Gating Of the Channel Pore of Ionotropic Glutamate Receptors with Bacterial Substrate Binding Proteins: Functional Coupling of the Ectoine Binding Protein Ehub to Glur0
Article Information
Max Bernhard1, Bodo Laube1,2*
1Department of Biology, Neurophysiology and Neurosensory Systems, Technical University of Darmstadt, Schnittspahnstrasse 3, 64287 Darmstadt, Germany
2Centre for Synthetic Biology, Technical University of Darmstadt, 64283 Darmstadt, Germany
*Corresponding author: Bodo Laube, Department of Biology, Neurophysiology and Neurosensory Systems, Technical University of Darmstadt, Schnittspahnstrasse 3, 64287 Darmstadt, Germany.
Received: 17 October 2023; Accepted: 25 October 2023; Published: 01 November 2023
Citation: Max Bernhard, Bodo Laube. Gating Of the Channel Pore of Ionotropic Glutamate Receptors with Bacterial Substrate Binding Proteins: Functional Coupling of the Ectoine Binding Protein Ehub to Glur0. Journal of Biotechnology and Biomedicine. 6 (2023): 554-564.
View / Download Pdf Share at FacebookAbstract
Mammalian neuronal tetrameric ionotropic glutamate receptors (iGluRs) are thought to have originally arisen from the fusion of a bacterial substrate binding protein (SBP) with an inverted potassium channel. This hypothesis is based on structural and sequential similarities between the ligand binding and channel domains of iGluR subunits with SBPs and potassium channels. Ligand binding occurs at the interface between two lobed domains in both ligand binding domains (LBDs) and leads to closure of the shell-like structure, which is considered to be a key element in the transition from ligand recognition to ion channel gating in iGluRs. Here we report the functional coupling of the ectoine-binding protein EhuB to the channel pore of the GluR0 receptor. Fusion of an unmodified EhuB-binding protein to the transmembrane domain of GluR0 did not result in activation of the channel pore. Only by stabilizing the inserted EhuB-binding domain with a dimerization interface the resulting chimera was activated by ectoine, resembling the activation properties of other iGluRs. These results demonstrate the functional compatibility of SBPs to the gate of the channel pore of an iGluR and highlight the role of LBD dimerization in the functional evolution of iGluRs. Based on the high specificity and affinity of SBPs for an incredible variety of substrates, our results demonstrate the competence of SBP/ion channel chimeras for the development of new Biosensors for specific recognition of analytes by functionally linking a bacterial binding protein to the channel pore of an iGluR.
amino articles amino Research articles amino review articles amino PubMed articles amino PubMed Central articles amino 2023 articles amino 2024 articles amino Scopus articles amino impact factor journals amino Scopus journals amino PubMed journals amino medical journals amino free journals amino best journals amino top journals amino free medical journals amino famous journals amino Google Scholar indexed journals C-terminal domain articles C-terminal domain Research articles C-terminal domain review articles C-terminal domain PubMed articles C-terminal domain PubMed Central articles C-terminal domain 2023 articles C-terminal domain 2024 articles C-terminal domain Scopus articles C-terminal domain impact factor journals C-terminal domain Scopus journals C-terminal domain PubMed journals C-terminal domain medical journals C-terminal domain free journals C-terminal domain best journals C-terminal domain top journals C-terminal domain free medical journals C-terminal domain famous journals C-terminal domain Google Scholar indexed journals Substrate binding articles Substrate binding Research articles Substrate binding review articles Substrate binding PubMed articles Substrate binding PubMed Central articles Substrate binding 2023 articles Substrate binding 2024 articles Substrate binding Scopus articles Substrate binding impact factor journals Substrate binding Scopus journals Substrate binding PubMed journals Substrate binding medical journals Substrate binding free journals Substrate binding best journals Substrate binding top journals Substrate binding free medical journals Substrate binding famous journals Substrate binding Google Scholar indexed journals Ionotropic Glutamate articles Ionotropic Glutamate Research articles Ionotropic Glutamate review articles Ionotropic Glutamate PubMed articles Ionotropic Glutamate PubMed Central articles Ionotropic Glutamate 2023 articles Ionotropic Glutamate 2024 articles Ionotropic Glutamate Scopus articles Ionotropic Glutamate impact factor journals Ionotropic Glutamate Scopus journals Ionotropic Glutamate PubMed journals Ionotropic Glutamate medical journals Ionotropic Glutamate free journals Ionotropic Glutamate best journals Ionotropic Glutamate top journals Ionotropic Glutamate free medical journals Ionotropic Glutamate famous journals Ionotropic Glutamate Google Scholar indexed journals Bacterial† articles Bacterial† Research articles Bacterial† review articles Bacterial† PubMed articles Bacterial† PubMed Central articles Bacterial† 2023 articles Bacterial† 2024 articles Bacterial† Scopus articles Bacterial† impact factor journals Bacterial† Scopus journals Bacterial† PubMed journals Bacterial† medical journals Bacterial† free journals Bacterial† best journals Bacterial† top journals Bacterial† free medical journals Bacterial† famous journals Bacterial† Google Scholar indexed journals nervous system articles nervous system Research articles nervous system review articles nervous system PubMed articles nervous system PubMed Central articles nervous system 2023 articles nervous system 2024 articles nervous system Scopus articles nervous system impact factor journals nervous system Scopus journals nervous system PubMed journals nervous system medical journals nervous system free journals nervous system best journals nervous system top journals nervous system free medical journals nervous system famous journals nervous system Google Scholar indexed journals chemical† articles chemical† Research articles chemical† review articles chemical† PubMed articles chemical† PubMed Central articles chemical† 2023 articles chemical† 2024 articles chemical† Scopus articles chemical† impact factor journals chemical† Scopus journals chemical† PubMed journals chemical† medical journals chemical† free journals chemical† best journals chemical† top journals chemical† free medical journals chemical† famous journals chemical† Google Scholar indexed journals mechanism† articles mechanism† Research articles mechanism† review articles mechanism† PubMed articles mechanism† PubMed Central articles mechanism† 2023 articles mechanism† 2024 articles mechanism† Scopus articles mechanism† impact factor journals mechanism† Scopus journals mechanism† PubMed journals mechanism† medical journals mechanism† free journals mechanism† best journals mechanism† top journals mechanism† free medical journals mechanism† famous journals mechanism† Google Scholar indexed journals periplasmic† articles periplasmic† Research articles periplasmic† review articles periplasmic† PubMed articles periplasmic† PubMed Central articles periplasmic† 2023 articles periplasmic† 2024 articles periplasmic† Scopus articles periplasmic† impact factor journals periplasmic† Scopus journals periplasmic† PubMed journals periplasmic† medical journals periplasmic† free journals periplasmic† best journals periplasmic† top journals periplasmic† free medical journals periplasmic† famous journals periplasmic† Google Scholar indexed journals ectoine† articles ectoine† Research articles ectoine† review articles ectoine† PubMed articles ectoine† PubMed Central articles ectoine† 2023 articles ectoine† 2024 articles ectoine† Scopus articles ectoine† impact factor journals ectoine† Scopus journals ectoine† PubMed journals ectoine† medical journals ectoine† free journals ectoine† best journals ectoine† top journals ectoine† free medical journals ectoine† famous journals ectoine† Google Scholar indexed journals
Article Details
Abbreviations:
AMPA: α-amino-3-hydroxy-5-methyl-4-isoaxazolepropionic acid; CTD: C-terminal domain; iGluR: Ionotropic glutamate receptor; LBD: Ligand binding domain; NTD: N-terminal domain; RMSD: Root-mean-square deviation; SBD: Substrate binding domain; SBP: Substrate binding protein; TMD: Transmembrane domain
Introduction
Ionotropic glutamate receptors (iGluRs) mediate the majority of rapid excitatory neurotransmission in the vertebrate central nervous system by converting a chemical signal (the release of the neurotransmitter glutamate) into the opening of a cation- permeable ion channel [1]. Based on their pharmacological properties, these iGluRs can be classified into four subfamilies: α-amino-3-hydroxy-5- methyl-4-isoaxazolepropionic acid (AMPA), kainate (KA), N-methyl-D-aspartate (NMDA), and δ-receptors. Other members of the iGluR family are found throughout the animal kingdom [2], in plants [3], and bacteria [4]. Despite their different physiological functions, all eukaryotic iGluRs share the same modular architecture (Fig. 1a). They consist of an extracellular N- terminal domain (NTD), an extracellular ligand-binding domain (LBD), a transmembrane domain (TMD; TM1, TM2, TM3) including the pore loop (P), and an intracellular C-terminal domain (CTD). The origin of this modular architecture is largely unknown. The common complex architecture of eukaryotic iGluRs suggests an ancient separation of the protein family [3,5] with a common ancestor that can be traced back to bacteria [4]. Prokaryotic iGluR subunits, such as GluR0 [4], have a simplified architecture, lacking the third transmembrane segment TM3 and an NTD, while exhibiting unique functional and pharmacological properties, such as a potassium selectivity filter and the ability to be activated by a wide range of amino acids. Interestingly, individual domain segments of iGluRs share structural similarities with other prokaryotic protein families. For example, the LBD of iGluRs is structurally homologous to bacterial class II solute-binding proteins (SBP) [6] and the TMD exhibits an inverted architecture of tetrameric K+ channels [7,8], leading to the proposal that iGluRs may have arisen from the fusion of a SBP and potassium channels [9]. Recent findings confirm the general relationship between K+ channels and iGluRs by coupling an iGluR LBD to a small viral K+ channel [10], suggesting a conserved activation mechanism of the channel pore in both protein families.
However, the sequence identities between the LBDs of iGluRs and SBPs are low [11] and the evolutionary link between the two remains unclear. SBPs mediate the uptake of a variety of substances across the cell membrane [12] and are also involved in chemotaxis and DNA regulation [13]. Despite low sequence similarity, SBPs, like iGluR LBDs, display a highly conserved three-dimensional shell-like structure consisting of two α/β-domains (D1 and D2) connected by a small hinge of connecting strands. Ligand binding occurs between the interface of the two-subdomains and initiates closure in a Venus fly trap mechanism [14], a key element of ligand recognition and signal transduction in all SBP-associated protein families. SBPs sometimes recognize even small targets in a highly specific manner through non-covalent protein-ligand interactions that result in KD values in the micromolar to nanomolar range. This specificity and the ability to bind even targets that are difficult for antibody-based detection have made these proteins attractive for use in sensing applications [15]. Based on their topological arrangement, SBPs can be classified into two classes [16] or more recently into seven clusters [17,18]. The LBD of iGluRs also forms a binding pocket in the cleft between two lobes (D1 and D2) of a shell-like conformation consisting of two extracellular regions, where two polypeptide segments on the amino- terminal side of TM1 (called S1) and between TM2 and TM3 (called S2) largely form domains D1 and D2, respectively. Many experimental works have shown that indeed the agonist binding site of soluble LBD constructs are similar to the binding sites and properties in intact receptors [19,20]. The LBDs, which are arranged as two dimers "back-to-back" in the tetrameric iGluR, are induced to close the two shell-like domains after ligand binding, which is directed to the TMD and leads to the permeation of Na+, K+, and Ca2+ ions across the cell membrane [20-22].
Although iGluR LBDs and SBPs share similarities in their overall structure and ligand recognition, the functional compatibility of SBPs to gate the channel pore of an iGluR has not been demonstrated. We selected the TMDs of GluR0 as a candidate for the functional compatibility of a bacterial binding protein with a channel pore of an iGluR. Here, we then used a proof-of-principle approach to couple the bacterial ectoine binding protein EhuB to the channel pore of GluR0 to construct a functional ectoine-activated receptor. With this approach, we provide the first experimental evidence that iGluRs may have arisen from the fusion of an amino acid SBP to an ion channel. Furthermore, our results highlight a conserved ligand binding mechanism in both protein families and the role of LBD dimerization in the functional evolution of iGluRs from SBPs. Furthermore, our results support the possibility of fusion of SBPs, a huge protein family with diverse pharmacological properties and binding ligands, for the targeted design of hybrid receptors as the basic module of a new generation of Biosensors.
Results
Identification of a potential SBP candidate for coupling to the channel pore of GluR0.
The functional coupling of a bacterial SBP with the ion channel of an iGluR has not been demonstrated to date, and thus the requirements of SBPs for successful coupling of ligand binding to the channel gating of iGluRs are still unclear. To investigate the structural basis of the coupling of SBPs with iGluR pore domains, we aimed to generate a functional chimeric receptor by replacing the ligand-binding domain (LBD) of the bacterial GluR0 receptor with a bacterial SBP. The bacterial GluR0 receptor was chosen because of its less complex architecture compared with eukaryotic iGluRs, characterized by the absence of the NTD, the TM3, and a CTD, and may represent the evolutionary link between potassium channels, SBPs, and iGluRs [4]. Because of the high number of SBPs and their low sequence identity, which is usually less than 20% [17], we decided to identify a suitable SBP based on its structural similarity to the LBD of iGluRs using the Vast (+) algorithm [23]. We used the structure of the glutamate-bound closed LBD conformation [24] (PDB ID: 1ii5) as a search template to allow for similar domain closure in ligand binding for sufficient receptor activation [25,26] (see also Materials and Methods section). The 391 identified structures with a total RMSD value of up to 4 Å were further manually inspected to remove duplicates and open or unliganded structures, as well as structures of enzymes and gene regulators. The remaining 94 structures included structures that were mostly part of the TRAP (tripartite ATP-independent periplasmic transporter family), TT- transporter (tripartite tricarboxylate transporter), and iGluR LBDs and SBPs for amino acids, sugars, ions, inorganic salts, and various organic compounds. To analyze the structural relationship between SBPs and iGluR LBDs, all remaining RMSD values were used to construct a phylogenetic tree using the Kitsch program (Figure 1b). The structures can be divided into 3 clusters according to Berntsson et al., [17]. Cluster F includes SBPs with two hinge segments (8-10 amino acids each) that bind a variety of substrates, including amino acids, trigonal planar anions, compatible solutes, and iron, as well as all iGluR LBDs (Figure 1b). Almost all SBPs closely related to iGluRs bind amino acids or derivatives. Structural feature analysis revealed that the amino acid-binding proteins have a short C-terminus similar to the length of the GluR0 LBD compared with nonamino acid-binding proteins (p < 0.0001, Student's two-tailed, unpaired t-test with Welch's correction; Figure 1c). Furthermore, all iGluR LBDs form a small subcluster with a branched order, supporting the current assumption that GluR0 may represent a bacterial iGluR archetype. Cluster D, which has only SBPs, is characterized by two short hinge segments (4-5 amino acids each) and binds various substances such as sugars, phosphates, iron, and molybdate (Figure 1b). Cluster E is located between clusters D and F and features exclusively SBDs of TRAP and TT transporters. Within the cluster, the ectoine-binding protein EhuB turned out to be the non-amino acid SBP most phylogenetically related to the GluR0 LBD. EhuB is part of the Ehu ABC transporter system from Sinorhizobium meliloti, which mediates uptake of the compatible solutes ectoin and hydroxyectoin with high affinity (Kd = 1.6 µM) under osmotic stress [27]. Despite a sequence identity of only 17%, EhuB shows a nearly similar overall structure as the GluR0 LBD with an RMSD of 2.2 Å and 216 of 258 overlapping amino acids (Figure 1d). In addition, both binding domains possess strong rotation of the two domains during the transition to the closed, ligated conformation. These favorable features formed the basis of a simple approach for us to combine the ectoine binding protein EhuB (PDB ID 2Q88) with the channel pore of GluR0 to prove, in a proof-of-concept approach, the general mechanistic and evolutionary ability of SBPs to gate an ion channel by converting chemical information into an electrical signal.
Replacement of the GluR0 LBD and optimization of the ectoine binding protein with stabilization of the dimer interface
In the first step, we wondered whether a simple fusion of the wt EhuB binding protein with the GluR0 TMD (referred to as GluR0EhuB) would be sufficient to open the channel pore. After superimposing the crystal structures of both binding domains (Figure 2a), the ends of the GluR0 LBD lobes (P139, A255) [24] were set as reference points and EhuB was fused to the TMD at positions K132 and N135 with the GluR0 S1-M1 and M2-S2 linkers in the form of two subdomains (Figure 2b). To retain the original GluR0 linker length, positions K133, G134 in EhuB were deleted in the GluR0EhuB receptor. In addition, the construct was equipped with a c-Myc tag, to allow efficient detection of the protein (see Materials and Methods). After heterologous expression, the GluR0Ehub construct unfortunately showed no function in electrophysiological measurements after application of ectoine (up to 1 mM) (data not shown). Analysis of total and surface expression of GluR0EhuB protein by surface biotinylation and Western blot (Figure 2c) revealed a single band with a molecular weight of approximately 40 kDa. We concluded that the GluR0EhuB chimera is efficiently expressed at the cell surface, suggesting that folding, assembly, and transport of the receptor to the cell membrane are unaffected in principle. Extensive intermolecular interactions within the dimerization interface of adjacent LBDs are crucial for the conversion of ligand binding into a channel opening in iGluRs [19, 28,29], interactions that may not be formed due to the monomeric nature between EhuB-SBPs. Furthermore, compared with the GluR0 LBD, EhuB has an elongated C-terminus consisting of three helices (10-12). We therefore speculated that a lack of LBD dimer interface and the large C-terminus in GluR0EhuB might prevent LBD dimerization, resulting in the nonfunctional receptor chimera GluR0EhuB. In GluR0, the dimerization interface between LBDs is characterized by multiple hydrogen bonds and van der Waals contacts formed mainly by 9 amino acids (V113, A118, E348, N349, Q353, K354, L361, N362, Y365) [24]. Therefore, we decided to design a receptor chimera containing the GluR0 LBD dimer interface in the ectoine binding domain (hereafter referred to as GluR0EhuBInt (Int for interface)). To reconstruct the GluR0 LBD dimerization interface, we identified by structural superposition the amino acid positions in EhuB that mediate LBD dimerization in GluR0 (Figure 2a,b) and substituted the corresponding amino acids in the GluR0EhuB LBD (positions M108V, C113A, K235E, E238N, R241Q, D242K, A249L, K250N, and E254Y). In addition, C-terminal helices 10-12 of the original EhuB binding protein were deleted in the GluR0EhuBInt receptor chimera. GluR0EhuBInt was expressed like the original GluR0EhuB construct and localized to the cell surface, as shown by western blot and surface biotinylation (Figure 2c). Even application of 10 µM ectoine (Figure 2d) resulted in ligand-dependent inward currents for GluR0EhuBInt, in contrast to the unmodified initial GluR0EhuB construct, after expression in both HEK293 cells and oocytes (Figure 2d and data not shown). These measurements clearly demonstrated that the ectoine- binding protein EhuB is able to open the channel pore of GluR0 with a maximal amplitude comparable to wt GluR0. However, the ectoin-induced currents showed marked desensitization with greatly reduced receptor currents after re-application of ectoin (run down), which prevented the generation of ectoin dose-response curves (Figure 2d). However, these results indicate that i) Ehub is able to form a functional LBD that can gate the ion channel of GluR0 and ii) that the formation of a GluR0-analog LBD dimerization interface is important to couple ectoin binding within the LBD with channel gating.
Our GluR0EhuBInt resulted in a chimera receptor that showed strong desensitization. We hypothesized that our modified dimerization interface in GluR0EhuBInt is not yet able to reorganize after activation. Although the cα-backbones of GluR0 LBD and EhuB are highly congruent, there are some minor structural differences at the exchanged positions, especially at positions C113A (1.9 Å), D242K (1.6 Å), A249L (1.6 Å), and E226Y (1.7 Å). We hypothesized that these side chains are unable to form intermolecular interactions, weakening the dimerization interface. To test this hypothesis, we decided to strengthen the LBD dimerization interface by introducing a disulfide bond at positions P119C and L376C to covalently couple the two LBD domains (GluR0EhuBInt,P119C,L376C). These positions (Figure 2a,b), when substituted with Cys in AMPA receptors, prevent desensitization of the receptor by stabilizing the LBD dimers [22,30,31]. Application of ectoine (Figure 2d) induced concentration-dependent activation after expression of GluR0EhuBInt,P119C,L376C with an EC50 value of 14.1 ± 7.3 nM (mean ± SEM, n= 4; Figure 2e). Receptor currents showed no desensitization upon prolonged application of ectoine, which is in good agreement with previous studies that stabilized LBD interactions through disulfide bonds within other members of the iGluR family. Apparently, by further stabilizing the dimerization interface, we were able to maintain receptor activity across multiple ectoine reapplications. Thus, our results demonstrate the functional coupling of an SBP to an iGluR channel pore through the exchange of specific amino acids within the LBD by mimicking an LBD dimerization interface of GluR0 and by introducing an additional disulfide bridge leading to ligand- and concentration-dependent currents that resemble general features of non-desensitizing iGluRs.
Discussion
Although it has long been suspected that the LBDs of iGluRs are derived from bacterial SBPs (6), the evolutionary and functional compatibility of SBPs as a module for ligand recognition in iGluRs has not yet been confirmed. In this study, we characterized the basic molecular requirements for functional coupling of an SBP to the channel pore of an iGluR, thereby demonstrating functional compatibility and strengthening a possible evolutionary relationship between the two protein families. All iGluRs consist of at least two domains: the LBD and the TMD forming an ion channel. The common molecular architecture is most likely the result of different protein precursors that fused during the evolution of iGluRs [9]. It has been clearly shown that potassium channels share similarities in overall structure, topology, and sequence with the pore domain of iGluRs [4,32,33] and share a common gating mechanism [10], suggesting a common origin. Felder and colleagues first proposed over 20 years ago [6] that SBPs represent a blueprint for modern iGluR LBDs because they share a common ligand-binding mechanism. However, due to their high functional diversity and low sequence identity, it is difficult to draw conclusions about a common origin of the two protein domains. By coupling the ectoine-binding protein EhuB to the channel pore of GluR0 in a semi-rational approach, we support the assumption of a common origin and ligand-binding mechanism of SBPs and iGluRs. For iGluRs, ligand binding can result in either partial closure of the LBD, as found in the preactive state, or complete closure associated with the active state. The extent of receptor activation depends on both the ability to effect complete closure of the LBD and the fraction of time that the LBD remains in this fully closed conformation. In contrast, incomplete closure of the domain results in conformations with reduced activity, as has been observed with partial agonists [34-39]. Interestingly, SBPs also exhibit a wide range of conformations of partially closed structures that are actively involved in substrate transport [40-42]. Therefore, it stands to reason that both the GluR0 LBD and EhuB must adopt a similar, fully closed conformation in the presence of the TMD to open the pore of the GluR0 channel. Furthermore, the functionality of our chimeric receptors suggests that the general mechanism of ligand binding and domain closure in EhuB and GluR0 must be similar to adopt the fully closed conformation. These results highlight the general modular architecture of iGluRs and support the common hypothesis that the LBD is derived from a bacterial SBP. They further suggest that the underlying mechanism of complete and partial agonism as well as competitive inhibition in iGluRs could be traced back to its bacterial precursors.
In addition, our data shed light on the minimal molecular requirements to preserve receptor function after a potential fusion event and to determine the physiological properties of modern iGluR LBDs. The formation and maintenance of the back-to-back dimer arrangement by the D1-D1 interface is critical for iGluR gating. During activation, the interface is responsible for keeping two adjacent LBDs attached to each other. This allows the D2 lobes within the LBD dimers to separate from each other to transfer the conformational change to the ion channel via the linkers [20-22]. In contrast, the same force that causes elongation of the connecting linker during ligand binding can lead to partial or complete rupture of the dimerization interface, which is referred to as desensitization. Breakage of the LBD dimerization interface leads to realignment of the LBD dimer, allowing the D2 lobes to adopt a conformation similar to the closed state, which uncouples the closed LBD conformation from channel activation [30,43-45]. Our data suggest that the mere fusion of an SBP with an ion channel pore is not sufficient to form a functional receptor. We suggest that the TMD of the GluR0EhuB receptor assembles as a tetramer because it is displayed at the plasma membrane and forms a putative functional channel pore. Because EhuB has not been reported to assemble as dimers or oligomers in solution [27], it is unlikely that functional dimer interactions form within the LBD layer of the GluR0EhuB receptor. Therefore, we hypothesize that ligand binding to the EhuB-binding domain is uncoupled from channel gating because the force generated by clam-shell closure cannot be used to separate the D2 lobes and be transmitted by the linkers. This is consistent with previous studies showing that complete disruption of the D1 dimerization interface by covalently linking the D2 lobes leads to uncoupling of agonist binding from ion channel gating [21]. Remarkably, alteration of a few amino acids within the putative LBD dimerization interface is sufficient to achieve basic receptor activity, as seen with GluR0EhuBInt. However, the GluR0 LBD and the EhuB protein are not completely homologous in their spatial structure. This is consistent with the fact that even minor perturbations of the dimerization interface by single point mutations enhance desensitization [30,46-48]. By covalently stabilizing the LBD dimerization interface in GluR0EhuBIntP119CL376C, we demonstrated that desensitization can be overcome. Moreover, the activation and deactivation kinetics of GluR0EhuBIntP119CL376C resemble the slow activation of GluR0 and constitutive activity upon prolonged ligand application observed in LBD-stabilized AMPA and kainate receptors. In summary, these results confirm that the dimerization interface is a key element in the molecular evolution of iGluRs that is critical for coupling the clamshell closure of the binding domain with ion channel gating. The fact that only minor adjustments are required to form an LBD dimerization interface to obtain a functional iGluR archetype is likely an advantage in functional evolution.
In conjunction with coupling to ion channels, the future of SBP-based sensing offers significant opportunities. SBPs can enable detection of molecules or ions that is not possible with antibodies, and can be tailored to specific applications through targeted mutagenesis of protein specificity and affinity. SBPs have consequently been used in a variety of platforms, e.g., those based on enzyme-linked immunosorbent assays, surface plasmon resonance, immunomagnetic separation, and fluorescence resonance energy transfer (reviewed in Edwards [15]). The tremendous intrinsic amplification of the signal of ligand-gated ion channels together with the unusual extreme sensitivity and selectivity of SBPs represents another major potential for the application of SBP-based biosensors. Due to their specific response, SBP-gated ion channels represent a new class of biosensors that can be used to perform sophisticated analytical measurements and will certainly attract interest in medicine, pharmaceuticals, biochemistry, and environmental analysis. Ion channels are also integral membrane proteins that play a special role in neuronal excitability and synaptic transmission. Using chimeric SBP-gated ion channels to modulate neuronal activity would also be an attractive possibility. There would be the possibility of developing SBP-derived ion channels that could be selectively introduced into neurons by genetic engineering methods and influence the activity of neuronal populations by tailoring ligands. Our work also allows a perspective on the evolutionary origin of iGluR LBDs. It is worth highlighting that the most closely structurally related SBPs in cluster F all bind amino acids or amino acid derivatives. This finding is less surprising because GluR0 can be activated by polar amino acids as a putative link between the evolution of modern metazoan iGluRs from SBPs and potassium channels [4]. Moreover, the LBD of AMPA receptors and GluR0 share weak amino acid sequence homology with glutamine-binding protein from Escherichia coli [4]. For this reason, it is very likely that the ancient SBP precursor belongs to class F SBPs. It is noteworthy that all of the identified amino acid and amino acid derivative class F SBPs have a relatively short C terminus compared with all of the nonamino acid- binding proteins of the more distant classes D and E. This is because of the length of the C terminus. These elongated C termini often pack onto the D1 lobe and obscure the same helices that mediate LBD dimerization in iGluRs. Presumably, in the evolution of iGluRs, it was advantageous that the putative LBD-dimer interface in amino acid-binding proteins was freely accessible, allowing rapid functional adaptation of dimer-dimer interactions without extensive changes in protein structure. Moreover, the short C-termini of amino acid-binding proteins could potentially serve as putative S2-TM3 linkers for coupling the additional TM3 helix in later evolutionary iGluR stages.
In summary, by functionally coupling the ectoine-binding protein EhuB to the GluR0 channel pore, we provide evidence for compatibility between the two protein families and shed light on the functional and molecular evolution of iGluRs from bacterial SBPs and potassium channels. Our results may contribute to the mechanistic understanding of ligand recognition in both protein families and to drug development. Finally, in combination with the diversity of recognized ligands and the structural adaptability of SBPs, our approach could be used as a versatile tool for the development of biosensors with high specificity and efficiency.
Materials and Methods
Identification of homologous SBP and structural distance tree
For structure identification and building a structural distance tree, we used the previously described method from Scheepers et al., [18] with some modifications. To identify SBPs that are structurally homologous to the LBD of GluR0, we used the ligand-bound LBD conformation (1ii5) as a search template against the Molecular Modeling Database (MMDB) using the Vector Alignment Search Tool (VAST+) [23]. The resulting 354 PDB codes were matched against the UniProt IDs to filter out multiple structures of the same protein. Additionally, unliganded structures, enzymes and gene regulators were filtered out by manual inspection. This search resulted in about 94 SBP- and iGluR LBD-structures similar to the GluR0 LBD. The remaining structures were pairwise aligned using the PDBeFold server [49]. To build the structural distance tree, the RMSD values obtained from PBDeFold were loaded into the KITSCH program of the PHYLIPS package with default parameters [50]. Protein clusters were verified by visual inspection using UCFS Chimera [51] according to Berntsson et al., 2010 [17] and the structural distance tree was visualized using iTOL (https://itol.embl.de/). For the determining the C-termini lengths of amino acid and non-amino acid SBP, structures were pairwise aligned to the GluR0 LBD (1ii5) and inspected using UCSF Chimera. The relative C-terminus was specified relative to position Ser366 in the GluR0 LBD. Statistical significance was determined at the p ≤0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***) and p ≤ 0.01 (****), levels using a Student’s two- tailed, unpaired t-test with Welch’s correction. Equality of variances was tested using a F-test.
Protein engineering
In this study, we used the sequences from GluR0 (UniProt ID P73797) and EhuB (UniProt ID Q92WC8). Signal peptide cleavage site was predicted using SignalP v. 4.1[52]. The intrinsic GluR0 (residues 1-19) or EhuB signal peptides (residues 1 – 27) were replaced by the GluR6 signal peptide (MRIICRQIVLLFSGFWGLAMG), followed by a c-myc tag (EQKLISEED) and a short linker (SGTPT). For GluR0EhuB, EhuB D1 domain (residues 28 – 132) was fused to the N-terminal and the D2 domain (residues 135 – 283) to the C-terminal end of the GluR0 TMD, including the linker (residues 139 – 255). For GluR0EhuBInt, the positions M81V, C86A, K208E, E211N, R214Q, D215K, A222L, K223N and E226Y were substituted and residues 266 – 283 were deleted (positions corresponding to the original EhuB protein). For receptor expression in eukaryotic expression systems, the sequences of all constructs were codon-optimized for Xenopus laevis. GluR0, GluR0EhuB and GluR0EhuBInt DNA strings were synthesized (GeneArt, Thermo Fisher Scientific, Regensburg, Germany) and subsequently cloned into the vector pCDNA3.1(+) via included NheI and XhoI restriction sites. For GluR0EhuBInt,P119C,L376C, the primers P119C forward (ATTGTTTGTCAAGTGTGAGAGAGCCGCT) and reverse (AGCGGCTCTCTCACACTTGACAAACAAT), as well L376C forward (TGATGTTGAACTCTGTAATCTGAAATACTCCG) reverse (CGGAGTATTTCAGATTACAGAGTTCAACATCA) were used for mutagenesis polymerase chain reaction (PCR). PCR reaction parameters were as follows: initial denaturation at 95 °C for 60 s; 30 cycles at 95 °C and 30 s, 56 °C for 15 s and 72 °C for 150 s; and one cycle at 72 °C for 180 s. All constructs were confirmed by sequencing (Seqlab, Göttingen, Germany).
Sequences
GluR0EhuB
MRIICRQIVLLFSGFWGLAMGEQKLISEEDLSGTPTDENKLEELKEQGFARIAIANEP PFTAVGADGKVSGAAPDVAREIFKRLGVADVVASISEYGAMIPGLQAGRHDAITAGL FMKPERCAAVAYSQPILCDAEAFALKGKPVSLWERFSPFFGIAALSSAGVLTLLLFLV GNLIWLAEHRKNPEQFSPHYPEGVQNGMWFALVTLTTVGYGDRSPRTKLGQLVAG VWMLVALLSFSSITAGLASAFSTALSEASNPLGLKSYKDIADNPDAKIGAPGGGTEE KLALEAGVPRDRVIVVPDGQSGLKMLQDGRIDVYSLPVLSINDLVSKANDPNVEVLA PVEGAPVYCDGAAFRKGDEALRDAFDVELAKLKESGEFAKIIEPYGFSAKAAMSTTR EKLCAAK
GluR0EhuBInt
MRIICRQIVLLFSGFWGLAMGEQKLISEEDLSGTPTDENKLEELKEQGFARIAIANEP PFTAVGADGKVSGAAPDVAREIFKRLGVADVVASISEYGAMIPGLQAGRHDAITAGL FVKPERAAAVAYSQPILCDAEAFALKGKPVSLWERFSPFFGIAALSSAGVLTLLLFLV GNLIWLAEHRKNPEQFSPHYPEGVQNGMWFALVTLTTVGYGDRSPRTKLGQLVAG VWMLVALLSFSSITAGLASAFSTALSEASNPLGLKSYKDIADNPDAKIGAPGGGTEE KLALEAGVPRDRVIVVPDGQSGLKMLQDGRIDVYSLPVLSINDLVSKANDPNVEVLA PVEGAPVYCDGAAFREGDNALQKAFDVELLNLKYSGEFAKIIEPYG
Heterologous expression in Xenopus laevis oocytes
Constructs were expressed in X. laevis oocytes as described previously [10]. In brief, all constructs in pCDNA3.1(+) were linearized with NotI and cRNA was synthesized using the AmpliCap-Max™ T7 High Yield Message Maker Kit (Cellscript, Madison, WI, USA). Surgically obtained oocytes from female X. laevis were enzymatically separated and defoliculated using 0.8 mg collagenase in ringer solution (96mM NaCl, 2 mM KCl, 1mM CaCl2, 1mM MgCl2, 5mM HEPES; pH 7.4) for 12 h and reaction was stopped with Ca2+-free ringer solution. 50 ng of capped and polyadenylated RNA with 50 nl water was microinjected in defoliculated stage IV – V oocytes and incubated 3 – 7 days in ND-96 solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, 50-mg ml-1 gentamycin; pH 7.5) at 18 °C. Related oocyte experiments (for example total and surface expression) were performed with the same oocyte batches on the same day.
Heterologous expression in HEK293 cells
HEK293 cells were cultured in minimum essential medium (MEM) supplemented with 10 % (v/v) FCS, 2 mM L-glutamine and streptomycin (100 μg/ml). Before transfection, 5x105 cells were reseeded into T25 flask and transfected using TurboFect (Thermo Fisher Scientific, Waltham, MA, USA) and 4 µg plasmid DNA per flask of the respective construct in pCDNA3.1(+) and pEGFP-N1 as a transfection control with a ratio of 4:1. After transfection, cells were incubated for 48-72 h at 37 °C and 5% CO2.
Surface biotinylation
For surface biotinylation, 20 oocytes per construct were injected as described above and incubated for 3 days in ND-96 solution at 18 °C. Oocytes were washed three times with PBS (100 mM phosphate buffer, 150 mM NaCl; pH 7.2) and surface proteins were biotinylated using 0.5 mg/ml Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) in PBS for 40 min at room temperature. Reaction was stopped with 50 mM Tris/HCl; pH 8.0. Oocytes were mechanically homogenized and membrane proteins were extracted using DDM (100 mM phosphate buffer, 150 mM NaCl 0.5 % n-dodecyl-β-D-maltoside, SIGMAFAST Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO, USA); pH 8.0) for 15 min at 4°C. Biotinylated membrane proteins were purified using Streptavidin High Performance Spintrap columns (GE Healthcare, Chicago, IL, USA).
Western blotting
For total protein expression, 10 oocytes per construct were injected and incubated for 3 days. Oocytes were washed with 0.1 M sodium phosphate buffer, pH 8.0 and mechanically homogenized. Membrane proteins were solubilized with 25 µl lysis buffer (0.1 M sodium phosphate buffer pH 8.0, 0.5 % DDM, 0.01% pefa block) for 10 min at 4°C and subsequently centrifuged with 1000 x g at 4°C. HEK293 cells were transfected by electroporation as described above and incubated 48 h. Cells were scraped, lysed with 200 µl lysis buffer and subsequently centrifuged 20 min with 16000 xg at 4°C. For SDS-PAGE and western blot, total and surface labeled proteins were separated using a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a PVDF membrane (Bio-Rad, Feldkirchen, Germany). The membrane was blocked for 1 h in PBS-T supplemented with 5% skim milk and afterward incubated with 1:600 primary c-Myc-tag Polyclonal antibody (sc-789, Santa Cruz Biotechnology, Dallas, TX, USA) in PBS-T containing 1% skim milk over night at 4 °C. After washing with PBS-T, the secondary goat anti-rabbit IgG-HRP (1:10000) (sc-2054, Santa Cruz Biotechnology) was incubated in TBS-T containing 1% skim milk for 1 h at room temperature. The membrane was washed three times in TBS-T and the signal was visualized using Pierce Western Blotting Substrate (Thermo Fisher Scientific) and detected with a CCD camera.
Electrophysiological recordings of Xenopus laevis oocytes using two-electrode voltage clamp 3-7 days after injection, whole-cell currents were recorded by two-electrode voltage-clamp at -80 mV using microelectrodes filled with 3M KCl (resistance 0.8–2.5 MΩ) as described previously [53]. Data were sampled at 5 kHz after low-pass filtering at 200 Hz using an Axoclamp 900A amplifier connected to a Digidata 1550A digitizer and recorded with Clampex 10.7 (Molecular Devices, San Jose, USA). Oocytes were placed in a perfusion chamber and rinsed with high potassium Ringer’s solution (100 mM KCl, 1 mM CaCl2, 5 mM HEPES, pH 7.4 with KOH). Ectoine (10 µM to 0.1 nM) or L-glutamate (1 mM to 1 µM) was applied to the oocytes in external solution. For dose-response analysis, normalized current responses were plotted against the agonist concentration and fitted with a sigmoidal Hill equation
l/lmax = 100 × xh + EC50k in GraphPad Prism 9 (GraphPad Software Inc., La Jolla, USA) where I/Imax is the normalized current, x the concentration, h the Hill coefficient and EC50 the agonist concentration resulting in a half-maximal response. Data and graphs presented in mean ± SEM.
Electrophysiological recordings using Port-a-patch
Electrophysiological recordings in HEK293 cells were performed using the Port-a-Patch system (Nanion Technologies, München, Germany). Adherent HEK293 cells were harvested 48-72 h after transfection using 1% accutase solution. The reaction was stopped with MEM and the cell suspension was centrifuged at 100 xg for 3 min. Cells were resuspended in extracellular solution (140 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM D-glucose, 10 mM HEPES; pH 7.4; osmolarity 298 mOsmol) with a final concentration of 2x106 - 3x106 cells/ml. Measurements were performed using a Port-a-Patch system with external perfusion system (Nanion Technologies) and NPC-1 chips (3-5 MΩ) according to the manufacturer instructions, connected to an EPC10 amplifier (HEKA, Ludwigshafen, Germany), PatchControl (Nanion Technologies) and Patchmaster software (HEKA). External solution and internal solution (50 mM KCl, 10 mM NaCl, 60 mM KF, 20 mM EGTA, 1 mM ATP, 10 mM HEPES; pH 7.2; osmolarity 285 mOsmol) were used for experiments and seal enhancer (80 mM NaCl, 3 mM KCl, 10 mM MgCl2, 35 mM CaCl2, 10 mM HEPES; pH 7.4; osmolarity 298 mOsmol) was used for sealing cells. For recording receptor currents, cells were clamped at -60 mV and continuously perfused with external solution during recording. Ectoine dilution series (10 µM – 0.1 nM) was dissolved in external solution. Agonist was applied for 3 s, followed by 10 s of wash. Dose-response analysis was performed as described above. Data and graphs presented in mean ± SEM.
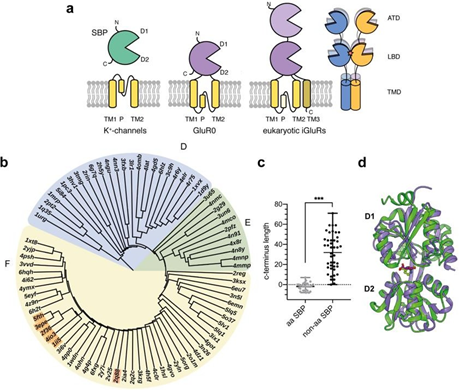
Figure 1: Structural relationship between iGluRs and SBPs. a Modular organization of SBPs, K+-channels and prokaryotic and eukaryotic iGluRs (left) and tetrameric organization of metazoan iGluRs (right) including the amino-terminal domain (ATD), the ligand binding domain (LBD) with the dimerization interface (red star) and the transmembrane domain (TMD). b Structural distance tree of iGluR LBDs and SBPs structural homologous to the GluR0 LBD (1ii5). Clusters were categorized following the nomenclature of Berntsson et al. [17]. The structural distance tree is subdivided into the following clusters: cluster F: two hinged SBPs including all iGluR LBDs (orange; GluR0:1ii5, AvGluR1:4io3, GluR5:2f36, GluR4:3epe, GluR2:5fth) and the ectoine binding protein EhuB (2q88), cluster E: SBDs from TRAP-transports cluster D: short hinged SBPs. c Comparison of the C-terminus lengths between amino acid (aa) or aa derivate SBPs and non-amino acid SBPs relatively measured to position Ser366 in the GluR0 LBD (p < 0.0001, Student’s two-tailed, unpaired t-test with Welch’s correction). d Structural superposition between the ligand-bound, closed conformations of the GluR0 LBD (purple, 1ii5) and the ectoine binding protein EhuB (green, 2q88).
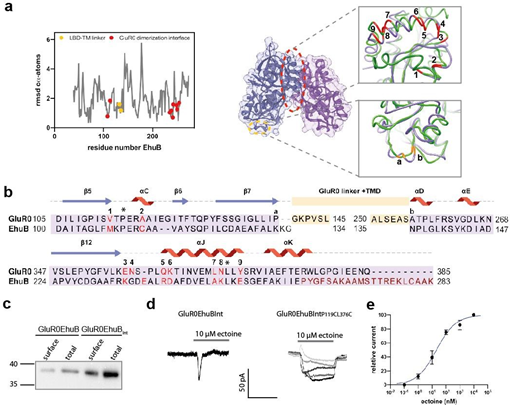
Figure 2: Design and expression of the chimeric GluR0EhuB and GluR0EhuBInt receptors. a Local RMSD of cα- atoms between the GluR0 LBD and EhuB (left) and structural overview (right) of the exchanged amino acid positions within the EhuB LBD in GluR0EhuB and GluR0EhuBInt. The connection points to the LBD-TMD linkers (orange) and amino acid positions that are responsible for LBD dimerization in GluR0 and are exchanged in GluR0EhuBInt (red) are highlighted. b Structural sequence alignment of EhuB and the GluR0 LBD, including the linker positions (orange a,b), the substituted amino acid positions of the dimerization interface (red 1-9) and the deleted helices 10-12 (dark red) in GluR0EhuBInt. Previously published amino acid positions substituted with Cys to covalently couple the D1 domain of each LBD-dimer in AMPA- and kainate recepors and stabilizing the LBD dimerization interface in GluR0EhuBInt,P119C,L376C by disulfide bonds are highlighted with asterisks. c Western blot of total and surface expression of GluR0EhuB (left) and GluR0EhuBInt (right) with a calculated molecular weight of 40 kDa and 38 kDa, respectively. Proteins were detected using a c-Myc tag located at the N-terminus after the signal peptide. d Representative traces of whole-cell currents of GluR0EhuBInt (left) and GluR0EhuBInt,P119C,L376C (right) in response from 1 nM to 10 µM ectoine in HEK293 cells. e Dose-response analysis of GluR0EhuBInt,P119C,L376C revealed an EC50 value of 14.1 ± 7.3 nM (mean ± SEM, n = 4).
Author contributions
M.B. designed and performed the experiments and analyzed the data. M.B. and B.L. wrote the paper.
Competing interests
The authors declare no competing interests.
Acknowledgements
This work has been supported in the frame of the LOEWE project iNAPO by the Hessen State Ministry of Higher Education, Research and the Arts. The authors acknowledge support by the German Research Foundation and the Open Access Publishing Fund of Technische Universität Darmstadt.
Data availability
All data supporting the findings of this study are available within the article or are available from the corresponding author upon reasonable request. All cDNA constructs are available from the corresponding author based on reasonable request.
Ethical approval
All procedures involving animals were in accordance with the guidelines and regulations of the local animal care and use committee. Methods were approved by the Technical University of Darmstadt (II25.3-19c20/15, RP Darmstadt, Germany).
Competing interests
The authors declare no competing financial interest.
References
- Traynelis SF, Wollmuth LP, McBain CJ, et al. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol Rev 62 (2010): 405-496.
- Ramos-Vicente D, Ji J, Gratacòs-Batlle E, et al. Metazoan evolution of glutamate receptors reveals unreported phylogenetic groups and divergent lineage-specific events. Elife 7 (2018): 1-36.
- Lam HM, Chiu J, Hsieh MH, et al. Glutamate- receptor genes in plants. Nature 396 (1998):125-126.
- Chen GQ, Cui C, Mayer ML, et al. Functional characterization of a potassium- selective prokaryotic glutamate receptor. Nature 402 (1999): 817-821.
- Price MB, Jelesko J, Okumoto S. Glutamate Receptor Homologs in Plants: Functions and Evolutionary Origins. Front Plant Sci 3 (2012): 1-10.
- Felder CB, Graul RC, Lee Y, et al. The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci 1 (1999): 7-26.
- Doyle D a, Morais Cabral J, Pfuetzner R a, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280 (1998): 69-77.
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462 (2009b): 745-756.
- Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. TINS 4 (1995): 161-168
- Schönrock M, Thiel G, Laube B. Coupling of a viral K+-channel with a glutamate- binding-domain highlights the modular design of ionotropic glutamate-receptors. Commun Biol 2 (2019).
- Tikhonov DB, Magazanik LG. Origin and molecular evolution of ionotropic glutamate receptors. Neurosci Behav Physiol 39 (2009): 763-773.
- Tam R, Saier MH. Structural, Functional, and Evolutionary Relationships among Extracellular Solute-Binding Receptors of Bacteria. Microbiol Rev 57 (1993): 320-346.
- Lewis M, Chang G, Horton NC, et al. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science (80- ) 271 (1996): 1247-1254.
- Mao B, Mccammon A. Structural Study of Hinge Bending in L-Arabinose-binding Protein. J Biol Chem 259 (1984): 4964-4970.
- Edwards KA. Periplasmic-binding protein-based biosensors and bioanalytical assay platforms: Advances, considerations, and strategies for optimal utility. Talanta Open 3 (2021)
- Fukami-Kobayashi K, Tateno Y, Nishikawa K. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J Mol Biol 286 (1999): 279-290.
- Berntsson RP a, Smits SHJ, Schmitt L, et al. A structural classification of substrate-binding proteins. FEBS Lett 584 (2010): 2606-2617.
- Scheepers GH, Lycklama a, Nijeholt JA, et al. An updated structural classification of substrate-binding proteins. FEBS Lett 590 (2016): 4393-4401.
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA- sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28 (2000): 165-181.
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462 (2009a): 745-756.
- Armstrong N, Jasti J, Beich-Frandsen M, et al. Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor. Cell 127 (2006): 85-97.
- Dürr K, Chen L, Stein R, et al. Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States. Cell 158 (2014): 778-792.
- Madej T, Lanczycki CJ, Zhang D, et al. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res 42 (2014): 297-303.
- Mayer ML, Olson R, Gouaux E. Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J Mol Biol 311 (2001): 815-836.
- Armstrong N, Gouaux E. Mechanisms for Activation and Antagonism of an AMPA- Sensitive Glutamate Receptor. Neuron 28 (2000): 165-181.
- Gill A, Birdsey-Benson A, Jones BL, et al. Correlating AMPA Receptor Activation and Cleft Closure across Subunits: Crystal Structures of the GluR4 Ligand-Binding Domain in Complex with Full and Partial 47 (2008): 13831-13841.
- Hanekop N, Höing M, Sohn-Bösser L, et al. Crystal Structure of the Ligand-Binding Protein EhuB from Sinorhizobium meliloti Reveals Substrate Recognition of the Compatible Solutes Ectoine and Hydroxyectoine. J Mol Biol 374 (2007): 1237-1250.
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462 (2009c).
- Yelshanskaya MV, Li M, Sobolevsky a.I. Structure of an agonist-bound ionotropic glutamate receptor. Science (80- ) 345 (2014): 1070-1074.
- Sun Y, Olson R, Horning M, et al. Mechanism of glutamate receptor desensitization. Nature 417 (2002): 245-253.
- Weston MC, Schuck P, Ghosal A, et al. Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol 13 (2006): 1120-1127.
- Kuner T, Seeburg PH, Guy HR. A common architecture for K+ channels and ionotropic glutamate receptors? Trends Neurosci 26 (2003): 27-32.
- Wood MW, Vandongen HMA, Vandongen AMJ. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc Natl Acad Sci USA 92 (1995): 4882-4886.
- Ahmed AH, Wang S, Chuang HH, et al. Mechanism of AMPA receptor activation by partial agonists: disulfide trapping of closed lobe conformations. J Biol Chem 286 (2011): 35257-35266.
- Lau AY, Roux B. The hidden energetics of ligand binding and activation in a glutamate receptor. Nat Struct Mol Biol 18 (2011): 283-287.
- Maltsev AS, Ahmed AH, Fenwick MK, et al. Mechanism of Partial Agonism at the GluR2 AMPA Receptor: Measurements of Lobe Orientation in Solution. Biochemistry 47 (2008): 10600-10610.
- Salazar H, Eibl C, Chebli M, et al. Mechanism of partial agonism in AMPA-type glutamate receptors. Nat Commun 8 (2017): 1-11.
- Twomey EC, Sobolevsky AI. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry 57 (2018): 267-276.
- Zhang W, Cho Y, Lolis E, et al. Structural and Single-Channel Results Indicate That the Rates of Ligand Binding Domain Closing and Opening Directly Impact AMPA Receptor Gating 28 (2008): 932-943.
- Boer M De, Gouridis G, Vietrov R, et al. Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. Elife 8 (2019): 1-28.
- Gouridis G, Schuurman-wolters GK, Ploetz E, et al. Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat Publ Gr 22 (2014): 57-64.
- Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR 449 (2007): 1078-1082.
- Meyerson JR, Kumar J, Chittori S, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature 57 (2014): 328-334.
- Plested AJR. Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat Struct Mol Biol 23 (2016): 494-502.
- Schauder DM, Kuybeda O, Zhang J, et al. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proc Natl Acad Sci U S A 110 (2013): 5921-5926.
- Fleck MW, Cornell E, Mah SJ. Amino-acid residues involved in glutamate receptor 6 kainate receptor gating and desensitization. J Neurosci 23 (2003): 1219-1227.
- Horning MS, Mayer ML. Regulation of AMPA Receptor Gating by Ligand Binding Core Dimers. Neuron 41 (2004): 379-388.
- Partin KM, Fleck MW, Mayer ML. AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci 16 (1996): 6634-6647.
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr Sect D Biol Crystallogr 60 (2004): 2256-2268.
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. http//www Evol gs washington edu/phylip html (2004).
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem 25 (2004): 1605-1612.
- Nielsen H, Engelbrecht J, Brunak S, et al. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10 (1997): 1-6.
- Laube B, Schemm R, Betz H. Molecular determinants of ligand discrimination in the glutamate-binding pocket of the NMDA receptor. Neuropharm 47 (2004): 994–1007.
