA |
|
|
NRTI |
Abacavir (ABC) |
HLR, 60 |
HLR, 120 |
HLR, 120 |
LLR, 15 |
S, 0 |
LLR, 15 |
|
Zidovudine (AZT) |
S, -10 |
S, -10 |
S, -10 |
S, -10 |
S, 0 |
S, -10 |
|
|
Emtricitabine (FTC) |
HLR, 60 |
HLR, 60 |
HLR, 60 |
HLR, 60 |
S, 0 |
HLR, 60 |
|
|
Lamivudine (3TC) |
HLR, 60 |
HLR, 60 |
HLR, 60 |
HLR, 60 |
S, 0 |
HLR, 60 |
|
|
Tenofovir (TDF) |
S, 5 |
S, 5 |
S, 5 |
S, -10 |
S, 0 |
S, -10 |
|
|
NNRTI |
Doravirine (DOR) |
IR, 30 |
PLLR, 10 |
LLR, 20 |
PLLR, 10 |
IR, 55 |
LLR, 15 |
|
Efavirenz (EFV) |
HLR, 130 |
IR, 30 |
HLR, 90 |
IR, 30 |
HLR, 85 |
HLR, 70 |
|
|
Etravirine (ETR) |
PLLR, 10 |
IR, 30 |
IR, 30 |
IR, 30 |
LLR, 20 |
S, 0 |
|
|
Nevirapine (NVP) |
HLR, 130 |
HLR, 60 |
HLR, 120 |
HLR, 60 |
HLR, 115 |
HLR, 75 |
|
|
Rilpivirine (RPV) |
IR, 45 |
IR, 45 |
IR, 45 |
IR, 45 |
LLR, 25 |
S, 0 |
|
DRI: Drug Resistance Interpretation, MS: Mutation Scoring, PI: Protease Inhibitor, NRTI: Nucleoside Reverse Transcriptase Inhibitors, NNRTI: Non-nucleoside Reverse Transcriptase Inhibitors. NA: Not applicable, HLR: High-Level Resistance, PLLR: Potential low-level resistance, LLR: Low-level resistance, IR: Intermediate resistance, S: Susceptible
Supplementary Table 1: Analysis of correlation of mutations to pharmacoresistance sequencing.
|
The ARV available for pediatric HIV in CAR are ABC, 3TC, Duovir-N (3TC/ZDV)-NVP), Kaletra (LPV/r) junior, EFV, NVP, ATV, Truvada (Emtricitabine/Tenofovir = FTC/TDF) and Ritonavir (r) |
|
|
- 1st line: |
Atripla (TDF + FTC + EFV) |
|
Duovir (AZT + 3TC) + EFV or NVP |
|
|
Abacavir + 3TC + EFV or NVP |
|
|
· Substitution 1st line 1 or Substitution 1st line 2 |
|
|
- 2nd line: |
Kaletra (LPV/r) + Abacavir (ABC) + 3TC (Lamivudine) |
|
Kaletra (LPV/r) + Truvada (TDF+FTC) |
|
|
Kaletra (LPV/r) + Duovir (AZT+3TC) |
|
|
Kaletra (LPV/r) + Atazanavir (ATV) |
|
|
· Substitution 2nd line 1 or Substitution 2nd line 2: |
|
|
- 3rd line: |
Introduct this year: Dolutegravir + Tenofovir + Lamivudine |
Supplementary Table 2: ART used in CAR National Program Against Aids.
The viral strains from our study are in Red Colour. 585 conserved sites were used to perform the phylogenetic reconstruction by maximum likelihood and with 1 000 bootstrap replicates. For each leaf, accession number of the sequence, number of replicated sequences, the serotype and the country location were indicated. Bootstrap score over 70% are shown in red with the characteristic amino acid mutations in blue. Five robust clusters are highlighted in color: in red for the cluster I: B, D and C, in blue for the cluster II: HIV-1 C, in pink for the cluster III: CRF11 and G-J, in green for the cluster IV: CRF02_AG and in orange for the cluster V: HIV-1 A.