Headway of miR200 Family as a Novel Biomarker in Head and Neck Cancers
Article Information
Shivani Kadarla1#, Rajesh Kumar Galimudi1*#, Surekha Rani Hanumanth2, Srimanth Kumar Barigela1, Durga NeeharikaRani1, Madhavi Jangala1, Pardha P Reddy1,3, Raja Meghanadh Koralla1,4
#Authors contributed equally
1MAA Research Foundation, Jubilee Hills, Hyderabad, Telangana, India
2Department of Genetics, Osmania University, Tarnaka, Hyderabad, Telangana, India
3Mahavir Hospital and Research Centre, Lakdikapul, Hyderabad, Telangana, India
4MAA ENT Hospitals, Jubilee Hills, Hyderabad, Telangana, India
*Corresponding Author: Rajesh Kumar Galimudi, Senior Scientist, Head of Research, MAA Research Foundation, Jubilee Hills, Hyderabad, Telangana, India.
Received: 23 September 2022; Accepted: 11 October 2022; Published: 18 November 2022
Citation: Shivani Kadarla, Rajesh Kumar Galimudi, Surekha Rani Hanumanth, Srimanth Kumar Barigela, Durga NeeharikaRani, Madhavi Jangala, Pardha P Reddy, Raja Meghanadh Koralla. Headway of miR200 Family as a Novel Biomarker in Head and Neck Cancers. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 392-410.
View / Download Pdf Share at FacebookAbstract
Head and neck squamous cell carcinoma (HNSCC) a varied disease, comprising variety of tumors that grow on the mucosa of the aerodigestive tract and arise in the regions of hypopharynx, oropharynx, salivary gland, oral cavity, nasopharynx, or larynx. About 650,000 new cases of HNSCC are diagnosed annually all over the world, with a ratio comprising of 3:1 between men and women. The role of microRNAs (miRNAs) in cancer development was known to be as oncogene or tumor suppressor. One of the miRNA family, miR-200 was mainly characterized as a tumor suppressor in head and neck cancers, it is composed of five highly conserved miRNAs, including miR-141, miR-200a/200b/200c, and miR- 429. Alterations observed in miR200 family genes concerning HNSCC can be well explained by epigenetic modifications like miRNA methylation and expression, In-silico analysis to facilitate the development of targeted therapies, leading to improved patient outcomes.
Keywords
Cancer Hallmarks; Epigenetics; Head and Neck Cancer; Micro RNAs; miR200 Family
Cancer Hallmarks articles; Epigenetics articles; Head and Neck Cancer articles; Micro RNAs articles; miR200 Family articles
Cancer Hallmarks articles Cancer Hallmarks Research articles Cancer Hallmarks review articles Cancer Hallmarks PubMed articles Cancer Hallmarks PubMed Central articles Cancer Hallmarks 2023 articles Cancer Hallmarks 2024 articles Cancer Hallmarks Scopus articles Cancer Hallmarks impact factor journals Cancer Hallmarks Scopus journals Cancer Hallmarks PubMed journals Cancer Hallmarks medical journals Cancer Hallmarks free journals Cancer Hallmarks best journals Cancer Hallmarks top journals Cancer Hallmarks free medical journals Cancer Hallmarks famous journals Cancer Hallmarks Google Scholar indexed journals Epigenetics articles Epigenetics Research articles Epigenetics review articles Epigenetics PubMed articles Epigenetics PubMed Central articles Epigenetics 2023 articles Epigenetics 2024 articles Epigenetics Scopus articles Epigenetics impact factor journals Epigenetics Scopus journals Epigenetics PubMed journals Epigenetics medical journals Epigenetics free journals Epigenetics best journals Epigenetics top journals Epigenetics free medical journals Epigenetics famous journals Epigenetics Google Scholar indexed journals Head and Neck Cancer articles Head and Neck Cancer Research articles Head and Neck Cancer review articles Head and Neck Cancer PubMed articles Head and Neck Cancer PubMed Central articles Head and Neck Cancer 2023 articles Head and Neck Cancer 2024 articles Head and Neck Cancer Scopus articles Head and Neck Cancer impact factor journals Head and Neck Cancer Scopus journals Head and Neck Cancer PubMed journals Head and Neck Cancer medical journals Head and Neck Cancer free journals Head and Neck Cancer best journals Head and Neck Cancer top journals Head and Neck Cancer free medical journals Head and Neck Cancer famous journals Head and Neck Cancer Google Scholar indexed journals Micro RNAs articles Micro RNAs Research articles Micro RNAs review articles Micro RNAs PubMed articles Micro RNAs PubMed Central articles Micro RNAs 2023 articles Micro RNAs 2024 articles Micro RNAs Scopus articles Micro RNAs impact factor journals Micro RNAs Scopus journals Micro RNAs PubMed journals Micro RNAs medical journals Micro RNAs free journals Micro RNAs best journals Micro RNAs top journals Micro RNAs free medical journals Micro RNAs famous journals Micro RNAs Google Scholar indexed journals miR200 Family articles miR200 Family Research articles miR200 Family review articles miR200 Family PubMed articles miR200 Family PubMed Central articles miR200 Family 2023 articles miR200 Family 2024 articles miR200 Family Scopus articles miR200 Family impact factor journals miR200 Family Scopus journals miR200 Family PubMed journals miR200 Family medical journals miR200 Family free journals miR200 Family best journals miR200 Family top journals miR200 Family free medical journals miR200 Family famous journals miR200 Family Google Scholar indexed journals hypopharynx articles hypopharynx Research articles hypopharynx review articles hypopharynx PubMed articles hypopharynx PubMed Central articles hypopharynx 2023 articles hypopharynx 2024 articles hypopharynx Scopus articles hypopharynx impact factor journals hypopharynx Scopus journals hypopharynx PubMed journals hypopharynx medical journals hypopharynx free journals hypopharynx best journals hypopharynx top journals hypopharynx free medical journals hypopharynx famous journals hypopharynx Google Scholar indexed journals oropharynx articles oropharynx Research articles oropharynx review articles oropharynx PubMed articles oropharynx PubMed Central articles oropharynx 2023 articles oropharynx 2024 articles oropharynx Scopus articles oropharynx impact factor journals oropharynx Scopus journals oropharynx PubMed journals oropharynx medical journals oropharynx free journals oropharynx best journals oropharynx top journals oropharynx free medical journals oropharynx famous journals oropharynx Google Scholar indexed journals nasopharynx articles nasopharynx Research articles nasopharynx review articles nasopharynx PubMed articles nasopharynx PubMed Central articles nasopharynx 2023 articles nasopharynx 2024 articles nasopharynx Scopus articles nasopharynx impact factor journals nasopharynx Scopus journals nasopharynx PubMed journals nasopharynx medical journals nasopharynx free journals nasopharynx best journals nasopharynx top journals nasopharynx free medical journals nasopharynx famous journals nasopharynx Google Scholar indexed journals oncogene articles oncogene Research articles oncogene review articles oncogene PubMed articles oncogene PubMed Central articles oncogene 2023 articles oncogene 2024 articles oncogene Scopus articles oncogene impact factor journals oncogene Scopus journals oncogene PubMed journals oncogene medical journals oncogene free journals oncogene best journals oncogene top journals oncogene free medical journals oncogene famous journals oncogene Google Scholar indexed journals tumor suppressor articles tumor suppressor Research articles tumor suppressor review articles tumor suppressor PubMed articles tumor suppressor PubMed Central articles tumor suppressor 2023 articles tumor suppressor 2024 articles tumor suppressor Scopus articles tumor suppressor impact factor journals tumor suppressor Scopus journals tumor suppressor PubMed journals tumor suppressor medical journals tumor suppressor free journals tumor suppressor best journals tumor suppressor top journals tumor suppressor free medical journals tumor suppressor famous journals tumor suppressor Google Scholar indexed journals
Article Details
1. Introduction
1.1 Overview of Head and Neck Cancers
Head and neck cancer usually begins at mucosal surfaces of the aero digestive tract (hypo pharynx, oropharynx, salivary gland, oral cavity, nasopharynx, or larynx)by forming the tumors, and these tumors are popularly known as malignant tumors between "dura matter and pleura" [1,2]. Head and neck cancer is also referred to as Head and Neck Squamous cell carcinoma (HNSCC) and is the sixth most common malignancy of humans and belongs to the most aggressive form of cancer [2,3]. The maximum number (90%) of HNSCCs are squamous cell carcinomas, in which 60-70% are oral cavity, and Laryngeal squamous cell carcinoma (LSCC) and other malignancy include nasal cavity, pharynx, middle ear, thyroid gland, salivary glands [3]. The highest frequency of LSCC is seen in persons of advancing age and occurs to a large extent in males. The incidence ratio of LSCC in males and females is 5:1 [4], Global epidemiology of LSCC is reported as 210,606 cases (2.76 new cases per100,000 inhabitants), and the mortality rate is 126,471 deaths (1.66 per 100,000 inhabitants), while the prevalence of LSCC in India is 3-6% and 0.2-1% in male and females respectively, the age-adjusted incidence rate of LSCC in western, central India is between 3.5 and 5 per 100,000 inhabitants [5]. The progression of LSCC is seen in four different stages. The early-stage cancers are seen in stage I and II (vocal cords become unstable) (3.8 months), advanced stage (extra nodal extension, metastasis) is seen in stages III and IV (4.7 months) and spreading of the tumor to thyroid cartilage, prevertebral space, carotid artery, and lungs is seen the final stage. Major symptoms of LSCC are associated with a higher tendency of lymph node metastasis [4].
1.1.1 Therapeutic Strategies of HNSCC: Head and neck squamous cell carcinomas (HNSCC) start to arise in the mucosal lining of the upper-aero digestive tract. HNSCC is mostly diagnosed at advanced stages in about 70% of affected individuals. Inspite of many intensive multimodal treatment etiquettes, an overall survival rate for patients with advanced stage of cancer in the past 5 years is 50–60% and seems to have reached a plateau [6]. Wong et.al. (2016), Moscow et.al. (2018) has revealed that during the last two decades, the therapeutic strategies in treating cancer have been expeditiously changing, and treatment for various types of malignancies is progressively based on tumor genetics to decrease the toxicity and to improve the efficacy of therapeutics [7,8]. Regardless of various efforts in the treatment strategies, surgery and radiotherapy are the mainstays in treating the HNSCC based on tumor site, stage, imaging and post-operative histological findings [6]. Of all the carcinomas, approximately 30% of tumors are diagnosed in the early stages [9,10,6] and the patients with early-stage tumors are treated with radiotherapy or surgical resection in the specific site of tumor growth. DeSantis et.al.,2014 have demonstrated that a complete cure is obtained only after the treatment process, and around 90% of 5- years survival rate is seen in HNSCC patients [11]. However, more than half of the patients (70%) are in the advanced stages with regional or distant lymph node metastasis [9,10] and Concomitant chemo radiotherapy, surgical salvage and post-operative chemo radiotherapy are the treatment measures used to treat these types of advance staged cancers [12,13]. A molecular view of head and neck cancers has been interpreted in great detail; identification of druggable targets can be achieved in the absence of oncogenic mutations to improve the therapeutical outcome of HNSCC. At present, functional genomic approaches are being prospected to discover potential therapeutic targets that consider the diagnosis and targeted therapy of oral premalignant (pre-HNSCC) changes, and that may further prevent the development of the tumors. Earlier it was revealed that the DNA damage response and cell cycle regulation pathways had been significantly altered in HNSCC, resulting in replication stress, which is an approach in further exploitation as an HNSCC susceptibility for treatment. Our aim is to review the current literature on micro RNA studies in HNSCC, which may further act as potential biomarkers in the early diagnosis of disease [6].
1.2 Micro RNAs (miRs)
Micro RNAs (miRs/ miRNA) are endogenousnon-coding single-stranded RNAs, interspersed between the introns and exons of protein-coding genes with the length of 18–25 nucleotides and a minor set of miRs were mapped to long interspersed nuclear elements (LINEs). Besides, miRs have also been found in many other areas (fragile sites, regions of amplification, and LOH regions) of the genome [14,15]. Lee et al., (1993) and Wightman et al., (1993) were the first to discover the Micro RNA (miR) lin-4 gene in Caenorhabditis elegans in 1993 [16,17]. These small non-coding miRs developed during the post-transcriptional regulation by messenger ribonucleic acid (mRNA) cleavage, which hangs on the complementarity of miR-mRNA. The cleavage of mRNA occurs only when there is a perfect match, while the flawed combination results in gene repression [18]. Many studies have identified the importance of miRs in the progression of various cancers and were found to be linked with proliferation, differentiation, apoptosis, metabolism, invasion, metastasis, and drug resistance. The pathological origin of cancer is directly related to the dysregulation of miRs. Furthermore, miRs are tissue-specific. Each tumor has its own miR expression profiles [19,20].
1.2.1 Genomic Organization of miRs: Currently, 46,554 miRs are registered in the miR database for eukaryotes.2300 true human mature miRs are known, of which 1115 were annotated according to bioinformatic data obtained from miRbase V22. Micro RNAs (miRs) are named as miRs- plus number, with few notable exceptions, while the miRs with a similar sequence is differentiated by an additional letter (such as a, b, c) followed by the miR number (e.g., miRs-200a). Identical mature miR sequences may find at several genomic loci with distinct precursor sequences. In such cases, the miR genes are represented by miRs-200a-1. Roughly one-third of the miRs of humans are arranged in clusters. Usually, the given cluster is found to be a single transcriptional unit, associated with the regulation of suggested miRs in the cluster. In silico analysis identified that most miRs of the same cluster were found to have a similar sequence [21]. However, it is scarce to find a duplicate sequence of the same miRs in a given cluster. It has been confirmed that the synchronized expression of similar miRs may possibly lead to combinable diversity and synergy in the biological ramification [22].
1.2.2 miRs Action and Mechanisms: miRs suppress the targeted mRNA expression through interaction with the 3'untranslated region (3' UTR). However, the mechanism for miRs on their target sites is still controversial. Most miRs targeted sites had bulges because of mismatches while the siRNA and mRNA show perfect complementarity sequence. Previous studies have suggested that the siRNA destabilizes mRNA, whereas the miR inhibits the mRNA translation without affecting the mRNA level [22]. Even though the translational repression mechanism still holds true for many miRs, studies from yesteryears have demonstrated that the miRs were associated with decreased levels of a targeted mRNA despite of imperfect complementarity sequence between the miRs and the mRNA target [23,24]. Degradation of mRNA by miRs is distinguished from Small interfering RNA (siRNA)-mediated mRNA cleavage, which is further explained by RNA processing bodies and location for RNA decay. Usually, miRs inhibit targeted mRNAs' translation, which was then sequestered to P-bodies and subjected to degradation [25]. While in few cases mRNA level remains similar in spite of miRs inhibition, and each individual miR& mRNAs have their unique interactions.
1.2.3 miR Expression Mechanisms: Transcription of miR genes is mediated by RNA polymerase II to form primary miR (Pri-miRs) with an altered nucleotide at the 5' end and a polyadenylated tail at the 3'end.The Microprocessor Complex, formed by Di George Syndrome Critical Region 8 and Drosha ribonuclease, convert primRNAs Molecules into pre-miR. Exportin-5 then transports this pre-miR into the cytoplasm, where Dicer cleaves it to form a miR duplex. In order to form the complex that exhibits gene silencing, the duplex needs to be separated into the functional strand and loaded along with argonaute proteins into the RNA-induced silencing complex (RISC). This final complex mediates gene expression by binding target mRNAs and inducing mRNA degradation [26] (as depicted in Figure 1).
miR expression can be measured by techniques like RNase protection assay or primer extension assay. Initially, the petite size of these micro RNAs hampered the PCR-based expression assays. Later on, due to their high sensitivity, qRT-PCR-based techniques have become widely used in miR expression profiling. Further Microarray techniques have become popular as they completely assay the entire miRNAome (global miR expression profile) in tissues or cell lines. If global gene expression profiles are compared between cancers and normal tissues, many miRs are found to be deregulated. Hence, it is very likely that tumorigenesis or progression of cancer may result from alterations in the entire miRNAome rather than from modifying a single miR that regulates an oncogenic or tumour-suppressive gene [27]. Deregulation of miRs in head and neck cancers indicating its role in tumorigenesis has accumulating evidence from studies comparing miR expression profiles in tumors and corresponding normal tissues. Alterations in miR expression in the context of head & neck cancers can be attained by abnormalities in chromosomes, transcription binding factors and other epigenetic alterations. Such epigenetic repression of tumor suppressor miRs can be potentially reversed as a strategy for future epigenetic therapies. However, the lack of efficient techniques is a significant hurdle in the effective use of this strategy [28].
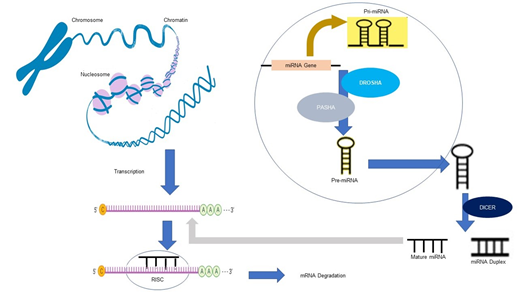
Figure 1: Involvement of miR in mRNA degradation. Formation of mature miR occurs by the Involvement of enzymes such as DROSHA (Ribonuclease), DICER (cleavage enzyme), and PASHA (a cofactor). The so formed mature miR associates with the RISC complex and binds to mature mRNA leading to mRNA degradation.
1.3 miRs as Key Regulators in Human Cancers
To understand the framework of neoplastic disease Hanahan and Weinberg, 2000 have proposed six hallmarks of cancers (sustaining proliferative signaling, evading growth suppressors, activating invasion and metastasis, enabling replicative immortality, inducing angiogenesis, resisting cell death), and they also discussed how normal cells get evolved to form neoplastic state [29]. During this evolution, the hallmarks' capabilities undergo multi-step processes to develop the traits that enable them to form malignant tumors. In addition to the above conventional hallmarks Negrini et al., (2010), Luo et al. (2009) and Colotta et al. (2009) have proposed reprogramming of energy metabolism and evading immune destruction hallmarks which also play a significant role in tumorigenesis [30-32]. Invasion and metastasis are clinically the most important hallmarks of cancers. Though the above mechanisms are studied extensively, their findings were very limited in cancer management [33]. Micro RNAs are considered vital points in the development of cancers. They are found to be linked with cancer stem biology, angiogenesis, epithelial-mesenchymal transition (EMT), metastasis and drug resistance, etc [34].
1.3.1 Evading Growth Suppressors and Sustained Proliferative Signaling: Cell proliferation is one of the most critical cancer hallmarks, and its generated abnormalities are associated with tumorigenesis. Usually, the cell cycle progression is controlled by an intra-extra cellular program signaling molecule to attain the balance between promoting cell proliferation and suppression [35]. It has been evident that some of the micro RNAs functionally incorporate into various critical cell proliferation pathways, and the dysregulation of such micro RNAs are associated with evading growth suppressors and sustaining proliferative signaling. The progression of the cell cycle depends on a variety of cyclins, cyclin-dependent kinases (CDKs) and their inhibitors which are highly regulated by miRs. Hatfield et.al. (2005) have provided the first evidence that germline stem cells of drosophila with dicer-1 knockout were blocked in the G1/S transition, suggesting that microRNAs act as crucial requirements for germline stem cells to pass the normal G1/S checkpoint [36]. It is also evident that dicer deficient germline stem cells are associated with increased expression of DACAPO (P21/P27 family of CDK inhibitors), suggesting that this protein is negatively controlled by microRNAs to promote the progression of the cell cycle. Yi et.al., (2012) demonstrated that miR-663 is upregulated in nasopharyngeal cancer, promoting the cellular transition of cells in invivo and in vitro by targeting p21CIP1 [37]. Peng et.al., (2013) have demonstrated that miR-486 is substantially downregulated in non-small-cell lung cancer, promoting cell migration and proliferation through P13K signaling pathways and insulin growth receptor (IGF) targeting IGF1, IGF1R and P85α [38].
1.3.2 Resisting Cell Death: The evasion of apoptosis is another important hallmark of cancer that is regulated by microRNAs [39,40]. Cancer cells emit a variety of scenarios in controlling apoptosis. A variety of signaling genes are involved in the apoptosis pathway where, the function of P53 is widely studied as an example in this process. The loss of P53 function against the tumor suppressor activity leads to the generation of the apoptotic pathway. Up regulation of anti-apoptotic regulators, suppression of pro-apoptotic factors and inhibition of death pathway induced by extrinsic ligands are the various regulatory pathways that play a significant role in controlling the function of apoptosis [35]. MicroRNAs regulated by P53 were found to have actively participated in P53 functions. Pichiorri et.al. (2010) described the functions of miR192, miR194, miR215 in multiple myeloma, which P53 transcriptionally activates to suppress MDM2 expression via binding to its mRNA [41]. Thereby, degradation of P53 is controlled, which ultimately restricts the development of multiple myeloma. The other novel target for P53 mediated transcriptional repression under hypoxia is the cluster of miR17-92, where the downregulation of microRNAs sensitizes the cells to undergo apoptosis, while the overexpression inhibits apoptosis. Hence, tumor cells with increased expression of miR17-92 may skip hypoxia-induced apoptosis [42].
1.3.3 Activating Invasion and Metastasis: Metastasis is a highly complex dynamic biological event. Epithelial-mesenchymal transition (EMT) is considered as a key step in the cascade of metastatic events, where it losses the cell adhesion through repression of E- Cadherin and activation of genes linked with motility and invasion. A variety of signaling pathways (Transforming growth factor-beta (TGF-β), Zinc finger E-Box (ZEB), Zinc finger protein SNAI1 (SNAIL), Twist- related protein (TWIST)) were regulated by the EMT pathway [43]. Growing shreds of evidence revealed that microRNAs are part of epithelial-mesenchymal transition and cancer metastasis. microRNAs regulated by TGF-β was associated with TGF-β signaling to induce EMT and promote metastasis in advanced malignancy. miR 155 is one among them involved in the regulation process [44].
1.3.4 Inducing Angiogenesis: Angiogenesis is an immensely interconnected process to grow new blood vessels from pre-existing ones to gratify the requirements for oxygen and food in tumor growth and metastasis [45]. As tumor tissues have considerably minimal oxygen concentration than the nearby normal tissues, hypoxia has an essential role in the tumor microenvironment by enabling the improvement and maintenance of cancer cells. Hypoxia-inducible factor (HIF) is a significant transcription factor regarding hypoxia, which impacts the expression of several genes, including miRs. Therefore, miRs that target HIF or VEGF signaling pathways probably hold a considerable impact on angiogenesis. It is now well explained that the process of angiogenesis is intricately regulated by miRs [46].
1.4 MicroRNA in the Regulation of Head and Neck Cancer
During sustained cell proliferation, altered microRNAs enhances cell proliferation by targeting the regulators of the cell cycle. miR- 137, miR- 193a participates in downregulating the expression of CDK6 and E2F6 transcription factors, respectively and results in reduced cell growth of oral squamous cell carcinoma [47]. In addition to that, miR- 125b, miR-100 also plays an essential role in suppressing the oral squamous cell region tumours. Another importantly studied hallmark of HNSCC is the evasion of apoptosis, wherein tongue squamous cell carcinoma (TSCC) inhibition of apoptosis is attributed to the overexpression of miR21 [48]. The study by Zhang et.al., (2012) suggested that microRNAs could have an association with reactive oxygen species (ROS), one such microRNA named as microRNA-21 is linked with ROS by suppressing the antioxidants (Superoxide dismutase family) activity which limits the production of TNFα and ultimately participates in tumorigenesis [49]. The upregulated miR- 24 in OSCC targets RNA binding protein dead end-1 (DND1), which directly contributes to the apoptosis resistance and proliferation by downregulating the cyclin-dependent Kinase inhibitor 1B (CDKN1B) [34,50]. Human telomerase reverse transcriptase (hTERT) is expressed in the early step of oral carcinogenesis, which maintains the length of telomerase and provides an escape from replicative senescence. Studies have found that miR-31 gets activated in hypoxia conditions and targets FIH (factor inhibiting HIF) gene. Therefore, during the state of hypoxia, telomerase gets activated by the transcriptional function of the hTERT enzyme, which is further regulated by a transcription factor called HIF-1α [51]. In vitro studies by Hung et.al. (2014) have demonstrated that the introduction of miR-31 along with hTERT causes the immortalization of oral keratinocytes [52]. In OSCC, it was reported that miR 124 inhibits metastasis by regulating the expression of ITGB1, which indicates a possible interrelation between miR 124 and hTERT. Another tumor suppressor miR called miR-512-5p suppresses tumor growth in HNSCC by targeting hTERT. Hypoxia-inducible factor (HIF) regulates a number of genes associated with tumor angiogenesis and metastasis [53]. C.-J. Liu et.al., (2010) Xiao et.al., (2012) and Hung et.al. (2014) demonstrated that the oncogenic miR31 along with its passenger strand (miR-31*) have been found to be upregulated in OSCC and oral leukoplakia (OLP) [52,54,55]. Differentially overexpressed miR 31 directly represses the expression of HIF in HNSCC. At the same time, it activates the HIF pathway in normal growth conditions. While the elevated expression of miR 31 is linked with vascular endothelial growth factor up regulation and downregulation of E- cadherin in oral premalignant disease [54]. In HNSCC patients, the spread of neoplastic cells to loco-regional lymph nodes is found to be very high. Routine clinical check-up has not provided the details regarding the advancement of tumor stages. Therefore, miRs are considered as a novel potential prognostic marker in classifying the various stages of tumor. One such miR called miR 205 is regarded as a marker to identify lymph node metastasis [56]. The elevated levels of miR -10b, reduced levels of miR- 138 and miR- 222 were found to be associated with metastasis of tongue squamous cell carcinoma (TSCC) [57]. Similarly, miR-29a is downregulated in OSCC tissues and inhibits the expression of MMP2 by its direct binding to the 3′-UTR regions. Active studies unveiled that transfection with miR-29a mimics increased apoptosis rate, attenuated invasive potential and increased chemosensitivity towards cis-platinum (CDDP) in OSCC cell lines [58]. Metabolic reprogramming of tumor cells is an essential step for tumorigenesis and metastasis, which is finely regulated by miRs [59]. Xu et.al., (2016) reported that decreased expression of miR 340 in OSCC is induced by the metabolic switch. The downregulated miR 340 was associated with increased glucose transporter -1 (GLUT-1) expression, which subsequently increases the lactate secretion and rate of glucose uptake [60]. Therefore, altered metabolism induced by miR 340 results in the rapid proliferation of oral cancer cells by regulating the glycolysis period. Further up regulation of miR 23a controls expression of SMAD- 4. Hence, the translocation of GLUT-4 indirectly regulates the glucose transport period [61]. Though sufficient oxygen is present malignant tumors acquire a high glycolytic effect (Warburg effect). To attain this, cancer cell dysregulates the inevitable glucose metabolism steps. Peschiaroli et.al., (2013) demonstrated that the expression of miR143 is inversely correlated with hexokinase-2 and the upregulated HK-2 promotes the metabolic shift to attain aerobic glycolysis in OSCC [62].
1.5 Role of miR200 Family in Cancers
miR200A, 200B, 200c, 141, 429 are 20- to 22-nucleotide noncoding RNAs which belongs to the miR200 family, which can be divided into 2 subfamilies based on a 1-nucleotide difference in seed sequences. miR200A and miR141have a seed sequence of AACACUG, whereas miR200B, miR200C and miR429 have a seed sequence of AAUACUG. Cytogenetic location of miR200A, miR200B, and miR429 are mapped to 1p36.33 which are clustered on chromosome 1 with the genomic sequences as 1:1,167,862-1,167,951; 1:1,167,103-1,167,197 and 1:1,169,004-1,169,086 respectively, whereas Cytogenetic location of miR200C and miR141 are mapped to 12p13.31 which are clustered on chromosome 12 with the genomic sequences as 12:6,963,698-6,963,765 and 12:6,964,096-6,964,190 respectively. These micro RNAs inhibit gene expression at the post transcriptional level by binding to complementary sequences in the 3-prime UTRs of target mRNAs. Expression of these miRs showed a positive correlation with expression of E-cadherin, an epithelial cell marker and a negative correlation with expression of vimentin, a mesenchymal cell marker. Aggressiveness in human cell lines is reduced due to the induction of mesenchymal-to-epithelial transition (MET) by increasing the levels of miR200A and miR200C.Conversely, induction of epithelial-to-mesenchymal transition (EMT) is observed by the reduction in the levels of miR200 [63] (as depicted in Figure 2).
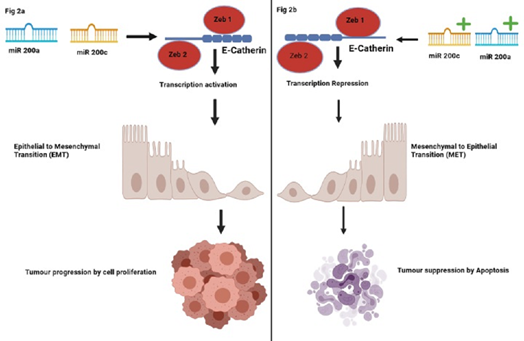
Figure 2: Correlation of miR200 family with EMT/MET pathways. Figure 2a depicts the positive Correlation of miR expression with E-cadherin (epithelial cell marker); low levels of these miR200 family genes induce epithelial to mesenchymal transition (EMT). Figure 2b depicts the Correlation of miR200 family genes with E- cadherin expression; high levels of miR200A and miR200C induces mesenchymal-to-epithelial transition (MET) in human cancer cell lines, reducing their aggressiveness.
Microarray analysis revealed a negative Correlation between miR200A expression and genes involved in MET, while it is shown a positive Correlation with oxidative stress genes.miR200c is one of the five members of the microRNA-200 family that regulates the epithelial-mesenchymal transition (EMT) by targeting EMT-related gene expression exhibiting tumour-suppressive properties [63]. miR200B and miR429 have their role in various physiological mechanisms like mediating myometrial transition to a contractile phenotype. miR200b and miR429 target the ZEB1 gene in the regulation of mammalian reproduction. Thus, the hypothalamus-pituitary-ovarian axis requires miR200b and miR429 to support ovulation.
1.6 MicroRNA200 Family in the Regulation of Head and Neck Cancer
Predominantly, miRs play a significant role in the normal and pathological processes, including cancer; they could stimulate the genes post-transcriptionally, which further participates in the development of tumor [64]. In various tumors, the levels of miR-141 expression are predominantly lesser than the healthy tissue, and its enhanced expression can prevent tumor progression. For instance, miR-141 has been shown to inhibit the growth of colorectal, liver, gastric and prostate cancers by restraining cell proliferation, metastasis and promoting cell apoptosis [65–67]. Liet.al., (2018) demonstrated that the levels of miR 141 are predominantly lesser in healthy tissue, and its enhanced expression can prevent tumor progression in HNSCC patients by increasing their survival rates [68].
EGFR plays a crucial role in the migration, angiogenesis, proliferation and apoptosis of cancer cells. Zhao et.al., (2019) illustrated that EGFR is greatly expressed in HNSCC tissues and miR-141 targets and bounds to EGFR. Therefore, higher expression of miR-141 could inhibit the expression of EGFR at both mRNA and protein levels [69]. Zhao et.al., (2019) reported that overexpression of miR-141 inhibits proliferation of HNSCC cells by inhibiting the expression of CDK4 and facilitates the apoptosis of HNSCC cells by inhibiting bcl-2. In addition, miR-141 inhibits the migration and invasion of HNSCC cells by inhibiting the expression of MMP2 [69]. Zhao et.al., (2019) has demonstrated that the tumor suppressor property of miR 141 suppresses tumor growth and metastasis by EGFR signaling. By this way, miR 141 appears to be a potentially beneficial therapeutic target for the treatment of HNSCC [69]. In this review, we have focused on the expression of miR141 and miR 200c in the regulation of tumorigenesis. miR-200c exhibits important roles in radio chemoresistance, regulating self-renewal and metastatic properties of HNSCC cancer stem cells, including the progression and metastasis of HNSCC. Higher expression of miR200c vitiates tumorigenic and metastatic oppression effects in HNSCC partially during the inhibition of ZEB1, ZEB2, BMI1 and EMT properties. Hence, miR200c could illustrate a new therapeutic attitude in treating advanced HNSCC [70]. miR expression profiling based on recent studies in the context of HNSCC are summarized in Table 1. The vast majority of studies included a sparse number of cases, and information's are not conclusive yet. miR200 family genes (miR200a, 200b, 200c, 141, 429) are selected because of their important role in tumorigenesis/ apoptosis and on the basis of previously found associations with cancer and has also been frequently observed in Head and Neck Squamous Cell Carcinoma as reported in Table 1. http://mircancer.ecu.edu/. In this review, we have outlined few microRNA genes and their role in different types of head and neck cancers, as shown in Table 1 and the number of miRs regulating various cancer hallmarks are depicted in Figure 3 based on the data referred from table 1. The databases and resources searched for articles included in Table I are PubMed Central. The inclusion criteria for articles search: (a) Published between 2010 and 2020(b) Published only in the English language. Keywords included in the literature search are epigenetic gene names, cancer, epigenetics, tumorigenesis, Head and neck cancer, microRNA. The literature search retrieved articles that met the set criteria. Head and Neck cancers were majorly studied in different ethnicities, but the data is not complete due to the vulnerability of the disease. The microRNA genes included in the Table 1 mainly was found to up regulate or downregulate the RNA expression of tumor suppressor or oncogenic genes. miRs play significant roles in the physiological and pathological processes of Head and neck cancer through regulating tumor cell proliferation, metastasis, invasion, and apoptosis. The expression of multiple mRNAs can be regulated by a single miR, and different miRs can modulate an individual mRNA. It is considered that miRs can regulate approximately 60% of genes in the human genome. One of the miR family, miR-200, was mainly characterized as a tumor suppressor in head and neck cancers, composed of five highly conserved miRs, including miR-141, miR-200a/200b/200c, and miR-429. They are located in two clusters of chromosomal locations: the miR-200ba/429 cluster on chromosome 1p36 and the miR-200c/141 cluster on chromosome 12p13 [149,150] as shown in Figure 4A& Figure 4B.
|
S.No. |
miRNAs of HNSCC |
Regulation of miRs |
References |
|
1. |
miR139-3p |
Down |
[71] |
|
2. |
miR363 |
Down |
[72] |
|
3. |
miR874 |
Down |
[73] |
|
4. |
miR93 |
Up |
[48,74] |
|
5. |
miR451 |
Down |
[75] |
|
6. |
miR205 |
Up |
[76] |
|
7. |
miR141 |
Down |
[69] |
|
8. |
miR204 |
Down |
[77] |
|
9. |
miR876-5p |
Down |
[78] |
|
10. |
miR34a |
Down |
[79] |
|
11. |
miR372 |
Up |
[80] |
|
12. |
miR375 |
Down |
[81] |
|
13. |
miR203 |
Down |
[82] |
|
14. |
miR422a |
Down |
[83] |
|
15. |
miR206 |
Down |
[84] |
|
16. |
miR1 |
Down |
[84] |
|
17. |
miR96-5p |
Up |
[85] |
|
18. |
miR9 |
Down |
[86] |
|
19. |
miR125a-5p |
Up |
[87] |
|
20. |
miR218 |
Down |
[87] |
|
21. |
miR25 |
Up |
[48] |
|
22. |
miR27a |
Down |
[88] |
|
23. |
miR29a |
Down |
[89] |
|
24. |
miR29c |
Down |
[89] |
|
25. |
miR300 |
Down |
[90] |
|
26. |
miR30a-5p |
Up |
[91] |
|
27. |
miR31 |
Up |
[92] |
|
28. |
miR184 |
Up |
[92] |
|
29. |
miR196a |
Up |
[93] |
|
30. |
miR200c |
Down |
[70] |
|
31. |
miR106b |
Up |
[48] |
|
32. |
miR107 |
Down |
[94,95] |
|
33. |
miR128 |
Down |
[96] |
|
34. |
miR133a |
Down |
[97] |
|
35. |
miR134 |
Up |
[98] |
|
36. |
miR138 |
Down |
[99,100] |
|
37. |
miR149 |
Down |
[101] |
|
38. |
miR16 |
Up |
[91] |
|
39. |
miR150-5p, miR150-3p |
Down |
[102] |
|
40. |
miR182 |
Up |
[103] |
|
41. |
miR675 |
Up |
[104] |
|
42. |
miR223-3p |
Up |
[105] |
|
43. |
miR192 |
Up |
[106] |
|
44. |
miR451a |
Down |
[106] |
|
45. |
miR203 |
Up |
[107] |
|
46. |
miR504 |
Down |
[108,109] |
|
47. |
miR217 |
Down |
[110] |
|
48. |
miR632 |
Up |
[111] |
|
49. |
miR23a |
Up |
[112] |
|
50. |
miR181a |
Down |
[113] |
|
51. |
miR885-5p |
Down |
[14] |
|
52. |
miR21-3p |
Up |
[14] |
|
53. |
miR525-5p |
Up |
[14] |
|
54. |
miR125a |
Down |
[114] |
|
55. |
miR27a |
Up |
[115] |
|
56. |
miR503 |
Down |
[116] |
|
57. |
miR26b |
Up |
[117] |
|
58. |
miR423-3p |
Up |
[118] |
|
59. |
miR379 |
Down |
[119] |
|
60. |
miR101 |
Down |
[120] |
|
61. |
miR129-5p |
Down |
[121] |
|
62. |
miR221 |
Up |
[122] |
|
63. |
miR195 |
Down |
[123] |
|
64. |
miR365a-3p |
Up |
[124] |
|
65. |
miR155 |
Up |
[125] |
|
66. |
miR301a-3p |
Up |
[126] |
|
67. |
miR19a |
Up |
[127] |
|
68. |
miR153 |
Down |
[128,129] |
|
69. |
miR148a |
Up |
[130] |
|
70. |
miR375 |
Up |
[130] |
|
71. |
miR4497 |
Down |
[131] |
|
72. |
miR7 |
Up |
[132] |
|
73. |
miR144 |
Down |
[133] |
|
74. |
miR194 |
Down |
[134] |
|
75. |
miR101 |
Down |
[135] |
|
76. |
miR145 |
Down |
[136,137] |
|
77. |
miR125b-5p |
Down |
[138] |
|
78. |
miR625 |
Down |
[139] |
|
79. |
miR145-5p |
Down |
[140] |
|
80. |
miR143-3p |
Down |
[141] |
|
81. |
miR218 |
Down |
[142] |
|
82. |
miR26a |
Down |
[143] |
|
83. |
miR613 |
Down |
[144] |
|
84. |
miR21-5p |
Up |
[145] |
|
85. |
miR181a |
Down |
[146] |
|
86. |
miR155 |
Up |
[147] |
|
87. |
miR370 |
Down |
[148] |
Table 1: MicroRNAs epigenetically regulated in Head & Neck Cancer are reported.
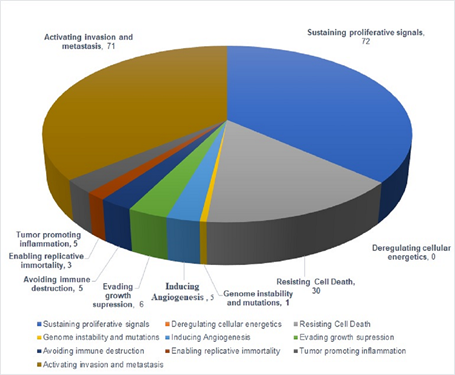
Figure 3: Pie chart showing the number of miRs regulating different hallmarks of HNSCC as reported in Table 1.
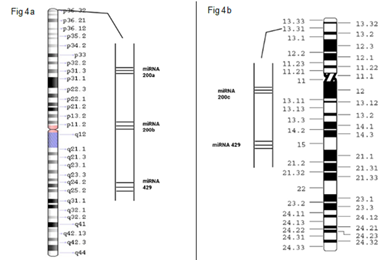
Figure 4: Figure 4a: depicts the cytogenic locations of miR200a, miR200b, mir429 mapped on chromosome 1(1p36.33). Figure 4b: depicts the cytogenic locations of miR200c, miR141 mapped on chromosome12 (12p13.31).
According to previous studies, abnormal miR expression is involved in many human diseases, especially cancers, and may act as potential prognostic biomarkers. Also, miRs play a significant role in the pathogenesis of cancers, acting as oncogenes, tumor suppressors, or modulators of cancer stem cells. miRs may act as oncogenes by inhibiting tumor suppressor genes and act as tumor suppressor genes by downregulating oncogenes [35].
1.7 Prediction and Identification of miRs
Each miRs exerts its function by regulating the targeted mRNAs; hence it is imperative to identify the miRs targets. The first eight nucleotides (seed sequence) of miRs play a vital role in the specificity of miRs-mRNA interaction [151]. Several computational algorithms studies have predicted the targeted mRNAs, but they were found to be very far from perfect. The gold standard experiment demonstrated that the luciferase reporter fused with 3′UTR of the predicted target was found to be repressed by the overexpression of the miRs, and the repression is revoked by the point mutation in the target sequence(s) at 3′UTR [152] (Yong et.al.,2009).
1.8 Role of Epigenetics in Cancer
Epigenetic mechanisms are necessary for the normal maintenance of tissue-specific gene expression patterns in humans. The precise epigenomic landscape present in normal cells undergoes extensive distortion in cancer. These epimutations, along with widespread genetic alterations, play a vital role in cancer initiation and progression. Advancements in the field of cancer epigenetics have shown rigorous reprogramming of every component of the epigenetic machinery in cancer, including DNA methylation, histone modifications and non-coding RNAs (microRNA expression). In addition to inactivating tumor suppressors, epimutations can also promote tumorigenesis by activating oncogenes. The events that lead to the initiation of these epigenetic abnormalities are still not fully understood [153].
1.8.1 Epigenetics and HNSCC: Epigenetic research aims to unearth how psychosocial factors, nutrition and environment may affect an individual's phenotypical expression of genetic information. In eukaryotes, different phenotypes may be expressed from a single genotype because of the targeting ability of inheritable epigenetic markers that appear during development. From previous research, it is already evident that the division and differentiation of single-cell during embryogenesis is strongly associated with epigenetic attributes. Hence, even though monozygotic twins have the same genetic information, they may differ in their epigenetic profiles, leading to potential differences in their health and disease phenotypes. Theoretically, Epigenetics plays a crucial role in cell division and differentiation. Therefore, it can answer variable phenotypes and their effects on different chronic diseases [154]. Increasing evidence suggests that epigenetic alterations play a critical part in the development of head and neck carcinoma. Compared with squamous cell carcinoma of other head and neck regions, studies on the epigenetic dysregulation of laryngeal cancers are relatively few. Most studies focused only on few epigenetic changes. With the recent advancement in technologies such as next-generation sequencing and microarray (e.g., methylation array and microRNA array), one could foresee those epigenetic studies on LSCC may shift from candidate gene approach to global high-throughput profiling. This contributes to further exploration of the disease from novel perspectives. Therefore, understanding the disease could identify suitable epigenetic markers [20,155].
1.8.2 DNA Methylation Aberrations: A cancer epigenome is marked by genome-wide hypomethylation and site-specific CpG island promoter hypermethylation. Even though the underlying mechanisms initiating these global changes are still under investigation, recent studies suggested that few of these alteration mechanisms may contribute to cancer initiation [156]. Hypomethylation of DNA promotes abnormal activation of genes and non-coding regions through various mechanisms that contribute to cancer development and progression. In contrast to hypomethylation, which increases genomic instability and activates proto-oncogenes, site-specific hypermethylation contributes to tumorigenesis by silencing tumor suppressor genes. DNA hypermethylation indirectly silences the gene expression by silencing transcription factors and DNA repair genes. In addition to this, it also directly inactivates the expression of tumor suppressor genes [156,157]. Studies in the context of HNSCC documented that in normal mammalian cells, promoter genes, LINEs & SINEs generally exhibit high methylation status, while during carcinogenesis, and they are hypomethylated, which contributes to activating transcription of sequences through genome destabilization. In normal cells, CpG islands of transcriptionally active genes are poorly methylated. Whereas hypermethylation in gene promoters is a characteristic for epigenomes of cancerous cells that may lead to transcriptional silencing of tumor suppressor genes, thus promoting malignant transformation [156]. Previous studies have shown that the group of proteins with methyl DNA binding activity, including Methyl-CpG binding domain protein 1 (MBD1), methyl CpG binding protein 2 (MeCP2), and Kaiso [also known as ZBTB 33 (Zinc finger and BTB domain-containing protein 33)] mediates the link between DNA methylation and histone modifications. These group of proteins localize to DNA methylated promoters and recruit a protein complex that contains histone deacetylases (HDACs) and histone methyltransferases [158-160]. Studies by Okitsu and Hsieh et.al., (2007), Weber et.al., (2007) suggest that chromatin structural changes are induced by DNA methylation with alteration of histone modifications [161,162]. It is known that DNA methylation inhibits H3K4me, which also acts as evidence that DNA methylation affects histone modifications. Despite that, earlier studies in fungi (Neurospora Crassa) demonstrated that mutations of histone H3K9 methyltransferase lowered DNA methylation, signifying a simple linear model. Where H3K9 methylation acts as an upstream epigenetic mark that signals to DNA methylation [163,164].
1.8.3 Histone Modifications: Histone protein modifications play a regulatory role in modifying chromatin structure and transcriptional activity of DNA. DNA tightly wrapped around the Histone octamer (two copies of H2A, H2B, H3, and H4) comprises the primary unit of chromatin known as a nucleosome. These are basic group proteins that consist of a C-terminal domain (globular form) and an N-terminal domain (tail form) [165]. The N-terminal domains of histone proteins undergo different post-translational modifications like acetylation, methylation, phosphorylation, ubiquitination, and ADP-ribosylation modulating the interactions between DNA and histone octamer. These post-translational histone modifications are mediated by enzymes that add or remove the chemical group on the amino-acids serine or lysine and arginine [165]. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) catalyze the process of acetylation and deacetylation, respectively. Acetylation of lysine results in chromatin structure relaxation, thus, facilitates gene transcription. Whereas deacetylation leads to gene silencing and decreased accessibility of DNA to the activity of transcription factors [165]. Methylation of lysine, histidine and arginine in histones results in changes in the structure of chromatin and gene activity without any alterations in the charge of histones. The methyl group is added to amino acid residues by histone methylases (HMT), while histone demethylases (HDMTs) reverse this process. The epigenetic effects of methylation depend on the location where the methyl group is added. Phosphorylation occurs by adding a phosphate group from ATP that modifies primarily threonine, serine and tyrosine located in the N-terminal histone tails and is regulated by phosphates and kinases (Figure 5). As a result, the histones will have a lower positive charge that may influence chromatin organization [165]. Recent advances in high-throughput sequencing have enabled genome-wide mapping of chromatin changes that occur during tumorigenesis. Studies have revealed that loss of histone acetylation mediated by HDACs results in gene repression. In addition to changes in histone acetylation, cancer cells also display widespread changes in histone methylation patterns that are associated with aberrant tumor suppressor gene silencing in various cancer forms. DNA methylation and histone modifications work independently and also in concert to alter gene expression during tumorigenesis. Therefore, deregulation of these histone protein alterations may regulate the transcription of various genes and, consequently, may lead to malignant transformations in various cancers like HNSCC [165].
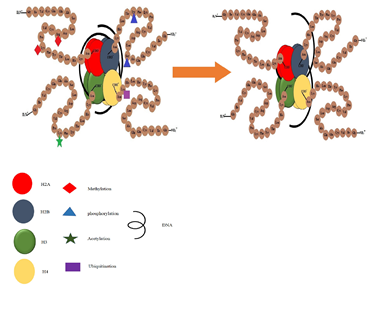
Figure 5: Histone Modifications.
1.8.4 miR Methylation: DNA methylation and histone modification at promoter regions and microRNA regulation at 3 prime UTRs are considered major epigenetic regulator mechanisms in eukaryotes. They predominantly regulate gene expression. Genes with low DNA methylation in the promoter regions are tend to be targeted by miRs. Epigenetic mechanism regulates the miRs; thereby, the expression of the given gene gets upregulated or downregulated in specific pathological conditions (tumor progression (HNSCC, Ovarian, Cervical cancer)) [166,167]. Iorio and Croce, 2012 have found that half of the miR genes tend to undergo DNA methylation by epigenetic regulation of miRs [166]. Malumbres, 2013 also reported that hypermethylation of miR 137 in OSCC ultimately targets the expression of CDK-6, E2F-6, NCoA2 and Lsd-1. miRs are found to have fine-tuned feedback systems where the first one controls the expression of vital epigenetic regulators, namely DNMTs, HDACs, PRC1 and PRC2, which are termed as epi-miRs, the other miR recruits the specific protein complexes directly to the promoter region of genomic DNA. Endogenous miRs directly participate in the induction or repression of specific genes; therefore, the gene regulatory mechanisms have their own function in the gene expression profile of specific cells. Disruption in the above two mechanisms may lead to cause various multifactorial disorders, including cancer [168].
1.8.5 miR200 Methylation in Cancers: In the stem-like phenotype, specifically miR200b, miR200a, miR429 cluster tends to get silenced primarily by histone modification (by poly comb) while the other two (miR 200c, miR 141) get repressed by DNA methylation [169]. Studies by Iliopoulos et al., (2010), Iliopoulos et al., (2009) and Shimono et al., (2009) have demonstrated that the re-expression of the miR 200 family in stem-like HMLE (sl- HMLE) have revealed that the miR200 family represses the stem-like properties. In addition to that, they have also found repression of miR-200 family in BCSCS isolated cells of metastatic breast cancer patients [170–172]. miR-200 family acts as a key regulator of the epithelial-mesenchymal transition (EMT). Studies on the regulation mechanism of miR-200 demonstrated that both the DNA methylation and histone modifications were considerably get altered in the stem-like and non-stem like phenotypes [169]. Especially in the stem-like phenotype, polycomb group-mediated histone modifications primarily silence the cluster of miR-429-200b-200a, whereas the miR-200c-141 cluster was suppressed by DNA methylation. The persistent decrease in miR-200 gene expression in stem-like -HMLE cells suggested that epigenetic changes might be involved in the maintenance and/or initiation of the stem-like sl-HMLE subpopulation [169]. CpG hypermethylation was identified across the promoter in the mesenchymal MDA-MB-231 and Hs578T cell lines that never again express the miR-200 genes. Interestingly, HMLE and sl-HMLE cells show a quite similar expression of CpG methylation profiles of the miR-429 -200b-200a gene; the TSS and promoter region was predominantly unmethylated except for region B, where a slight increase in DNA methylation of sl-HMLE cells is seen [173]. conversely, miR200c-141 gene silencing and primary transcript and CpG promoter methylation were positively correlated in the HMLE and sl-HMLE cell types.
1.8.6 Methylation of miR 200 in HNSCC: All human cancers, along with OSCC, are polygenic and poly epigenetic by nature [47]. Inactivation of tumor suppressor genes is the most common detectable epigenetic alteration [174]. In determining the expression of miRs in cancers, hypermethylation of CpG island plays an essential role in the transcriptional inactivation of miRs, which propagates epigenetic modifications [175]. P53 directly targets the family of miR 34 gene, and their ectopic expression in cancer cells initiates the arrest of the cell cycle followed by apoptosis by targeting MYC [176]. Kozaki et.al., (2008) identified the promoter regions of miR 34b, miR 34c that are found to have hypermethylated in oral and other cancer malignancies. Inactivation of tumor suppressor genes is the most common detectable alteration seen in the mechanism of poly epigenetics [47]. However, DNA hypermethylation epigenetically activates miR-200 in tumors, which is repressed in the absence of hypermethylation, specifically in CD44 high OSCCs. Members of the miR-200 gene family (miR-200a, miR-200b, miR200c, miR141 and miR429) are direct targets of CD44, and their ectopic expression in head and neck cancer cells stimulate apoptosis and cell cycle arrest by targeting BMI1 [34].
1.8.7 miRs as a Biomarker for HNSCC: Due to the miRs unique stability and their availability in body fluids (plasma, serum, sputum, urine, semen and milk), they have grabbed multiple attentions to be used as a biomarker for detecting cancer progression. miRs influence many biological processes such as proliferative signaling, cell death, and metastasis [177]. In carcinogenesis, the expression profile of miRs get altered, and such alterations can be identified very easily by accessible specimens like peripheral blood and saliva (Especially for HNCC) instead of a surgical tumour biopsy, and they have minimal invasiveness [178]. In the last two decades, nearly 20,000 publications have identified that the miRs are a potential biomarker for the prediction of disease progression because of their extreme stability. Studies have been performed to determine the efficacy rate of miRs as a biomarker in various forms of specimens like fresh, frozen, plasma, serum and formalin-fixed paraffin-embedded (FFPE) samples stored for many years, and it has been revealed that the expression of miRs found to be stable and unaffected by storage duration and the conditions. Most of the circulating miRs are not encapsulated in vesicles. Still, they allied with protein complexes containing the Argonaute 2 (Ago2), a vital effector of miRs-mediated silencing [178,179]. The potentiality of extracellular miRs is well established in cancer diagnosis, prognosis, and therapy compared to its actual function in cellular signaling [180]. Shreds of evidence have suggested that the altered expression profile contemplate pathogenesis link with tumor initiation, progression, and metastasis [181]. Consequently, the use of miRs as a biomarker in the clinical system may help to predict the patient's survival rate, loco-regional relapse and make up clinical decisions based on the anticipated responsiveness to a specific mode of treatment. Therefore, miRs as a biomarker would be highly useful in head and neck squamous cell carcinomas (HNSCCs) because of their high heterogenic nature [181].
Future prospects and Challenges
The epigenetic revolution that has come about in the field of biology during the last few decades has challenged the conventional view of the genetic code as the key determinant of cellular gene function and its alteration being the major cause of human diseases. Advances in cancer epigenetics have led to understanding genome packaging as potentially important for preserving cellular identity and giving rise to disease states like cancer. Deeper insights into the global patterns of epigenetic modifications and their corresponding changes in cancer may enable designing novel treatment strategies. A combinatorial approach utilizing different epigenetic therapeutics along with standard chemotherapy holds significance for successful treatment of cancer in future. Further understanding of cancer stem cells and the development of more specific epigenetic drugs may hold the key to successfully resetting the abnormal cancer epigenome. Our understanding indicates that various miRs were found to be significantly associated with poor prognosis in HNSCC patients. Even though many miRs are identified to be deregulated in cancers, their pathological consequences are yet to be explored. We envision that in the near future advanced miR-based studies may become a key component of tumor diagnosis. Therefore, our review highlights the emerging potential of miRs as the key players in cancer management as diagnostic or prognostic tools and for monitoring therapeutic responses.
Author Acknowledgements
Not Applicable.
Conflicts of Interest Statement
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Funding Statement
No funding was received to assist with the preparation of this manuscript.
References
- MACCOMB WS. Treatment of cancer of the head and neck. Postgrad Med 17 (1955): 498-500.
- Lo Nigro C, Denaro N, Merlotti A, et al. Head and neck cancer: Improving outcomes with a multidisciplinary approach. Cancer Manag Res 9 (2017): 363-371.
- Heroiu AD, Danciu CE, Popescu CR. Multiple Cancers of the Head and Neck. Maedica A Journal of Clinical Medicine 8 (2013).
- Obid R, Redlich M, Tomeh C. The Treatment of Laryngeal Cancer. Vol. 31, Oral and Maxillofacial Surgery Clinics of North America. W.B. Saunders (2019): 1-11.
- Bobdey S, Jain A, Balasubramanium G. Epidemiological review of laryngeal cancer: An Indian perspective. Indian J Med Paediatr Oncol 36(3): 154-160.
- van Harten AM, Brakenhoff RH. Targeted Treatment of Head and Neck (Pre)Cancer: Preclinical Target Identification and Development of Novel Therapeutic Applications. Cancers (Basel) 13 (2021): 2774.
- Wong KM, Capasso A, Eckhardt SG. The changing landscape of phase I trials in oncology. Nat Rev Clin Oncol 13 (2016): 106-117.
- Moscow JA, Fojo T, Schilsky RL. The evidence framework for precision cancer medicine. Nat Rev Clin Oncol 15 (2018): 183-192.
- Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 18 (2018): 269-282.
- Kobayashi K, Hisamatsu K, Suzui N, et al. A Review of HPV-Related Head and Neck Cancer. J Clin Med 7 (2018): 241.
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 64 (2014): 252-271.
- Koyfman SA, Ismaila N, Crook D, et al. Management of the Neck in Squamous Cell Carcinoma of the Oral Cavity and Oropharynx: ASCO Clinical Practice Guideline. J Clin Oncol 37 (2019): 1753-1774.
- Szturz P, Wouters K, Kiyota N, et al. Weekly Low-Dose Versus Three-Weekly High-Dose Cisplatin for Concurrent Chemoradiation in Locoregionally Advanced Non-Nasopharyngeal Head and Neck Cancer: A Systematic Review and Meta-Analysis of Aggregate Data. Oncologist 22 (2017): 1056-1066.
- Cybula M, Lukasz W, Józefowicz-Korczynska M, et al. New miRNA expression abnormalities in laryngeal squamous cell carcinoma. Cancer Biomarkers 16 (2016): 559-568.
- Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol 10 (2010): 543-550.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (1993): 843-854.
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 (1993): 855-862.
- Bartel DP. MicroRNAs. Cell 116 (2004): 281-297.
- Hsieh JC, Wang H, Wu M, et al. Review of emerging biomarkers in head and neck squamous cell carcinoma in the era of immunotherapy and targeted therapy. Head Neck 41 (2019): 19-45.
- Cui M, Wang H, Yao X, et al. Circulating MicroRNAs in Cancer: Potential and Challenge. Front Genet 10 (2019).
- Yu J, Wang F, Yang GH, et al. Human microRNA clusters: Genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun 349 (2006): 59-68.
- Lee YS, Dutta A. MicroRNAs in Cancer. Annu Rev Pathol Mech Dis 4 (2009): 199-227.
- Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 (2005): 769-773.
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21 (2007): 1025-1030.
- Chu C, Rana TM. Translation Repression in Human Cells by MicroRNA-Induced Gene Silencing Requires RCK/p54. Carrington J, editor. PLoS Biol 4 (2006): e210.
- Wong KY, Huang X, Chim CS. DNA methylation of microRNA genes in multiple myeloma. Carcinogenesis 33 (2012): 1629-1638.
- Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med 14 (2016): 143.
- Leoncini E, Ricciardi W, Cadoni G, et al. Adult height and head and neck cancer: a pooled analysis within the INHANCE Consortium. Eur J Epidemiol 29 (2014): 35-48.
- Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell 100 (2000): 57-70.
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11 (2010): 220-228.
- Luo J, Solimini NL, Elledge SJ. Principles of Cancer Therapy: Oncogene and Non-oncogene Addiction. Cell 136 (2009):823-837.
- Colotta F, Allavena P, Sica A, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30 (2009): 1073-1081.
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell 144 (2011): 646-674.
- Manasa V, Kannan S. Impact of microRNA dynamics on cancer hallmarks: An oral cancer scenario. Tumor Biol 39 (2017): 101042831769592.
- Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1 (2016): 15004.
- Hatfield SD, Shcherbata HR, Fischer KA, et al. Stem cell division is regulated by the microRNA pathway. Nature 435 (2005): 974-978.
- Yi C, Wang Q, Wang L, et al. MiR-663, a microRNA targeting p21WAF1/CIP1, promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene 31 (2012): 4421-4433.
- Peng Y, Dai Y, Hitchcock C, et al. Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc Natl Acad Sci 110 (2013): 15043-15048.
- Lima RT, Busacca S, Almeida GM, et al. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer 47 (2011): 163-174.
- Li C, Hashimi SM, Good DA, et al. Apoptosis and microRNA aberrations in cancer. Clin Exp Pharmacol Physiol 39 (2012): 739-746.
- Pichiorri F, Suh SS, Rocci A, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 Impairs the p53/MDM2 Autoregulatory Loop in Multiple Myeloma Development. Cancer Cell 18 (2010): 367-381.
- Yan H, Xue G, Mei Q, et al. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28 (2009): 2719-2732.
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 119 (2009): 1420-1428.
- Kong W, Yang H, He L, et al. MicroRNA-155 Is Regulated by the Transforming Growth Factor β/Smad Pathway and Contributes to Epithelial Cell Plasticity by Targeting RhoA. Mol Cell Biol 28 (2008): 6773-6784.
- Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med 6 (2000): 389-395.
- Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer 2 (2002): 795-803.
- Kozaki K, Imoto I, Mogi S, et al. Exploration of Tumor-Suppressive MicroRNAs Silenced by DNA Hypermethylation in Oral Cancer. Cancer Res 68 (2008): 2094-2105.
- Hui ABY, Lenarduzzi M, Krushel T, et al. Comprehensive MicroRNA Profiling for Head and Neck Squamous Cell Carcinomas. Clin Cancer Res 16 (2010): 1129-1139.
- Zhang X, Ng WL, Wang P, et al. MicroRNA-21 Modulates the Levels of Reactive Oxygen Species by Targeting SOD3 and TNF α. Cancer Res 72 (2012): 4707-4713.
- Liu X, Wang A, Heidbreder CE, et al. MicroRNA-24 targeting RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS Lett 584 (2010): 4115-4120.
- Reis PP, Tomenson M, Cervigne NK, et al. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol Cancer 9 (2010): 238.
- Hung PS, Tu HF, Kao SY, et al. miR-31 is upregulated in oral premalignant epithelium and contributes to the immortalization of normal oral keratinocytes. Carcinogenesis 35 (2014): 1162-1171.
- Li J, Lei H, Xu Y, et al. miR-512-5p Suppresses Tumor Growth by Targeting hTERT in Telomerase Positive Head and Neck Squamous Cell Carcinoma In Vitro and In Vivo. Fan G-C, editor. PLoS One 10 (2015): e0135265.
- Liu CJ, Tsai MM, Hung PS, et al. miR-31 Ablates Expression of the HIF Regulatory Factor FIH to Activate the HIF Pathway in Head and Neck Carcinoma. Cancer Res 70 (2010): 1635-1644.
- Xiao W, Bao ZX, Zhang CY, et al. Upregulation of miR-31* Is Negatively Associated with Recurrent/Newly Formed Oral Leukoplakia. Lo K-W, editor. PLoS One 7 (2012): e38648.
- Fletcher AM, Heaford AC, Trask DK. Detection of Metastatic Head and Neck Squamous Cell Carcinoma Using the Relative Expression of Tissue-Specific Mir-205. Transl Oncol 1 (2008): 202-IN2.
- Liu X, Yu J, Jiang L, et al. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics 6 (2009): 131-139.
- Lu L, Xue X, Lan J, et al. MicroRNA-29a upregulates MMP2 in oral squamous cell carcinoma to promote cancer invasion and anti-apoptosis. Biomed Pharmacother 68 (2014): 13-19.
- Arora A, Singh A, Bhatt AN, et al. Interplay Between Metabolism and Oncogenic Process: Role of microRNAs. Transl Oncogenomics 7 (2015): 11-27.
- Xu P, Li Y, Zhang H, et al. MicroRNA-340 Mediates Metabolic Shift in Oral Squamous Cell Carcinoma by Targeting Glucose Transporter-1. J Oral Maxillofac Surg 74 (2016): 844-850.
- Hatziapostolou M, Polytarchou C, Iliopoulos D. miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol Metab 24 (2013): 361-373.
- Peschiaroli A, Giacobbe A, Formosa A, et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene 32 (2013): 797-802.
- Arunkumar G, Deva Magendhra Rao A, Manikandan M, et al. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol Lett (2017).
- Swick AD, Prabakaran PJ, Miller MC, et al. Cotargeting mTORC and EGFR Signaling as a Therapeutic Strategy in HNSCC. Mol Cancer Ther 16 (2017): 1257-1268.
- Xu S, Ge J, Zhang Z, et al. miR-141 inhibits prostatic cancer cell proliferation and migration, and induces cell apoptosis via targeting of RUNX1. Oncol Rep (2018).
- Wang H, Li H, Zhang L, et al. Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis. Biomed Pharmacother 106 (2018): 1607-1615.
- Sha M, Lin M, Wang J, et al. Long non-coding RNA MIAT promotes gastric cancer growth and metastasis through regulation of miR-141/DDX5 pathway. J Exp Clin Cancer Res 37 (2018): 58.
- Li X, Tian Z, Jin H, et al. Decreased c-Myc mRNA Stability via the MicroRNA 141-3p/AUF1 Axis Is Crucial for p63α Inhibition of Cyclin D1 Gene Transcription and Bladder Cancer Cell Tumorigenicity. Mol Cell Biol 38 (2018).
- Zhao Z, Gao D, Ma T, et al. MicroRNA-141 suppresses growth and metastatic potential of head and neck squamous cell carcinoma. Aging (Albany NY) 11 (2019): 921-932.
- Lo WL, Yu CC, Chiou GY, et al. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol 223 (2011): 482-495.
- Sannigrahi MK, Sharma R, Singh V, et al. Role of Host miRNA Hsa-miR-139-3p in HPV-16–Induced Carcinomas. Clin Cancer Res 23 (2017): 3884-3895.
- Chapman BV, Wald AI, Akhtar P, et al. MicroRNA-363 targets myosin 1B to reduce cellular migration in head and neck cancer. BMC Cancer 15 (2015): 861.
- Nohata N, Hanazawa T, Kinoshita T, et al. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer 108 (2013): 1648-1658.
- Li G, Ren S, Su Z, et al. Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumor Biol 36 (2015): 3949-3956.
- Wang H, Zhang G, Wu Z, et al. MicoRNA-451 is a novel tumor suppressor via targeting c-myc in head and neck squamous cell carcinomas. J Cancer Res Ther 11 (2015): 216.
- Kolenda T, Guglas K, Teresiak A, et al. Low let-7d and high miR-205 expression levels positively influence HNSCC patient outcome. J Biomed Sci 26 (2019): 17.
- Wu Q, Zhao Y, Wang P. miR-204 inhibits angiogenesis and promotes sensitivity to cetuximab in head and neck squamous cell carcinoma cells by blocking JAK2-STAT3 signaling. Biomed Pharmacother 99 (2018): 278-285.
- Dong Y, Zheng Y, Wang C, et al. MiR-876-5p modulates head and neck squamous cell carcinoma metastasis and invasion by targeting vimentin. Cancer Cell Int 18 (2018): 121.
- Kumar B, Yadav A, Lang J, et al. Dysregulation of MicroRNA-34a Expression in Head and Neck Squamous Cell Carcinoma Promotes Tumor Growth and Tumor Angiogenesis. Lebedeva I V., editor. PLoS One 7 (2012): e37601.
- Yeh LY, Liu CJ, Wong YK, et al. miR-372 inhibits p62 in head and neck squamous cell carcinoma in vitro and in vivo. Oncotarget 6 (2015): 6062-6075.
- Jimenez L, Sharma VP, Condeelis J, et al. MicroRNA-375 Suppresses Extracellular Matrix Degradation and Invadopodial Activity in Head and Neck Squamous Cell Carcinoma. Arch Pathol Lab Med 139 (2015): 1349-1361.
- Obayashi M, Yoshida M, Tsunematsu T, et al. microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget 7 (2016): 8223-8239.
- Bonnin N, Armandy E, Carras J, et al. MiR-422a promotes loco-regional recurrence by targeting NT5E/CD73 in head and neck squamous cell carcinoma. Oncotarget 7 (2016): 44023-44038.
- Koshizuka K, Hanazawa T, Fukumoto I, et al. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum Genet 62 (2017): 113-121.
- Vahabi M, Pulito C, Sacconi A, et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J Exp Clin Cancer Res 38 (2019): 141.
- Hersi HM, Raulf N, Gaken J, et al. Micro RNA -9 inhibits growth and invasion of head and neck cancer cells and is a predictive biomarker of response to plerixafor, an inhibitor of its target CXCR 4. Mol Oncol 12 (2018): 2023-2041.
- Jin S, Liu M, Wu H, et al. Overexpression of hsa-miR-125a-5p enhances proliferation, migration and invasion of head and neck squamous cell carcinoma cell lines by upregulating C-C chemokine receptor type�7. Oncol Lett (2018).
- Wu X, Bhayani MK, Dodge CT, et al. Coordinated Targeting of the EGFR Signaling Axis by MicroRNA-27a*. Oncotarget 4 (2013): 1388-1398.
- Kinoshita T, Nohata N, Hanazawa T, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin–integrin signalling in head and neck squamous cell carcinoma. Br J Cancer 109 (10): 2636-2645.
- Yu J, Xie F, Bao X, et al. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer 13 (2014): 121.
- Kimura S, Naganuma S, Susuki D, et al. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol Rep 23 (6).
- Kao SY, Tsai MM, Wu CH, et al. Co-targeting of multiple microRNAs on factor-Inhibiting hypoxia-Inducible factor gene for the pathogenesis of head and neck carcinomas. Head Neck 38 (2016): 522-528.
- Suh YE, Raulf N, Gäken J, et al. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. Int J Cancer 137 (2015): 1021-1034.
- Datta J, Smith A, Lang JC, et al. microRNA-107 functions as a candidate tumor-suppressor gene in head and neck squamous cell carcinoma by downregulation of protein kinase C?. Oncogene 31 (2012): 4045-4053.
- Piao L, Zhang M, Datta J, et al. Lipid-based Nanoparticle Delivery of Pre-miR-107 Inhibits the Tumorigenicity of Head and Neck Squamous Cell Carcinoma. Mol Ther 20 (2012): 1261-1269.
- Hauser B, Zhao Y, Pang X, et al. Functions of MiRNA-128 on the Regulation of Head and Neck Squamous Cell Carcinoma Growth and Apoptosis. Ray RB, editor. PLoS One 10 (2015): e0116321.
- Kinoshita T, Nohata N, Fuse M, et al. Tumor suppressive microRNA-133a regulates novel targets: Moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun 418 (2012): 378-383.
- Liu CJ, Shen WG, Peng SY, et al. miR-134 induces oncogenicity and metastasis in head and neck carcinoma through targeting WWOX gene. Int J Cancer 134 (2014): 811-821.
- Jin Y, Chen D, Cabay RJ, et al. Role of microRNA-138 as a Potential Tumor Suppressor in Head and Neck Squamous Cell Carcinoma. In (2013): 357-385.
- Islam M, Datta J, Lang JC, et al. Down regulation of RhoC by microRNA-138 results in de-activation of FAK, Src and Erk1/2 signaling pathway in head and neck squamous cell carcinoma. Oral Oncol 50 (2014): 448-456.
- Tu HF, Liu CJ, Chang CL, et al. The Association between Genetic Polymorphism and the Processing Efficiency of miR-149 Affects the Prognosis of Patients with Head and Neck Squamous Cell Carcinoma. Christensen BC, editor. PLoS One 7 (2012): e51606.
- Koshizuka K, Hanazawa T, Kikkawa N, et al. Antitumor miR-150-5p and miR-150-3p inhibit cancer cell aggressiveness by targeting SPOCK1 in head and neck squamous cell carcinoma. Auris Nasus Larynx 45(2018): 854-865.
- Wang L, Jiang H, Li W, et al. Overexpression of TP53 mutation-associated microRNA-182 promotes tumor cell proliferation and migration in head and neck squamous cell carcinoma. Arch Oral Biol 73 (2017): 105-112.
- Guan G, Zhang D, Wen L, et al. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int J Med Sci 13 (2016): 914-922.
- Bozec A, Zangari J, Butori-Pepino M, et al. MiR-223-3p inhibits angiogenesis and promotes resistance to cetuximab in head and neck squamous cell carcinoma. Oncotarget 8 (2017): 57174-57186.
- Doukas SG, Vageli DP, Sasaki CT. NF -κB inhibition reverses acidic bile-induced miR-21, miR-155, miR-192, miR-34a, miR-375 and miR-451a deregulations in human hypopharyngeal cells. J Cell Mol Med 22 (2018): 2922-2934.
- Wang R, Fang J, Ma H, et al. Effect of microRNA-203 on tumor growth in human hypopharyngeal squamous cell carcinoma. Mol Cell Biochem 405 (2015): 97-104.
- Kikkawa N, Kinoshita T, Nohata N, et al. microRNA-504 inhibits cancer cell proliferation via targeting CDK6 in hypopharyngeal squamous cell carcinoma. Int J Oncol 44 (2014): 2085-2092.
- Guan Y, Chen L, Bao Y, et al. Downregulation of microRNA-504 is associated with poor prognosis in high-grade glioma. Int J Clin Exp Pathol 8 (2015): 727-734.
- Miao S, Mao X, Zhao S, et al. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget 8 (2017): 62143-62153.
- Zhou Z, Zhang Z, Tao Z, et al. miR-632 Promotes Laryngeal Carcinoma Cell Proliferation, Migration, and Invasion Through Negative Regulation of GSK3β. Oncol Res Featur Preclin Clin Cancer Ther 28 (2020): 21-31.
- Zhang XW, Liu N, Chen S, et al. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn Pathol 10 (2015): 22.
- Wang H, Tong X, Zhang Z, et al. MYCT 1 represses apoptosis of laryngeal cancerous cells through the MAX /miR-181a/ NPM 1 pathway. FEBS J 286 (2019): 3892-3908.
- Liu J, Tang Q, Li S, et al. Inhibition of HAX-1 by miR-125a reverses cisplatin resistance in laryngeal cancer stem cells. Oncotarget 7 (2016): 86446-86456.
- Tian Y, Fu S, Qiu GB, et al. MicroRNA-27a promotes proliferation and suppresses apoptosis by targeting PLK2in laryngeal carcinoma. BMC Cancer 14 (2014): 678.
- Ma H, Lian R, Wu Z, et al. MiR-503 enhances the radiosensitivity of laryngeal carcinoma cells via the inhibition of WEE1. Tumor Biol 39 (2017): 101042831770622.
- Wang S, Guo D, Li C. Downregulation of miRNA-26b inhibits cancer proliferation of laryngeal carcinoma through autophagy by targeting ULK2 and inactivation of the PTEN/AKT pathway. Oncol Rep 38 (2017): 1679-1687.
- Guan G, Zhang D, Zheng Y, et al. microRNA-423-3p promotes tumor progression via modulation of AdipoR2 in laryngeal carcinoma. Int J Clin Exp Pathol 7 (2014): 5683-5691.
- Wei J, Zhang L, Zhao Z, et al. MicroRNA-379 inhibits laryngeal carcinoma cell proliferation and invasion through directly targeting TCF-4. Kaohsiung J Med Sci 35 (2019): 731-738.
- Li M, Tian L, Ren H, et al. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J Transl Med 13 (2015): 271.
- Li M, Tian L, Wang L, et al. Down-Regulation of miR-129-5p Inhibits Growth and Induces Apoptosis in Laryngeal Squamous Cell Carcinoma by Targeting APC. Teh M-T, editor. PLoS One 8 (2013): e77829.
- Hussein S, Mosaad H, Rashed HE, et al. Up-regulated miR-221 expression as a molecular diagnostic marker in laryngeal squamous cell carcinoma and its correlation with Apaf-1 expression. Cancer Biomarkers 19 (2017): 279-287.
- Shuang Y, Li C, Zhou X, et al. MicroRNA-195 inhibits growth and invasion of laryngeal carcinoma cells by directly targeting DCUN1D1. Oncol Rep 38 (2017): 2155-2165.
- Geng J, Liu Y, Jin Y, et al. MicroRNA-365a-3p promotes tumor growth and metastasis in laryngeal squamous cell carcinoma. Oncol Rep 35 (2016): 2017-2026.
- Wang J, Wang X, Yang D, et al. The Expression of MicroRNA-155 in Plasma and Tissue Is Matched in Human Laryngeal Squamous Cell Carcinoma. Yonsei Med J 57 (2016): 298.
- Lu Y, Gao W, Zhang C, et al. Hsa-miR-301a-3p Acts as an Oncogene in Laryngeal Squamous Cell Carcinoma via Target Regulation of Smad4. J Cancer 6 (2015): 1260-1275.
- Wu TY, Zhang TH, Qu LM, et al. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int J Clin Exp Pathol 7 (2014): 56-63.
- Liu Jy, Lu JB, Xu Y. MicroRNA-153 inhibits the proliferation and invasion of human laryngeal squamous cell carcinoma by targeting KLF5. Exp Ther Med 11 (2016): 2503-2508.
- Zhang B, Fu T, Zhang L. MicroRNA-153 suppresses human laryngeal squamous cell carcinoma migration and invasion by targeting the SNAI1 gene. Oncol Lett (2018).
- Wu Y, Yu J, Ma Y, et al. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol Lett 12 (2016): 871-878.
- Chen X, Zhang L, Tang S. MicroRNA-4497 functions as a tumor suppressor in laryngeal squamous cell carcinoma via negatively modulation the GBX2. Auris Nasus Larynx 46 (2019): 106-113.
- Zhang J, Hu H, Zhao Y, et al. CDR1as is overexpressed in laryngeal squamous cell carcinoma to promote the tumour’s progression via miR-7 signals. Cell Prolif 51 (2018): e12521.
- Wu X, Cui CL, Chen WL, et al. miR-144 suppresses the growth and metastasis of laryngeal squamous cell carcinoma by targeting IRS1. Am J Transl Res 8 (2016): 1-11.
- Li P, Yang Y, Liu H, et al. MiR-194 functions as a tumor suppressor in laryngeal squamous cell carcinoma by targeting Wee1. J Hematol Oncol 10 (2017): 32.
- Chen L, Jia J, Zang Y, et al. MicroRNA-101 regulates autophagy, proliferation and apoptosis via targeting EZH2 in laryngeal squamous cell carcinoma. Neoplasma 66 (2019): 507-515.
- Zhao X, Zhang W, Ji W. MYO5A inhibition by miR-145 acts as a predictive marker of occult neck lymph node metastasis in human laryngeal squamous cell carcinoma. Onco Targets Ther 11 (2018): 3619-3635.
- Zhu X, Zhu R. Curcumin suppresses the progression of laryngeal squamous cell carcinoma through the upregulation of miR-145 and inhibition of the PI3K/Akt/mTOR pathway. Onco Targets Ther 11 (2018): 3521-3531.
- Hui L, Zhang J, Guo X. MiR-125b-5p suppressed the glycolysis of laryngeal squamous cell carcinoma by down-regulating hexokinase-2. Biomed Pharmacother 103 (2018): 1194-1201.
- Li Y, Tao C, Dai L, et al. MicroRNA-625 inhibits cell invasion and epithelial–mesenchymal transition by targeting SOX4 in laryngeal squamous cell carcinoma. Biosci Rep 39 (2019).
- Gao W, Zhang C, Li W, et al. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Mol Ther 27 (2019): 365-379.
- Han L, Tang M, Xu X, et al. MiR-143-3p suppresses cell proliferation, migration, and invasion by targeting Melanoma-Associated Antigen A9 in laryngeal squamous cell carcinoma. J Cell Biochem 120 (2019): 1245-1257.
- Zhang Y, Hu H. Long non-coding RNA CCAT1/miR-218/ZFX axis modulates the progression of laryngeal squamous cell cancer. Tumor Biol 39 (2017): 101042831769941.
- Wu Z, Lu B, Li X, et al. MicroRNA-26a inhibits proliferation and tumorigenesis via targeting CKS2 in laryngeal squamous cell carcinoma. Clin Exp Pharmacol Physiol 45 (2018): 444-451.
- Wang J, Yang S, Ge W, et al. MiR-613 suppressed the laryngeal squamous cell carcinoma progression through regulating PDK1. J Cell Biochem 119 (2018): 5118-5125.
- Re M, Magliulo G, Gioacchini FM, et al. Expression Levels and Clinical Significance of miR-21-5p, miR-let-7a, and miR-34c-5p in Laryngeal Squamous Cell Carcinoma. Biomed Res Int 2017 (2017): 1-9.
- Zhao X, Zhang W, Ji W. miR-181a targets GATA6 to inhibit the progression of human laryngeal squamous cell carcinoma. Futur Oncol 14 (2018): 1741-1753.
- Zhao X, Zhang W, Ji W. YB-1 promotes laryngeal squamous cell carcinoma progression by inducing miR-155 expression via c-Myb. Futur Oncol 14 (2018): 1579-1589.
- Yungang W, Xiaoyu L, Pang T, et al. miR-370 targeted FoxM1 functions as a tumor suppressor in laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother 68 (2014): 149-154.
- Tejero R, Navarro A, Campayo M, et al. miR-141 and miR-200c as Markers of Overall Survival in Early Stage Non-Small Cell Lung Cancer Adenocarcinoma. Chellappan SP, editor. PLoS One 9 (2014): e101899.
- Huang GL, Sun J, Lu Y, et al. MiR-200 family and cancer: From a meta-analysis view. Mol Aspects Med 70 (2019): 57-71.
- Kedde M, Strasser MJ, Boldajipour B, et al. RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA. Cell 131 (2007): 1273-1286.
- No Title.
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 31 (2010): 27-36.
- Zhang L, Lu Q, Chang C. Epigenetics in Health and Disease. In (2020): 3-55.
- Yu X, Li Z. The role of MicroRNAs expression in laryngeal cancer. Oncotarget 6 (2015): 23297-232305.
- Gazdzicka J, Golabek K, Strzelczyk JK, et al. Epigenetic Modifications in Head and Neck Cancer. Biochem Genet 58 (2020): 213-244.
- Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics 2 (2011): 123-150.
- Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393 (1998): 386-389.
- Hendrich B, Bird A. Identification and Characterization of a Family of Mammalian Methyl-CpG Binding Proteins. Mol Cell Biol 18 (1998): 6538-6547.
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16 (2002): 6-21.
- Okitsu CY, Hsieh CL. DNA Methylation Dictates Histone H3K4 Methylation. Mol Cell Biol 27 (2007): 2746-2757.
- Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39 (2007): 457-466.
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 (2001): 277-283.
- Kondo Y. Epigenetic Cross-Talk between DNA Methylation and Histone Modifications in Human Cancers. Yonsei Med J 50 (2009): 455.
- Gazdzicka J, Golabek K, Strzelczyk JK, et al. Epigenetic Modifications in Head and Neck Cancer. Vol. 58, Biochemical Genetics. Springer (2020): 213-244.
- Malumbres M. miRNAs and cancer: An epigenetics view. Mol Aspects Med 34 (2013): 863-874.
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4 (2012): 143-159.
- John K, Wu J, Lee BW, et al. MicroRNAs in Head and Neck Cancer. Int J Dent 2013 (2013): 1-12.
- Lim Y, Wright JA, Attema JL, et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem cell-like state. J Cell Sci 126 (2013): 2256-2266.
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, et al. Loss of miR-200 Inhibition of Suz12 Leads to Polycomb-Mediated Repression Required for the Formation and Maintenance of Cancer Stem Cells. Mol Cell 39 (2010): 761-772.
- Iliopoulos D, Polytarchou C, Hatziapostolou M, et al. MicroRNAs Differentially Regulated by Akt Isoforms Control EMT and Stem Cell Renewal in Cancer Cells. Sci Signal 2 (2009): ra62–ra62.
- Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c Links Breast Cancer Stem Cells with Normal Stem Cells. Cell 138 (2009): 592-603.
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10 (2008): 593-601.
- Herman JG, Baylin SB. Gene Silencing in Cancer in Association with Promoter Hypermethylation. N Engl J Med 349 (2003): 2042-2054.
- Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 31 (2012): 1609-1622.
- He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 447 (2007): 1130-1134.
- Larrea E, Sole C, Manterola L, et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int J Mol Sci 17 (2016): 627.
- Nowicka Z, Stawiski K, Tomasik B, et al. Extracellular miRNAs as Biomarkers of Head and Neck Cancer Progression and Metastasis. Int J Mol Sci 20 (2019): 4799.
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci 108 (2011): 5003-5008.
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101 (2010): 2087-2092.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68 (2018): 394-424.
