Genetic Diversity of Plasmodium falciparum before and after intensive and massive relocation of populations into Yaoundé, Cameroon
Article Information
Akindeh Mbuh Nji1,2,3*, Aristid Herve Ekollo Mbange1,2, 8 , Abdel Aziz Selly-Ngaloumo 1,2,3, Peter Thelma Ngwa Niba 1,2,3, Jean Paul Kengne Chedjou1,2,3, Ngum Lesley Ngum 1,2,9,10, Tsiambom Monju1,2,4, Lorraine Matchio Fokou-Kuignou 2,3, Olivia Achonduh-Atijegbe2, Innocent Mbulli Ali1,2,5, Cyrille Mbanwi Mbu’u1,2,7, Calvino Tah Fumbah 1,2,3, Sebastien Kenmoe11,12, Jude Bigoga1,2,3, Wilfred Fon Mbacham1,2,3,6,*
1MARCAD-DELTAS Programme, Laboratory for Public Health Research Biotechnologies, University of Yaounde I, Yaounde, Cameroon
2The Biotechnology Centre, University of Yaounde I, Yaounde, Cameroon
3Department of Biochemistry, Faculty of Science, University of Yaounde I, Yaounde, Cameroon
4Department of Biochemistry, Faculty of Medicine and Biomedical Sciences, University of Yaounde I, Yaounde, Cameroon
4School of Health Sciences, Catholic University of Central Africa, Yaounde, Cameroon
5Department of Biochemistry, Faculty of Science, University of Dschang, Dschang, Cameroon
6Malaria Consortium-Cameroon Coalition Against Malaria, Yaounde, Cameroon
7Department of Microbiology, Faculty of Science, University of Yaounde I, Yaounde, Cameroon
8University Institute of Technologies, University of Ngaoundere, Ngaoundere, Cameroon
9Department of Biochemistry, Faculty of Medicine and Biomedical Sciences,University of Yaoundé I, Yaoundé, Cameroon
10Institute of Medical Research and Medicinal Plant Studies/IMPM/ Yaoundé, Cameroon
11Virology Department, Centre Pasteur of Cameroon, Yaounde, Cameroon
12Department of Microbiology and Parasitology, University of Buea, Buea, Cameroon
*Corresponding author: Wilfred Fon Mbacham, Akindeh Mbuh Nji
Received: 08 June 2022; Accepted: 15 June 2022; Published: 24 June 2022
Citation: Akindeh Mbuh Nji, Aristid Herve Ekollo Mbange , Abdel Aziz Selly-Ngaloumo, Peter Thelma Ngwa Niba, Jean Paul Kengne Chedjou, Ngum Lesley Ngum, Tsiambom Monju, Lorraine Matchio Fokou-Kuignou, Olivia Achonduh-Atijegbe, Innocent Mbulli Ali, Cyrille Mbanwi Mbu’u, Calvino Tah Fumbah, Sebastien Kenmoe, Jude Bigoga, Wilfred Fon Mbacham. Genetic Diversity of Plasmodium falciparum before and after intensive and massive relocation of populations into Yaounde, Cameroon. Fortune Journal of Health Sciences 5 (2022): 334-351
View / Download Pdf Share at FacebookAbstract
The introduction of new, genetically diverse and divergent populations of Plasmodium falciparum parasites into a given location, due to population influx, may have serious public health consequences. This study compared the genetic diversity (GD) of P. falciparum amongst Cameroonian populations sampled before (2014) and after (2018) intensive and massive relocations due to civil unrest, from affected regions with different malaria dynamics to the capital city Yaounde. For each time point, a subset of 50 samples was retrieved from a pool of samples collected from participant children aged between 6 months and 18 years and adults attending the hospital for outpatient consultation. To precisely assess the breadth of P. falciparum diversity, genotyping was performed using 2 PCR-based techniques which were further evaluated for their performance: nested-PCR targeting the merozoite surface protein 2 (msp2) gene and Random Amplified Polymorphic DNA (RAPD-PCR). Three of the 6 RAPD primers used (R8, E8, L12) yielded useful polymorphic patterns with higher genotyping rates (91-95%) than nested-PCR (67%). There was a significant difference between the 4 (3RAPD and msp2) primers used (ANOVA; P<0.001). Based on msp2, the Multiplicity of Infection in 2014 was greater than in 2018 (2.28 vs. 1.97). The calculated mean of GD parameters across years and markers showed RAPD-R8 had the highest index of GD (Shannon’s Index, Unbiased Nei-GD) except for the Percentage of Polymorphic Loci. These indices, based on RAPD-R8, confirmed the comparatively higher trend of GD in 2014 compared to 2018, respectively - 0.294 vs 0.246; 0.179 vs 0.156; 88.64 vs 75.0. Clustering analysis was also used as a proxy for population structure, with the msp2-based UPGMA distance-tree depicting intermixed and epidemiologically related clonal populations of 2014 and 2018 across sub-trees. Interestingly, the distance-tree based on RAPD-R8 primer revealed 2 clearly distinct unrelated clusters for 2014
Keywords
genetic diversity, MSP2, Plasmodium falciparum RAPD-PCR and population relocation
Article Details
1. Introduction
Malaria is a major human disease that affects people all over the world, especially in tropical and sub-tropical areas. Malaria is a preventable and curable disease, with an estimated 229 million infections and 409000 deaths worldwide in 2019 [1]. The African area of the World Health Organization (WHO) bears a disproportionately large percentage of the global malaria burden [1]. In order to alleviate this burden, WHO has refocused attention on the malaria-endemic nations and established a variety of malaria-reduction programs [2]. However, it is important not to exclude the chance that these treatments will only have a minor impact on the expected outcomes. Especially in the context of civil unrest, disasters, and other situations where health-care provision is disrupted, as well as an increase in human population migration from endemic areas, the epidemiological landscape of malaria burden can be drastically altered, with serious public-health consequences. It has been established that genetic relatedness exists. It has been documented that genetic relatedness and population structure can be used to determine P. falciparum and P. vivax transmission levels. This conclusion is based on growing genetic relatedness across infections as parasite populations diminish due to inbreeding [3]. Shrinking populations tend to generate moderately distinct pockets (or foci) of infection, resulting in population structure alterations that are typically influenced by external variables [3]. The population structure of P. falciparum, depends on the local epidemiology and demography, such as the incidence of infected people, the vector transmission intensity and movement of inhabitants (i.e., interchange across geo-graphical areas) [4, 5]. As a result, pathogen genetic diversity is an important component that can be used to detect small changes in the pathogen population, such as treatment resistance or evasion of the host immune system through antigenic variation [6]. The genotyping of malaria parasite populations is an important method in molecular epidemiological investigations of malaria to assess the types and number of parasite clones in an infection [7]. In places of high transmission intensity, P. falciparum parasites are known to have a lot of genetic diversity, and infected people in those communities often have a lot of different parasite clones with diverse genetic traits [8].The level of genetic variation of P. falciparum parasites in the micro-environment, on the other hand, remains largely unknown, posing a challenge to malaria control and elimination efforts. Analyzing the structure of P. falciparum populations on a broad scale, such as continents, or using markers that are subject to non-neutral selection can obscure and misunderstand the effective transmission process [4]. As a result, understanding the genetic structure and organization of P. falciparum populations in a limited location, such as Yaounde, Cameroon's political capital, could be more useful. For nearly five years, this cosmopolitan metropolis has seen massive population displacement from regions (Far North, South West, and North West) with differing malaria prevalence than Yaounde [9], which has been plagued by civil unrest. The social crisis affecting the country's north-west and south-west regions, as well as civil unrest in the country's far north, which has a displaced population, could have an impact on malaria epidemiology in the area, posing a huge challenge to malaria elimination and control efforts in these areas and neighboring regions where the displaced are relocated [9]. According to data from Cameroon's demographic and health survey (DHS) and malaria indicator survey (MIS), vegetation and altitude are major predictors of malaria's geographic distribution [10]. If internally displaced people (IDPs) are not well monitored or expected, they may increase the risk of transmission of a population of P. falciparum parasites with a separate or diversified genetic background. Estimating parasite population connectivity can help determine the danger of parasite spread across geographical areas. This information is critical for detecting large reservoirs ("sources") that support infections in other locations ("sinks") [3]. Human mobility data, such as patient travel histories or mobile phone data, can be used to estimate connectedness, but Noviyanti et al. (2013) cite two studies that demonstrate the potential of combining these data with parasite genetic data to reveal both local and long-distance parasite transmission routes [3]. In contrast to mobile phone data, which provides information on human population movement, genetic data provide insight into parasite gene flow, which may take different paths than people [3]. P. falciparum has a flexible genome, which allows researchers to measure the parasite's variety. Size polymorphism is caused by the loss of either coding or repetitive regions. Random amplified polymorphic DNA (RAPD) is a simple and quick procedure that uses very little genomic DNA and no sequencing, cloning, or hybridization, offering considerable advantages over other molecular techniques commonly employed in genomic characterization [11]. Furthermore, RAPDs markers have a very high genomic yield, can identify more polymorphisms, and are scattered randomly throughout the genome [12]. RAPD has been used to identify taxonomic groups as well as obtain genetic markers for various organisms [11, 13, 14]. However, in the realm of malaria, RAPD-PCR has been rarely used [11, 15-17]. For genotyping P. falciparum, several approaches based on DNA fingerprinting for microorganisms are used, including restriction fragment length polymorphism (RFLP) (msp1), nested PCR (msp2 and glurp), microsatellites, and single-stranded confirm polymorphism (SSCP), among others (11). In this investigation, we used msp2 and RAPD-PCR techniques to genotype P. falciparum field isolates found in Yaounde, Cameroon. The purpose of this study was to compare the genetic diversity of P. falciparum before and after intensive population relocation into Yaounde from malaria-endemic regions using merozoite surface protein 2 (msp2) and Random Amplified Polymorphic DNA (RAPD)-PCR, and to evaluate the performance of the methods used. Because more genetic variations can be reflected by utilizing a wide range of molecular marker systems [18].
2. Materials and Methods
2.1. Study design and Study site
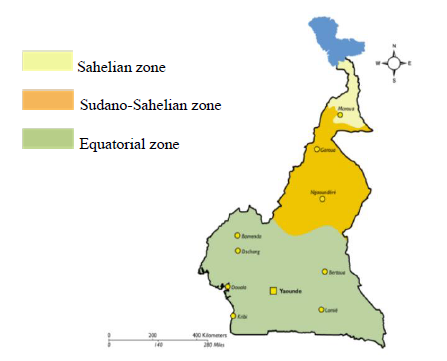
The samples of 2014 were archive samples gotten from a hospital based cross-sectional study that was conducted at the Etoug-Ebe Baptist Health Centre, Yaounde in 2014 while in 2018 a hospital based cross-sectional study was carried out at the Cite Verte District Hospital, Yaounde. Cameroon is made of 3 main malaria ecological zones which are the Sahelian zone, the Equatorial zone, and the Sudano-Sahelian zone (Figure 1B). The Sahelian area encompasses the Far North, which is typified by hot, dry weather with annual rainfall never surpassing 700 mm.The Far North is classified as a hyperendemic malaria stratum, with seasonal malaria parasite transmission prone to cyclic outbreaks, according to malaria stratification.With an estimated 3.9 million residents occupying a surface area of 34,263 km2, the Far North is one of the country's most densely populated regions [19].However, according to the NMCP annual report, the region still had the greatest number of malaria cases in the country in 2015 [9]. During the rainy season, the entomological inoculation rate (EIR) was estimated to range between 2.4-24.0 infective bites/person/month, with An. arabiensis as the primary vector species [9].
The West and North-West regions which are characterized by a temperate climate with up to 8 months of rainfall per year has a vegetation dominated by grasslands. They are both situated in highland areas (> 1000 m above sea level) and are considered to be areas of very low level of seasonal malaria parasite transmission (hypo-endemic pattern). Entomological inoculation rate in the North-West region was found to vary from 4.9 to 11 infective bites/person/year with Anopheles gambiae being the main vector here. The North-West covers a surface area of 17,300 km2 with over 1.9 million inhabitants [9]. In the highlands of the North-West region, current entomological investigations found parasite inoculation rates ranging from 4.9 to 11 infective bites/person/year. An. gambiae, An. coluzzii, and An. funestus are the main vectors in the area [9]. The forest domain encompasses the littoral, central, south-west, east, and south areas. A sequence of vegetation, including mangrove, deep equatorial evergreen forest, and humid savannah, characterizes these regions, which stretches from the Atlantic coast to the Central African Republic's border. The climate is divided into four seasons, two rainy and two dry, with annual rainfall ranging from 1500 mm/year inland to 4000 mm/year towards the coast. These areas are thought to be part of a holoendemic stratum with strong and long-term malaria parasite transmission. The most densely inhabited regions are the Littoral, Centre, and South-West, with populations ranging from 4.09 million in the Centre, 3.3 million in the Littoral, and 1.5 million in the South-West. Prior to the implementation of LLINs, the prevalence rate in children aged 6 months to 15 years varied between 35 and 85.4 percent (n = 109-1690) [19, 20]. Studies conducted in the South-West regionafter the scale-up of LLINs mass campaigns on children aged one month to 14 years old revealed a prevalence ranging from 9 to 41.5 percent during the rainy season. There was also a high parasite incidence of 41.7 to 56.2 percent along the slope of Mount Cameroon [9]. Malaria parasite prevalence decreased in the majority of settings in the South and Central regions, with estimates ranging from 6.6 to 29.5 percent (n = 2525). EIR values ranging from 0 to 100 infective bites/person/year were reported after scale-up in the Central region.
2.2. Study participants and ethical considerations
Targeted eligible participants who voluntarily consented to participate to the study were children aged between 6 months and 18 years and adults visiting the hospital for outpatient consultations. Enrolled participants were those presenting with a history of fever (four the last two days at least), who had a positive malaria test by rapid diagnostic test (RDT) and microscopy and respected the inclusion criteria. The parent or legal guardian of the children provided a written and signed informed consent. Ethical clearance (CE Nº 00832/ CRERSHC/ 2018) was obtained from the Centre Regional Ethics Committee for Human Health in Yaounde and administrative authorisation from both hospitals. A subpopulation of samples was randomly selected from the pool of samples collected from 2014 (50) and 2018 (50).
2.3. Rapid diagnostic testing for malaria
The SD Bioline Malaria Ag P.f/Pan kit was used for the diagnosis of malaria and contains a coated strip of monoclonal antibodies for the capture of plasmodium antigen in the blood. It is a one-step, qualitative and rapid immunochromatographic test based on the differential diagnosis of histidine-rich protein II (HRP-II) antigen specific to P. falciparum and antigenic isoforms of the enzymelactate dehydrogenase (LDH) associated with Pan species of Plasmodium (PspLDH). Briefly, 5μl of blood was dispensed into the sample well of the RDT cassette immediately followed by 3 drops of buffer into the buffer well. The liquid then migrates across the test strip transporting the test sample. Samples positive by RDT were further tested by microscopy for confirmation.
2.4. Sample collection and processing
Two (2) milliliters of venipuncture blood were collected and dispensed in pre-labelled EDTA tubes that were transported within 4 to 5 days after collection on ice pack to the Laboratory for Public Health Research and Biotechnologies of the University of Yaounde-I. Blood specimens were used to prepare slides and in case of a positive diagnosis by microscopy, 10μl of blood was spotted onto corresponding filter papers (Whatman 3, United Kingdom) which were allow to air-dry overnight at room temperature 20 - 25 ? before molecular analyses. Dried-blood spots (DBS) were then packaged into pre-labeled envelopes containing a desiccant and stored at room temperature in a clean environment. The idea of preserving blood using the dried blood spots (DBS) for diagnostic purposes was first to facilitate long-term conservation of the samples and also to avoid the incontinency of wet blood samples such as; it must be transported refrigerated or frozen and must remain in cold storage to prevent bacterial growth. And that strict regulation governs the transportation of liquid blood samples from one country to another. Whereas dried blood sample collection doesn't require cold chains for transportation or cold storage [21, 22].
2.5. Molecular Genotyping
2.5.1. Plasmodium DNA extraction
Plasmodial DNA was extracted from dried-blood spots using the Chelex®-100 (Bio-Rad Laboratories, Sigma) boiling methods as described by Plowe et al., (1995) [23]. Treatment with Chelex removes hemoglobin and other inhibitors of the Taq polymerase enzyme. Briefly, DBS were carefully stripped of the filter paper with a sterilized scissor and soaked into a lysing solution of 0.5% saponin (1.5ml) containing 1x Phosphate Buffer Saline (PBS). The tubes were inverted about 2-3 times and incubated a 4ºC overnight. Upon aspiration of the brown solution, the strips were rinsed with an additional 1ml solution of PBS inverted and allowed to incubate for further 15-30 min at 4°C. During this step a 50μl stock solution of 20% Chelex-100 was prepared by adding 150μl of DNAase free water in a 1.5ml Eppendorf tube which was then placed on a heating plate set a 100°C. The strips were then transferred into the corresponding tubes containing the hot chelex and vortexed for 30sec, replaced in the heating plate for 10 min followed by another brief vortex during and once after the incubation. After two rounds of centrifugation of 10000xg for 2 min, first after incubation and second after recovering the supernatant, the final DNA solution was stored at -20°C until molecular analyses.
2.5.2. Randomly Amplified Polymorphic DNA (RAPD)-PCR of P.falciparum
The application of the RAPD-based PCR method for genotyping does not require any specific knowledge of the DNA sequence of interest. This technique randomly amplifies segments of the genomic DNA under low annealing temperature using a random and short (approximately 10 nucleotide) primer [24]. Random portions of the Plasmodium falciparum genome were genotyped using 6 random primers (Table 1) and following a slightly adjusted protocol by Farooq et al., (2012b) and Howard et al., (1996). Briefly, 3μl of plasmodial DNA extract was subjected to a single-round PCR reaction of 25μl containing nuclease-free water (16.25μl), 10x of 2.5μl Thermopol Buffer, 10mM of 0.5μl dNTPs, 1μl RAPD primer, 0.25 Units Taq DNA polymerase (Bangalore Genei, Pvt. Ltd., India). Applied thermal cycling conditions were as follows: pre-denaturation 94°C for 3 min, followed by 47 cycles of denaturation at 94°C for 1 min, annealing 36°C for 90 secs, extension 72° C for 2 min, and final extension 72° C for 10 min.
2.5.3. Nested-PCR for the amplification of pfmsp2 gene
The msp2 gene was amplified in two-round nested PCR reactions following the protocol described by (12). First-round PCR mixture specifically consisted of 0.1μM of primers S2 and S3 (Table 1) and 400ng of plasmodial DNA extract in a final reaction volume of 25μl, while the second round specifically consisted of 0.1μM of primers S1 and S4 (Table 1) and 1μl of DNA amplicons from the first round in a total reaction volume of 25μl. In addition, all two reaction mixtures contained Thermopol Buffer (10X), dNTPs (200μM), and Taq Polymerase (0.25 units). Thermal cycling conditions for the 2 PCR reactions were: - pre-denaturation 94°C for 3 min (1st reaction) - followed by 30 cycles of denaturation at 94°C for 30s, annealing 42°C (1st reaction) / 50°C (2nd reaction) for 60s, elongation at 65°C (1st reaction) / 72°C (2nd reaction) for 2 min and, - final elongation 72°C for 3 min. All primers were collected from previous literature, and quality testing for specificity was done on the NCBI database using the Primer Blast and Primer 3 software. This was done to ensure that all of the RAPD and msp2 primers were specific to P. falciparum.
Table 1: Primers Sequences
MSP2: merozoite surface protein 2, RAPD: Random Amplification of Polymorphic deoxyribonucleic acid, single RAPD primers (E-4, E-8, R-8, L-10, L-8 and L-12)
MSP2: merozoite surface protein 2, RAPD: Random Amplification of Polymorphic deoxyribonucleic acid, single RAPD primers (E-4, E-8, R-8, L-10, L-8 and L-12)
2.5.4. Agarose gel electrophoresis for the verification of PCR products
A qualitative analysis was performed by electrophoresis on an agarose (Seakem Nusieve) gel to verify all PCR products. Briefly, the casting tray containing the gel initially mixed with 2.5μl of the DNA interchelator Ethidium Bromide (EtBr), was inserted in the electrophoretic chamber of a minigel tank filled with 1X Tris-Borate EDTA buffer. Five (5.0μl) microliters of PCR products were gently mixed with 2.5μl of loading dye to monitor the migration of samples and, allowed to sink into the corresponding sample wells. For good separation, samples were allowed to run for approximately 45 min alongside molecular weight markers to determine the actual size of DNA bands of interest. We used a UV light trans-illuminator (High Performance) to visually detect in positive samples DNA bands, which fluoresce under UV light in the presence of the EtBr stain.
2.6. Data analysis
The presence or absence of reproducible bands on the gels was graded. Each band was thought of as a locus with two possible alleles. The msp2 and RAPD data were converted into a binary data matrix as discrete variables (1 = presence and 0 = absence). Analysis of diversity was performed using the combination of msp2 and RAPD data to uncover the extent of P. falciparum DNA polymorphisms and increase the reliability of the results. We used the GenAIEx 6.51 software, to determine parameters of genetic diversity: the percentage of polymorphic loci (PPL), Shannon's information index (SI), and the unbiased Nei's gene diversity index (H) [25]. Determination of the total genetic variation among samples was done using the phi-statistic for the Analysis of Molecular Variance (AMOVA). The total genetic variation is divided into two categories: within populations (Phi-PT) and among populations (per year) [25]. Clustering analysis was implemented with MEGA10, by computing the distance-tree based on the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) algorithm which uses Nei’s Genetic Distance. Mean genetic distances were compared in SPPS (v.25), by the independent sample T-test, and the Cohen's effect size was obtained [26]. The performances of the primer-based genotyping techniques used were checked with the One-Way ANOVA statistics and a multiple test comparison applied to verify the direction of differences if any.
2.7. Multiplicity of infection (MOI).
The mean number of P. falciparum genotypes per infected individual was used to calculate the multiplicity of infection (MOI). The MOI was determined by dividing the total number of P. falciparum genotypes for the same gene by the number of PCR positive isolates. Polyclonal infections were defined as isolates with more than one genotype after msp2 amplification, whereas monoclonal infections were defined as the presence of a single allele [27]. MOI was only determined for msp2 primers because RAPD-PCR typically underestimates within-population variance while yielding comparable or somewhat greater population differentiation values [28-30].
3. Results
Of the 100 participants sampled, females were predominant (58%) compared to males (42%). The median age of the population was 31 years [range: 6 months to 744 months].
3.1. Molecular Genotyping
3.1.1. nested-PCR of msp2gene
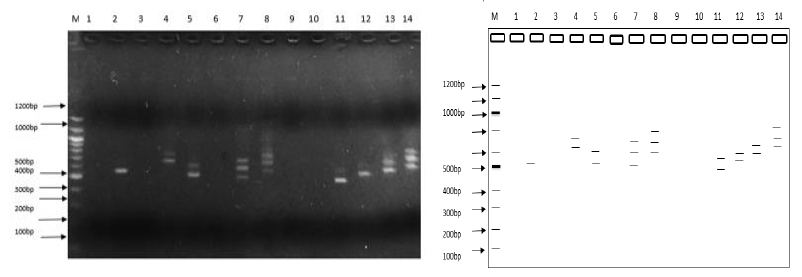
Several DNA bands were obtained after nested-PCR with size ranging from 401bp to 861bp, characteristic of the msp2 gene. The overall genotyping success rate was 67% (67/100) Mixed P. falciparum infections, defined as the presence of more than one msp2 allele or band on the electrophoregram, were detected in 26 of the 38 samples effectively genotyped for 2018 (68.42%) and 17/29 (58.62%) for 2014.
Double infections were the most dominant in 2018 (56%), while single infections were the most dominant in 2014 (42%).. The overall mean multiplicity of infection (MOI) for 2018 was 1.97 (75/38) and 2.28 (66/29) for 2014.
Table 2: Parameters of genetic diversity by population (2014 and 2018)
|
Genetic Markers |
||||
|
Parameters |
msp2 |
RAPD-R8 |
RAPD-E8 |
RAPD-L12 |
|
Genotyping Success Rate for 100 isolates (%) |
67 |
91 |
95 |
93 |
|
Range of (bp) |
401 - 861 |
361 - 1341 |
161 - 1341 |
261 - 1341 |
|
Percentage of Single Infection 2018 (%) |
31 |
- |
- |
- |
|
Percentage of Single Infection 2014 (%) |
42 |
- |
- |
- |
|
Percentage of Mixed Infection 2018 (%) |
69 |
- |
- |
- |
|
Percentage of Mixed Infection 2014 (%) |
58 |
- |
- |
- |
|
Multiplicity Of Infection (2018) |
1.973 |
- |
- |
- |
|
Multiplicity Of Infection (2014) |
2.276 |
- |
- |
- |
|
Percentage of Polymorphic Loci for 2018 (%) |
80.95 |
75 |
97.3 |
75.56 |
|
Percentage of Polymorphic Loci for 2014 (%) |
85.71 |
88.64 |
54.05 |
84.44 |
|
Shannon’s Information Index (2018) |
0.248 |
0.246 |
0.0379 |
0.254 |
|
Shannon’s Information Index (2014) |
0.247 |
0.294 |
0.153 |
0.265 |
|
Unbiased Nei’s Genetic Diversity (2018) |
0.143 |
0.156 |
0.238 |
0.157 |
|
Unbiased Nei’s Genetic Diversity (2014) |
0.14 |
0.179 |
0.089 |
0.161 |
|
Pairwise Population matrix of Nei Genetic Diversity Nei (%) |
1 |
4.2 |
2.5 |
2.6 |
|
Mean % of Polymorphic Loci (%) |
83.3 |
81.82 |
75.65 |
80 |
|
Mean Shannon’s Information Index |
0.25 |
0.27 |
0.096 |
0.26 |
|
Mean unbiased Genetic Diversity |
0.142 |
0.168 |
0.164 |
0.159 |
3.2. RAPD-PCR genotyping
While nested-PCR technique uniquely targets the msp2 gene, RAPD markers bind and amplify at random segments of genomic DNA without prior information about the DNA. Of the 6 primers used for RAPD analysis, 4 primers (R8, E4, E8 and L12) generated PCR products, with 3 of them (E4, E8, and R8) having useful polymorphic patterns. Genotyping with the other 2 primers (L8, and L10) resulted in failure.
3.2.1. Parameters of Genetic Diversity and Differences between RAPD-primers
Random amplification of P. falciparum with the R8 primer yielded bands of varying sizes between 361bp and 1341bp; a total of 232 bands for 2018 and a total of 236 bands for 2014. Supplementary materials (3 and 4) have been provided that illustrate the electrophoretic profiles of amplified DNA bands for primers L12, E4, and E8. When the mean of parameters of genetic diversity was determined for the two study time points and across markers, primer R8 gave the highest genetic indices (Table 2). Except for the percentage of polymorphic loci (PPL) which had the highest value with the msp2 marker. Contrary to the 2018-population, the 2014-population had the highest genetic diversity for all the parameters measured: PPL, Shannon Index, Unbiased Nei-genetic diversity (Table 2). R8 was therefore considered as the most discriminatory of the 4 markers considered in the table. Besides multiple test comparison for the one-way ANOVA test demonstrated a significant difference in the performance of each of the primer (Tukey Post-hoc test; P<0.001) (Table 3).
Table 3: Genetic distance One-Way ANOVA: Post Hoc Multiple Comparison Tukey Ba, b
|
Primer |
N |
Subset for alpha = 0.05 |
|||
|
1 |
2 |
3 |
4 |
||
|
MSP2 |
1095 |
.159 |
|||
|
RAPD L12 |
2172 |
.170 |
|||
|
RAPD R8 |
2027 |
.177 |
|||
|
RAPD E8 |
2209 |
.205 |
|||
Means for groups in homogeneous subsets are displayed
- Uses Harmonic Mean Sample Size = 1724.375
- At the 0.001 level, the correlation is significant (2-tailed)
3.3. Genetic differentiation between populations and levels of gene flow
The Analysis of Molecular Variance (AMOVA) indicated that there were considerable genetic differences between the groups. Specifically, 90% of the overall genetic diversity was identified within the individual populations, with the remaining 10% genetic variance derived from both populations. According to the permutation analyses, both levels contributed significantly to the overall genetic variance (Table 4).
Table 4: Summary AMOVA Table
|
Source |
(df) |
SS |
MS |
Est. Var. |
Per % |
Phi Statistic |
Value |
P |
|
Among Pops |
1 |
73.230 |
73.230 |
1.247 |
10% |
Phi’PT |
0.120 |
0.01 |
|
Within Pops |
98 |
1068.580 |
10.904 |
10.904 |
90% |
PhiPT |
0.1025 |
0.01 |
|
Total |
99 |
1141.810 |
12.150 |
100% |
d.f. stands for degree of freedom. ss stands for the sum of squared observations. MS stands for mean of squared observations. Phi'PT, the proportion of total genetic variance owing to variance between populations; PhiPT, the proportion of total genetic variance attributable to variance among individuals within specific populations (each period).
3.4. Clustering analysis
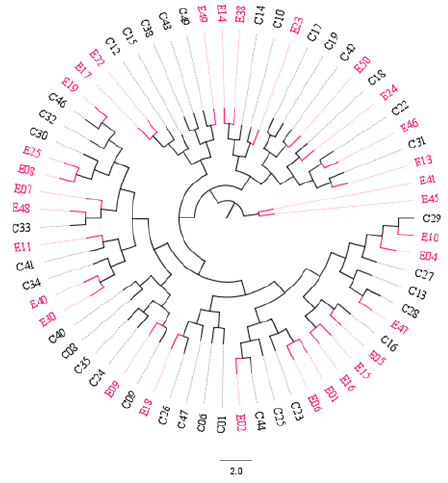
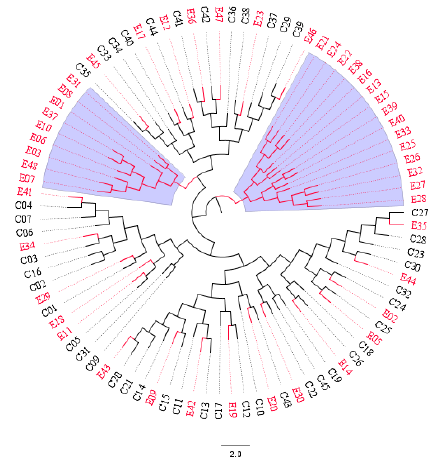
The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) algorithm was implemented to compute two distance-trees for clustering analysis based on msp2 and RAPD-R8 markers. As shown in Figure 3, isolates collected in both time points intermix with each other across sub-trees based on the msp2 gene: 2014 (red) and 2018 (black). However, a cluster pair was seen (E41 and E45). When the RAPD-R8 marker was used to compute the dendogram clusters were also obtained that depict intermingled isolates for both 2014 (red) and 2018 (black). Interestingly, two specific clusters (blue-highlighted) were obtained with only parasites populations samples in 2014 (red). These are characteristic of parasites populations with different genetic structures.
Figure 3: Distance-tree based on the msp2 gene depicting intermixed clonal Plasmodium falciparum parasites population across sub-trees. E = samples collected from the Etoug-Ebe hospital in 2014 before population intensive and massive relocation to Yaounde, the capital city of Cameroon; and C = samples collected from Cite-Verte in 2018 after intensive and massive relocation to Yaoundé. All samples were collected in Yaounde. The circular graphical representation is for visual purposes
Figure 4: Distance-tree based on the RAPD-R8 marker depicting: (i) intermixed clonal Plasmodium falciparum parasites population from 2014 (red) and 2018 (black) across sub-trees, and (ii) two sub-clusters highlighted in blue with only isolates from 2014. Parasites populations isolates were sampled before (2014) and after (2018) the intensive and massive relocations of the population into Yaounde, the capital city of Cameroon. The circular graphical representation is for visual purposes.
4. Discussion
Some of the 100 P. falciparum isolates from 2014 and 2018 were eliminated because good amplification bands were not obtained using the RAPD or msp2 primers. Allelic genotyping revealed that P. falciparum in Yaounde samples was polymorphic in terms of msp2, RAPD-R8, RAPD-E8, and RAPD-L12. A better understanding of P. falciparum genetic variation provides insight into the pathogenesis of malaria, the process of building anti-malarial immunity, mechanisms of resistance to therapy, and transmission settings. In 2018, Zhong and colleagues proposed that the number of co-infections inside a host could be a significant predictor of transmission intensity [31]. In this study, the RAPD dominant molecular markers that produced useful polymorphic patterns (E8, R8, and L12) were utilized in conjunction with the co-dominant marker (msp2) to assess the genetic diversity of the P. falciparum populations in Yaounde [29]. For the years 2014 and 2018, sixty-eight percent (45/67) and fifty-eight percent (39/67) of the samples included multiple msp2 alleles (mix / polyclonal infection). This was consistent with the findings of Doumbe-Belisse and colleagues in 2018, who concluded that Yaounde was a malaria transmission hotspot. This investigation revealed that the majority of malaria-infected samples in Yaounde are polyclonally infected. Our research population contained a high number of parasite genotypes, with an average MOI of 2.28 in 2014 and 1.97 in 2018. These findings are consistent with the findings of Koumbo and colleagues in 2019, who discovered a comparable MOI of 2.0 in Mfou, Centre region, Cameroon, as in 2018. We obtained 14.3 percent genetic diversity in 2018, which was lower than the 18 percent obtained by Koumbo and colleagues in 2019 in Mfou, Cameroon's Centre province [32].
Despite the fact that both settlements are in the same malaria ecozone (equal inoculation rate). This may be because the two places have very distinct urban settings. It could also be explained by the fact that transmission in Yaounde is high and sustained by malaria importation, while transmission in Mfou is not sustained by malaria importation. The genotyping success rate for the different primers in this investigation was (67 percent, 95 percent, 91 percent, and 93 percent for msp2, RAPD-E8, RAPD-R8, and RAPD-L12, respectively). This is consistent with the findings of a study conducted by Collard et al., in 2009 [33] that suggested that RAPD markers are easier to utilize for genotyping genetic variants than msp2 primers. When compared to co-dominant markers (msp2), dominant markers (RAPD primers) typically underestimate within-population variation while producing equivalent or somewhat greater population differentiation values [28-30]. Because dominant markers cannot differentiate between homozygotes and heterozygotes, these differences were expected [34]. Despite these drawbacks, dominant markers are distinguished by their ability to estimate unbiased genetic variation and lack of a sequence requirement, making them suitable for investigations of species with little or no genetic information [34]. High levels of genetic variability are typically observed in natural populations of P. falciparum that reproduce sexually and asexually, have diverse ecological niches, and/or have a large geographical range [35]. Our findings revealed a statistically significant difference in the level of genetic polymorphism between the two populations. Similar variation levels were previously seen in Eswatini isolates with substantial parasite diversity, corresponding with high rates of malaria importation and little local transmission [36]. Because of the relatively tiny impact size, the statistically significant difference between the two populations from the Independent Sample T-test studies that showed a significant difference between the two populations was inconsequential (-0.14). Table 1 (Supplemental) Even though the impact size is statistically significant, the difference seen is inconsequential if the effect size is less than 0.2 standard deviations [37]. The distance-trees obtained with both markers msp2 and RAPD-R8, suggest clonal parasites populations for 2014 and 2018 with similar genetic background as they intermix across subtrees. The second tree revealed a homogeneous clonal population individualized and separated from the rest of intermixed parasites in other sub-trees, suggesting a different genetic background. These may have consequences in the detectability and diagnosis of new clones which may come with higher virulence. These observations warrant in-depth investigation into how human migration redistribute malaria diversity and the interest of targeting populations with different genetic background for diagnostic purpose. The natural populations of P. falciparum in Yaoundé for both periods were composed of mostly a homogenous population as seen on the phylogenetic trees (Figure 3 and 4). These could signify that 2018 isolate could be clonal strain created from the inbreeding between the imported isolates with 2014 isolate, which resulted in shrinking populations (clonal population) of P. falciparum, forming moderately distinct pockets from the 2014 isolates that clustered separately (Figure 3). This support the result of the Cohen’s effect size that suggested that the difference in genetic polymorphism observed between the populations are negligible. It was inferred that the degree of differentiation between populations could be affected by the level of gene flow [38]. The population structure was coordinated in various ways by two forces: gene flow and genetic drift. A higher degree of gene flow would result in a more uniform population structure with less genetic differentiation. The gene flow between P. falciparum populations was clearly greater than that between populations from both eras [38]. Although, in theory, the degree of gene flow may not be sufficient to counteract the local genetic differentiation between some populations, the differentiation generated by genetic drift may be avoided [39]. The cluster analysis results supported the idea that the genetic relationships between populations were distinct to some extent (Figure 3 and 4). One of the study's drawbacks was that only two technics were utilized to assess P. falciparum genetic diversity, that the great majority of surface markers were not included, and that the sample size was small. Because of these constraints, the findings of this study are not the greatest for interpreting the relationship that exists between heavy human migration and genetic diversity. As a result, the findings of this study should be interpreted with caution. Our study reported low genetic diversity between the parasites isolates of 2014 and 2018 of Yaounde, Cameroon. However, the findings of this study do not corroborate the previous finding in the same region of Cameroon(Metoh et al, 2020), but is consistent with finding of Wanji et al., 2012.
5. Conclusions
Findings from this study highlight the importance of genetic diversity and how it can be altered if ecological conditions so permit. P. falciparum genetic diversity was higher in 2014, which probably increased the genetic split of the 2014 parasites population. This may lead eventually to a different phenotype requiring different measures of interventions. However, similar genetic structures seen across the sub-trees may be beneficial for generalize interventions. Therefore, molecular diagnosis remains crucial and must be fine-tuned to decipher complex genetic structures of parasites for a better surveillance.
Author contributions
Conceptualization, WFM, AMN, PTNN, JPC, JDB and OAA. Data curation, AMN, AHEM and AASN, Formal analysis, AASN, LMFK and SK. Investigation, AASN and TM. Methodology, PTNN and JPC. Software, AMN, AHEM, Supervision, WFM and AMN. Validation, WFM, AMN, PTNN and AHEM. Writing - original draft, AASN and AHEM. Writing - review & editing, WFM, AMN,CTF, LNN PTNN, JPC, JDB, OAA, IMA, AHEM, AASS and CMM. All authors contributed in the revision of the manuscript and approved the final version of the manuscript prior to submission.
Funding
This research was funded by the Malaria Research Capacity Development in West and Central Africa (MARCAD) Consortium through the Developing Excellence in Leadership, Training and Science (DELTAS) Africa Initiative (grant # DEL-15-010) to the University of Yaounde I. The DELTAS Africa Initiative is an independent funding scheme of the Alliance for Accelerating Excellence in Science in Africa (AESA) of the African Academy of Sciences (AAS) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (grant # 107741/A/15/Z) and the United Kingdom (UK) government. WFM, AMJ, IMA .and PTNN are supported by MARCAD.
Institutional Review Board Statement
The ethical clearance (CE No 00832/CRERSHC/ 2018) was obtained from the Centre Regional Ethics Committee for Human Health Research in Yaounde. The study was conducted according to the guidelines of the Declaration of Helsinki.
Informed Consent Statement
Prior to enrolment, written informed consent was obtained from parents and/or legal guardians of children and adult participants involved in this study.
Acknowledgments
We thank the administrative staffs of the Cité-Vert health district and Etoug-Ebe health district for their humble contributions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World malaria report 2019 [Internet]. [cited 2021 Aug 23].
- Anthony TG, Conway DJ, Cox-Singh J, Matusop A, Ratnam S, Shamsul S, et al. Fragmented Population Structure of Plasmodium falciparum in a Region of Declining Endemicity. J Infect Dis [Internet] 191 (2005): 1558-64.
- Noviyanti R, Miotto O, Barry A, Marfurt J, Siegel S, Thuy-Nhien N, et al. Implementing parasite genotyping into national surveillance frameworks: Feedback from control programmes and researchers in the Asia-Pacific region. In: Malaria Journal [Internet]. BioMed Central (2020): 1-20.
- Pumpaibool T, Arnathau C, Durand P, Kanchanakhan N, Siripoon N, Suegorn A, et al. Genetic diversity and population structure of Plasmodium falciparum in Thailand, a low transmission country. Malar J [Internet] 8 (2009): 155.
- Knudson A, González-Casabianca F, Feged-Rivadeneira A, MF P, Aponte S, Olaya A, et al. Spatio-temporal dynamics of Plasmodium falciparum transmission within a spatial unit on the Colombian Pacific Coast. [Internet] 10 (2020): 3756.
- Cui L, Escalante AA, Imwong M, Snounou G. The genetic diversity of Plasmodium vivax populations [Internet]. Vol. 19, Trends in Parasitology (2003): 220-6.
- Churcher TS, Cohen JM, Novotny J, Ntshalintshali N, Kunene S, Cauchemez S. Measuring the Path Toward Malaria Elimination. Science [Internet] 344 (2014): 1230.
- SI O, PU B, SA O, Ojurongbe O, HO A. Genetic diversity and complexity of Plasmodium falciparum infections in the microenvironment among siblings of the same household in North-Central Nigeria. Malar J [Internet] 19 (2020): 338.
- Antonio-Nkondjio C, Ndo C, Njiokou F, Bigoga JD, Awono-Ambene P, Etang J, et al. Review of malaria situation in Cameroon: Technical viewpoint on challenges and prospects for disease elimination. Parasites and Vectors [Internet] 12 (2019): 1-23.
- Massoda Tonye SG, Kouambeng C, Wounang R, Vounatsou P. Challenges of DHS and MIS to capture the entire pattern of malaria parasite risk and intervention effects in countries with different ecological zones: The case of Cameroon. Malar J [Internet] 17 (2018): 1-14.
- Farooq U, Dubey ML, Shrivastava SK, Mahajan RC. Genetic polymorphism in plasmodium falciparum: Differentiation of parasite isolates of high & low virulence by RAPD. Indian J Med Res [Internet] 136 (2012): 292-5.
- Sonnier L. Techniques de biologie moleculaire pour la caractérisation des moustiques (1999): 1-84.
- Feikin DR, Njenga MK, Bigogo G, Aura B, Aol G, Audi A, et al. Viral and bacterial causes of severe acute respiratory illness among children aged less than 5 years in a high malaria prevalence area of Western Kenya 32 (2007-2010).
- HADRYS H, BALICK M, SCHIERWATER B. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol 1 (1992): 55-63.
- Howard J, Carlton JMR, Walliker D, Jensen JB. Use of random amplified polymorphic DNA (RAPD) technique in inheritance studies of Plasmodium falciparum. J Parasitol 82 (1996): 941-6.
- Lescuyer P, Picot S, Bracchi V, Burnod J, Austin J, Pérard A, et al. Detection of RAPD markers correlated with chloroquine resistance in Plasmodium falciparum. Genome Res 7 (1997): 747-53.
- Waghmare VN, Bagde US. Random amplified polymorphic DNA based genetic characterization of four important species of Bamboo, found in Raigad district, Maharashtra State, India. African J Biotechnol 12 (2013): 4446-52.
- Luo C, He X hua, Chen H, Ou S jin, Gao M ping, Brown JS, et al. Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem Syst Ecol [Internet] 39 (2011): 676-84.
- Nyasa RB, Zofou D, Kimbi HK, Kum KM, Ngu RC, Titanji VPK. The current status of malaria epidemiology in Bolifamba, atypical Cameroonian rainforest zone: An assessment of intervention strategies and seasonal variations. BMC Public Health [Internet] 15 (2015): 1-12.
- Teh RN, Sumbele IUN, Meduke DN, Ojong ST, Kimbi HK. Malaria parasitaemia, anaemia and malnutrition in children less than 15 years residing in different altitudes along the slope of Mount Cameroon: Prevalence, intensity and risk factors 11 Medical and Health Sciences 1117 Public Health and Health Services. Malar J [Internet] 17 (2018): 1-13.
- Guthrie R. why use dried blood tests vs. wet blood tests? (2021): 1-7.
- Grüner N, Stambouli O, Ross RS. Dried Blood Spots - Preparing and Processing for Use in Immunoassays and in Molecular Techniques (2015): 1-9.
- Plowe C V., Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: Polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 52 (1995): 565-8.
- Panigrahi P, Panigrahi KK, Kalai- B, Gram B. Analysis of Genetic Diversity in Black Gram ( Vigna mungo L . Hepper ) Genotypes using RAPD Markers Name of the Crops 10 (2017): 2570-7.
- Peakall R, Smouse PE. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28 (2012): 2537-9.
- Becker LA. Effect Size (ES) (2000).
- AA F, Nate E, GL AH, Barnadas C, MW H, Iga J, et al. Nationwide genetic surveillance of Plasmodium vivax in Papua New Guinea reveals heterogeneous transmission dynamics and routes of migration amongst subdivided populations. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis [Internet] 58 (2018): 83-95.
- Lynch M, Milligan BG. Analysis of population genetic structure with RAPD markers. Mol Ecol 3 (1994): 91-9.
- H N. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol [Internet] 13 (2004): 1143-55.
- Soldati MC, Fornes L, Van Zonneveld M, Thomas E, Zelener N. An assessment of the genetic diversity of Cedrela balansae C. DC. (Meliaceae) in Northwestern Argentina by means of combined use of SSR and AFLP molecular markers los Reseros y N. Repetto (ex Las Cabañas) s. Biochem Syst Ecol [Internet] 47 (2013): 45-55.
- Zhong D, Koepfli C, Cui L, Yan G. Molecular approaches to determine the multiplicity of Plasmodium infections. Malar J [Internet] 23 (2018): 172.
- Xu J. Fundamentals of fungal molecular population genetic analyses. Vol. 8, Current Issues in Molecular Biology (2006): 75-90.
- Collard BCY, Mackill DJ. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol Biol Report 27 (2009): 86-93.
- Xu J. Fundamentals of fungal molecular population genetic analyses [Internet] 8 (2006): 75-90.
- James TY PD. (1999) Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune. [Internet] (2021) 1665-77.
- Roh ME, Roh ME, Tessema SK, Murphy M, Nhlabathi N, Mkhonta N, et al. The Journal of Infectious Diseases High Genetic Diversity of Plasmodium falciparum in the Low-Transmission Setting of the Kingdom of Eswatini. J Infect Dis ® [Internet] 220 (2019): 1346-54.
- McLeod S. What does effect size tell you? Simply psychology. Retriviewed 8 March, 2021 from (2019): 2-5.
- Wright S. THE GENETICAL STRUCTURE OF POPULATIONS.
- Slatkin M. the Geographic Structure of. Science (80- ) 236 (1987): 787-92.