Discovery of Potential RAGE inhibitors using Receptor-Based Pharmacophore Modeling, High Throughput Virtual Screening and Docking Studies
Article Information
Harbinder Singh, Devendra K Agrawal*
Department of Translational Research, College of the Osteopathic Medicine of the Pacific, Western University of Health Sciences, Pomona, California USA
*Corresponding author: Devendra K Agrawal, Department of Translational Research, College of the Osteopathic Medicine of the Pacific, Western University of Health Sciences, Pomona, California USA.
Received: 27 September 2023; Accepted: 12 October 2023; Published: 25 October 2023
Citation: Harbinder Singh, Devendra K Agrawal. Discovery of Potential RAGE inhibitors using Receptor-based Pharmacophore Modeling, High Throughput Virtual Screening and Docking Studies. Journal of Biotechnology and Biomedicine. 6 (2023): 501 - 513.
View / Download Pdf Share at FacebookAbstract
Receptor for Advanced Glycation End products (RAGE) is a transmembrane receptor that can bind to various endogenous and exogenous ligands and initiate the inflammatory downstream signaling pathways. So far RAGE has been involved in various disorders including cardiovascular and neurodegenerative diseases, cancer, and diabetes. Blocking the interactions between RAGE and its ligands is a therapeutic approach to treat these conditions. In this context, we effectively utilized the receptor-based-pharmacophore modeling to discover structurally diverse molecular compounds having potential to effectively bind with RAGE. Two pharmacophore models were developed on V-domain of RAGE using Phase application of Schrodinger suite. The developed pharmacophoric features were used for screening of 1.8 million drug-like molecules downloaded from ChEMBL database. The molecules were scrutinized according to their molecular weight as well as clogP values. Phase screening was performed to find out the molecules that matched the developed pharmacophoric features that were further selected to analyze their binding modes using high-throughput virtual screening, extra precision docking studies and MM-GBSA ΔG binding calculations. These analyses provided ten hit RAGE inhibitory molecules that can bind to two different shallow binding sites on the V-domain of RAGE. Among the obtained compounds two compounds ChEMBL501494 and ChEMBL4081874 were found with best binding free energies that proved their receptor-ligand complex stability within their respective binding cavity on RAGE. Therefore, these molecules could be utilized for further designing and optimizing the future class of potential RAGE inhibitors.
Keywords
ChEMBL database; Inflammation; MM-GBSA; Pharmacophore modeling; Phase screening; RAGE inhibitors; Virtual screening
ChEMBL database articles ChEMBL database Research articles ChEMBL database review articles ChEMBL database PubMed articles ChEMBL database PubMed Central articles ChEMBL database 2023 articles ChEMBL database 2024 articles ChEMBL database Scopus articles ChEMBL database impact factor journals ChEMBL database Scopus journals ChEMBL database PubMed journals ChEMBL database medical journals ChEMBL database free journals ChEMBL database best journals ChEMBL database top journals ChEMBL database free medical journals ChEMBL database famous journals ChEMBL database Google Scholar indexed journals Inflammation articles Inflammation Research articles Inflammation review articles Inflammation PubMed articles Inflammation PubMed Central articles Inflammation 2023 articles Inflammation 2024 articles Inflammation Scopus articles Inflammation impact factor journals Inflammation Scopus journals Inflammation PubMed journals Inflammation medical journals Inflammation free journals Inflammation best journals Inflammation top journals Inflammation free medical journals Inflammation famous journals Inflammation Google Scholar indexed journals MM-GBSA articles MM-GBSA Research articles MM-GBSA review articles MM-GBSA PubMed articles MM-GBSA PubMed Central articles MM-GBSA 2023 articles MM-GBSA 2024 articles MM-GBSA Scopus articles MM-GBSA impact factor journals MM-GBSA Scopus journals MM-GBSA PubMed journals MM-GBSA medical journals MM-GBSA free journals MM-GBSA best journals MM-GBSA top journals MM-GBSA free medical journals MM-GBSA famous journals MM-GBSA Google Scholar indexed journals Pharmacophore modeling articles Pharmacophore modeling Research articles Pharmacophore modeling review articles Pharmacophore modeling PubMed articles Pharmacophore modeling PubMed Central articles Pharmacophore modeling 2023 articles Pharmacophore modeling 2024 articles Pharmacophore modeling Scopus articles Pharmacophore modeling impact factor journals Pharmacophore modeling Scopus journals Pharmacophore modeling PubMed journals Pharmacophore modeling medical journals Pharmacophore modeling free journals Pharmacophore modeling best journals Pharmacophore modeling top journals Pharmacophore modeling free medical journals Pharmacophore modeling famous journals Pharmacophore modeling Google Scholar indexed journals Phase screening articles Phase screening Research articles Phase screening review articles Phase screening PubMed articles Phase screening PubMed Central articles Phase screening 2023 articles Phase screening 2024 articles Phase screening Scopus articles Phase screening impact factor journals Phase screening Scopus journals Phase screening PubMed journals Phase screening medical journals Phase screening free journals Phase screening best journals Phase screening top journals Phase screening free medical journals Phase screening famous journals Phase screening Google Scholar indexed journals RAGE inhibitors articles RAGE inhibitors Research articles RAGE inhibitors review articles RAGE inhibitors PubMed articles RAGE inhibitors PubMed Central articles RAGE inhibitors 2023 articles RAGE inhibitors 2024 articles RAGE inhibitors Scopus articles RAGE inhibitors impact factor journals RAGE inhibitors Scopus journals RAGE inhibitors PubMed journals RAGE inhibitors medical journals RAGE inhibitors free journals RAGE inhibitors best journals RAGE inhibitors top journals RAGE inhibitors free medical journals RAGE inhibitors famous journals RAGE inhibitors Google Scholar indexed journals Virtual screening articles Virtual screening Research articles Virtual screening review articles Virtual screening PubMed articles Virtual screening PubMed Central articles Virtual screening 2023 articles Virtual screening 2024 articles Virtual screening Scopus articles Virtual screening impact factor journals Virtual screening Scopus journals Virtual screening PubMed journals Virtual screening medical journals Virtual screening free journals Virtual screening best journals Virtual screening top journals Virtual screening free medical journals Virtual screening famous journals Virtual screening Google Scholar indexed journals kidneys articles kidneys Research articles kidneys review articles kidneys PubMed articles kidneys PubMed Central articles kidneys 2023 articles kidneys 2024 articles kidneys Scopus articles kidneys impact factor journals kidneys Scopus journals kidneys PubMed journals kidneys medical journals kidneys free journals kidneys best journals kidneys top journals kidneys free medical journals kidneys famous journals kidneys Google Scholar indexed journals lungs articles lungs Research articles lungs review articles lungs PubMed articles lungs PubMed Central articles lungs 2023 articles lungs 2024 articles lungs Scopus articles lungs impact factor journals lungs Scopus journals lungs PubMed journals lungs medical journals lungs free journals lungs best journals lungs top journals lungs free medical journals lungs famous journals lungs Google Scholar indexed journals nervous articles nervous Research articles nervous review articles nervous PubMed articles nervous PubMed Central articles nervous 2023 articles nervous 2024 articles nervous Scopus articles nervous impact factor journals nervous Scopus journals nervous PubMed journals nervous medical journals nervous free journals nervous best journals nervous top journals nervous free medical journals nervous famous journals nervous Google Scholar indexed journals
Article Details
Introduction
RAGE is a 45-55 kDa transmembrane receptor which is present in very low concentrations in healthy human tissues such as liver, kidneys, lungs, nervous, cardiovascular, and immune systems [1, 2]. Under inflammatory conditions, or in the presence of elevated amounts of RAGE ligands such as lipopolysaccharide (LPS), low density lipoproteins (LDL), high-mobility group box-1 (HMGB1), advanced glycation end products (AGE), β-amyloids and others, RAGE expression is significantly upregulated in various cellular systems [3-9]. The binding of these ligands to RAGE activates multiple signaling pathways, including ERK, STAT3, MAPK and JNK that result in the augmentation of transcription factors, including nuclear factor kappa B (NF-kB) [9, 10]. This further leads to leukocyte infiltration, oxidative stress, and inflammatory response by secreting the pro-inflammatory cytokines (TNF-α, IL-1β, and IL-6) contributing to atherosclerosis [11, 12]. Elevated serum concentration of RAGE is also an emerging biomarker of atherosclerotic inflammation [13-15]. Recently, we also confirmed the elevated expression of RAGE in the harvested carotid arteries from angioplasty-induced atherosclerotic Yucatan microswine [16]. Immunohistochemical findings revealed that angioplasty with lipopolysaccharide (LPS) induced significant expression of RAGE compared to other treatment groups. These results were further confirmed by the mRNA expression of RAGE in VSMCs isolated from the control (contralateral side) carotid arteries of the microswine. The mRNA expression of RAGE was found significantly increased with lipopolysaccharide as a RAGE ligand [16]. These findings and other studies from our laboratory further suggest the role of RAGE in various inflammatory diseases [17-25]. Structurally, full-length of human RAGE consists of three major domains as depicted in Figure 1. First, the extracellular domain with amino acid residues 23-342, second is hydrophobic transmembrane domain with amino acid residues 343-363, and third one is intracellular cytoplasmic domain with residues 464-404 [26]. An extracellular region further subdivided into three immunoglobulin like domains named: variable domain (V-domain) having amino acid residues 23-116 which is connected to two constant domains C1 (residues 124-221) and C2 (residues 227-317) (Figure 1). Integrated structural unit of V-domain is primarily responsible for interactions with various ligands such as HMGB1, AGE, β-amyloid, S100 proteins etc. [27-32]. Therefore, directly targeting RAGE to block its crosstalk with various ligands is the best approach to attenuate the adverse effects of inflammation in inflammatory disease conditions.
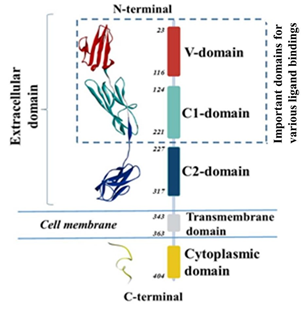
Figure 1: Various domains of RAGE with their number of amino acid residues along with its ribbon structure.
In this study, to effectively block the interactions of RAGE with other proteins or ligands, to our knowledge we used first time the receptor-based-pharmacophore modeling to find out structurally diverse therapeutic compounds with potential to block RAGE binding sites. Pharmacophore modeling is the tool of Maestro Schrodinger by which we can predict various features of the binding site available on the protein/receptor/or enzyme that can help in designing the molecules that can perfectly bind with these binding sites [33]. Two pharmacophore models were developed using the crystal structure of RAGE VC1 domain (PDB: 6XQ3) [34]. These developed features were used for the screening of a library of drug-like molecules which was downloaded from online ChEMBL database. Glide virtual screening and extra precision docking filtered these molecules that use a series of hierarchical filters to search for possible locations of the ligand in the binding-site region of a receptor. Further binding free energies were calculated for potential compounds and two best molecules were discovered that could potentially inhibit the RAGE interactions with its ligands. Overall, the workflow used to identify the potential molecules is shown in Figure 2.
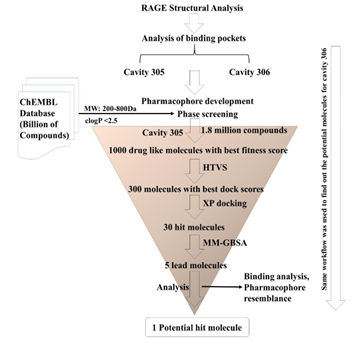
Figure 2: Schematic representation of the procedure followed to find out the potential molecules.
Materials and Methods
Preparation of RAGE structure
The co-crystallized structure of RAGE (VC1 domains) complexed with molecules (same molecule with different assignments) was retrieved from Protein Data Bank (PDB) with accession number 6XQ3 (resolution 1.71Å) [34]. The structure was downloaded in .pdb format and was further prepared using the Protein Preparation Wizard of Schrodinger suite. The protein was first pre-processed by assigning the bond orders and hydrogen, converting selenomethionines to methionines, creating zero order bonds to metals and adding disulphide bonds. The missing side chains and loops were filled using Prime Module of Schrodinger. All the water molecules beyond 5Å were deleted, protein structure was pre-processed, and hydrogen bonds were assigned which was followed by energy minimization by OPLS 2005 force field [35]. Ramachandran plot was obtained after the preparation of protein to validate the preparation protocol which justifies that all the amino acids lie in favorable region. The final structure obtained was saved in.pdb format.
Pharmacophore models development
There are two types of methodologies for the generation of pharmacophore models viz. ligand- based pharmacophore model and Receptor-based pharmacophore model [33]. In the current study, we have performed pharmacophore methodologies to generate the pharmacophore models on RAGE receptor particularly on V-domain of RAGE, that can be correlated with the specific chemical features of the molecules significant for the necessary biological activity. Pharmacophore development hypothesis in Schrodinger suite: release 2021 (in Phase application) was employed for seeking the different chemical features present on two shallow binding sites available on the V-domain of RAGE. The analysis of both the developed pharmacophoric models revealed seven combined features such as Ring Aromaticity, Hydrogen Bond Acceptor, Hydrogen Bond Donor, and Negative Ionizable [36, 37]. The distance between these features was also determined. The developed pharmacophore models were selected from the 10 different hypotheses based on the root mean square deviation (RMSD). These features were selected for further screening of the molecules.
Preparation of compounds
A complete set of drug-like molecules (1.8 million) were downloaded from ChEMBL database according to their molecular weight ranging from 200-800 Da. The molecular weight selection was done on the basis of molecular compounds already bound to the shallow binding sites on RAGE (PDB: 6XQ3). Compounds were further scrutinized according to their clogP values. Only those molecules with clogP values less than 2.5 were selected to download, as we just need the molecules that can bind extracellularly (extracellular V-domain of RAGE). The structures were downloaded in 2D format and further optimized by minimizing the energy using OPLS 2005 force field through Ligprep module of Schrodinger Suite 2021 [38]. The ionization of the structures of compounds were retained in the original state and optimized structures were saved in .sdf format for further screening procedures.
Ligand and database screening
Screening of library of molecules (1.8 million) was done to find out the best match molecules with the developed pharmacophoric models: using ‘Ligand & database screening’ tool. Validated Pharmacophore Hypothesis were utilized as a 3D query for screening ChEMBL Database containing a library of molecules. Top 1000 molecules were selected that have best fitness scores with both the screened pharmacophore models. Best fitness score provides the measurement of features that perfectly matched with the developed pharmacophoric models screening method was set to provide the molecules having at least four features perfectly matched with the models.
High-throughput virtual screening (HTVS) and docking studies
Grid generation and validation
The girds at two binding cavities 305 and 306 (on V-domain of RAGE) on the basis of pre- bound ligands were generated using the Receptor Grid Generation module of the Schrodinger suite, which was defined by simply picking any atom of the co-crystallized ligands within the binding pocket that defined the grid coordinates as X: -43.36, Y: -5.01, and Z: 25.93 for cavity no. 305 and X: -47.58, Y: -20.69, and Z: 26.40 for cavity no. 306 using PDB: 6XQ3. Rigid docking was performed with bound ligands at the defined binding sites of RAGE to validate the protocol and root mean square deviation was computed using Superposition tool of Schrodinger, which was found to be in acceptable range (RMSD: 0.0968Å).
Virtual screening and Extra Precision docking (XP)
The Virtual Screening Workflow in Maestro (Schrodinger) was used to dock and to score the lead-like compounds to identify potential ligands. It provides the different level of docking precision, namely High Throughput Virtual Screening (HTVS), Standard Precision (SP) and Extra Precision (XP) [39-41]. The compounds selected from the pharmacophore phase screening against a set of pharmacophoric features were selected to perform HTVS on both the grids generated on V-domain of RAGE, i.e., at 305 and 306 cavities. After performing HTVS on two sets of 1000 molecules with each cavity, XP mode was further used for refinement of good ligand poses on top 300 molecules for each cavity that exhibited best binding glide or dock scores.
MM-GBSA binding energy calculations
After performing the docking studies the best fit top 30 molecules at their respective binding sites were selected to predict their actual binding energies with RAGE. The binding energies are the sum of the energies obtained from the intermolecular forces between the ligand and protein/receptor. It generally denoted by ΔG. Top five molecules at both the binding sites were selected with best binding energies or affinities (ΔG values) and those molecules were selected for explaining the binding mechanisms with both the cavities of RAGE and pharmacophoric features that are responsible for their best binding behavior.
Results and Discussion
Pharmacophore modeling and ligand screening
From the crystal structure of RAGE (PDB: 6XQ3), we found that there are two shallow binding sites available on V-domain that are very close to each other and amino acids present in these cavities have been reported to be involved in various RAGE ligand interactions. The names to these two cavities were assigned on the basis of the ligands bound. The cavity at which ligand (chemical compound) no. 305 is bound was named as cavity 305 and similarly, cavity at which compound no 306 is bound was named as cavity 306 (Figure 3). However, the structure of both the compounds are same which represents that a same molecule (3-(3-(((3-(4- Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid) can bind to two different shallow binding sites that are very close to each other (Figure 3). These shallow cavities offered us to develop two pharmacophoric models on V-domain of RAGE.
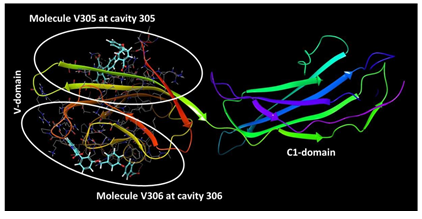
Figure 3: Two different binding sites available on V-domain of RAGE offers the development of two pharmacophore models
In order to develop pharmacophores, the binding interactions of the molecule at both the cavities 305 and 306 were analyzed and amino acids involved in these interactions were noted down. By using Pharmacophore development hypothesis, the amino acids that are involved in the interactions at 305 cavity (residues are mentioned in table 1) were incorporated into the hypothesis to generate receptor-based model. Same procedure was used for the development of model at cavity 306. Seven features were obtained at both the cavities having acceptor groups, donor groups, ring aromatic groups, and negative ionic groups (Figure 4). Figure 4A also represents the distance constraints present between the two features which means that the chemical structure containing chain or any linker having the same distance of 7.37Å could behave better as compared to the compounds with shorter or longer linker length. After performing pharmacophore-based ligand screening at both the cavities with a set of ChEMBL molecules we selected top 1000 molecules that have at least four best matched pharmacophoric features with fitness score above 0.8 to 1.5. These molecules were found nicely mapped on the chemical features present in the developed models (explained in section 3.3.2). Selected molecules were further subjected to high-throughput virtual screening to find out their binding scores (dock scores) with their respective binding cavities.
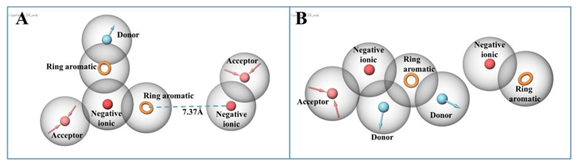
Figure 4: Developed Pharmacophore model Hypothesis. (A) Seven chemical features present in pharmacophore model at cavity 305, spatial arrangement and the distance constraints between the features; (B) Seven chemical features present in developed model at cavity 306 and their spatial arrangement.
|
Receptor |
Cavity Number |
Coordinates |
Amino acid residues |
|
V-domain of RAGE |
305 |
X = -43.36 Y= -5.01 Z= 25.93 |
Ala21, Met22, Ala23, Gln24, Glu50, Trp51, Lys52, Arg98, Cys99, Gln100 |
|
V-domain of RAGE |
306 |
X= -47.58 Y= -20.69 Z= 26.40 |
Pro45, Pro46, Gln47, Pro66, Gln67, Asp73, Ser74, Arg77, Val78, Leu79, Pro80, Phe85, Pro87 |
Table 1: Amino acid residues at two different shallow binding cavities and their coordinates
High-throughput virtual screening (HTVS)
Initially for the shallow binding cavity 305, we generated a grid by selecting the respective amino acids of the cavity 305. The coordinates and particular amino acids for grid generation are mentioned in Table 1. Selected best 1,000 molecules from the phase screening were docked in the generated grid and virtual screening was performed to determine their bindings in terms of their dock or glide scores. Resulted molecules were ranked according to their docking scores and the binding modes, molecular interactions with the active site, binding energy and dock score. Docking scores were considered as important components in selecting the best poses of the docked compounds. Analysis of virtual screening results of all filtered molecules put forth 300 molecules with good to excellent dock scores ranging from -4.00 to -5.911 having pharmacophoric fitness score >1 at cavity 305 of RAGE. The same workflow was used to virtually screen the molecules on the cavity 306 and top 300 hit molecules were found that could have potential to effectively bind with RAGE which was further evaluated and filtered using extra precision docking (XP) and by MM-GBSA free energy calculations.
Extra precision docking (XP) and MM-GBSA calculations
Two different set of 300 molecules were further selected to investigate their binding mode within the binding sites 305 and 306 of RAGE. The ligands were docked with the active sites using the ‘extra precision’ glide docking (Glide XP) which docks ligands flexibly. In XP docking, only active compounds show available poses that receive favorable scores for appropriate hydrophobic contact between the binding cavities of RAGE, hydrogen-bonding interactions, and so on. The purpose of the XP method is to weed out false positives and to provide a better correlation between excellent poses and good scores. Among the two set of 300 molecules 30 molecules were further selected with their best XP score (glide XP dock score) ranging from -4.93 to 6.23 that bind with cavity 305. Similarly, a set of 30 different compounds were selected on the basis of XP dock score ranging from -5.47 to -8.48 that bind with cavity 306. According to the range of XP dock scores, it can be concluded that compounds can tightly and more efficiently bind at the 306 binding site of RAGE as compare to the binding of compounds at cavity 305. The overall extra precision glide docking results with both the binding cavities 305 and 306 are provided in Table 2 and Table 3, respectively. For these selected set of 30 compounds the evaluation of docking procedure with a related post-scoring approach, MM-GBSA was performed against both the cavities on RAGE and two sets of five hit compounds were obtained from free energy binding prediction. (Figure 5 and 7). Compounds that found to bind with cavity 305 showed ΔG values ranging from -53.05 to -65.80 kcal/mol (Figure 5) and molecules that were found hit for cavity 306 exhibited ΔG values ranging from -51.81 to -68.53 kcal/mol. (Figure 7) These ten compounds (five against cavity 305 and five against cavity 306) were selected to elaborate the binding mechanisms in detail with their respective binding cavities.
|
Serial No. |
ChEMBL ID |
Glide XP Docking Score |
Glide XP energy (kcal/mol) |
Glide XP Emodel (kcal/mol) |
|
1 |
CHEMBL249147 |
-6.233 |
-49.28 |
-63.36 |
|
2 |
CHEMBL3656092 |
-5.952 |
-48.568 |
-81.481 |
|
3 |
CHEMBL4126863 |
-5.933 |
-41.703 |
-52.487 |
|
4 |
CHEMBL1889231 |
-5.89 |
-49.916 |
-65.278 |
|
5 |
CHEMBL3912163 |
-5.781 |
-50.858 |
-74.412 |
|
6 |
CHEMBL206726 |
-5.758 |
-48.294 |
-61.142 |
|
7 |
CHEMBL3677668 |
-5.745 |
-47.765 |
-60.614 |
|
8 |
CHEMBL3143543 |
-5.736 |
-50.202 |
-68.457 |
|
9 |
CHEMBL231404 |
-5.694 |
-45.213 |
-60.716 |
|
10 |
CHEMBL551572 |
-5.685 |
-40.537 |
-54.852 |
|
11 |
CHEMBL455260 |
-5.513 |
-45.8 |
-57.355 |
|
12 |
CHEMBL120659 |
-5.459 |
-44.095 |
-54.75 |
|
13 |
CHEMBL263749 |
-5.435 |
-47.014 |
-67.791 |
|
14 |
CHEMBL3326332 |
-5.385 |
-37.606 |
-52.192 |
|
15 |
CHEMBL144301 |
-5.233 |
-42.469 |
-58.204 |
|
16 |
CHEMBL501494 |
-5.191 |
-58.84 |
-84.308 |
|
17 |
CHEMBL3393957 |
-5.176 |
-36.155 |
-47.953 |
|
18 |
CHEMBL4303537 |
-5.153 |
-48.034 |
-60.15 |
|
19 |
CHEMBL1093942 |
-5.132 |
-51.877 |
-72.009 |
|
20 |
CHEMBL471413 |
-5.129 |
-38.827 |
-54.631 |
|
21 |
CHEMBL2314004 |
-5.129 |
-47.315 |
-64.261 |
|
22 |
CHEMBL3718691 |
-5.123 |
-39.514 |
-49.356 |
|
23 |
CHEMBL3457424 |
-5.123 |
-41.284 |
-52.002 |
|
24 |
CHEMBL1460147 |
-5.12 |
-39.916 |
-53.185 |
|
25 |
CHEMBL3908491 |
-5.051 |
-52.985 |
-74.374 |
|
26 |
CHEMBL358539 |
-5.042 |
-37.78 |
-49.263 |
|
27 |
CHEMBL2012851 |
-5.03 |
-40.63 |
-52.187 |
|
28 |
CHEMBL2172634 |
-5.026 |
-45.983 |
-64.311 |
|
29 |
CHEMBL1075716 |
-4.936 |
-57.264 |
-78.804 |
|
30 |
CHEMBL388067 |
-4.935 |
-39.858 |
-51.825 |
Table 2: Extra precision (XP) Glide docking results within the cavity 305 of RAGE.
|
Serial No. |
ChEMBL ID |
Glide XP Docking Score |
Glide XP energy (kcal/mol) |
Glide XP Emodel (kcal/mol) |
|
1 |
CHEMBL463170 |
-8.486 |
-54.556 |
-76.294 |
|
2 |
CHEMBL1160402 |
-7.431 |
-48.776 |
-69.68 |
|
3 |
CHEMBL543644 |
-7.32 |
-35.862 |
-38.901 |
|
4 |
CHEMBL543644 |
-7.251 |
-36.372 |
-41.58 |
|
5 |
CHEMBL1160402 |
-6.934 |
-50.678 |
-58.881 |
|
6 |
CHEMBL2179043 |
-6.914 |
-43.613 |
-56.631 |
|
7 |
CHEMBL130120 |
-6.706 |
-49.734 |
-70.446 |
|
8 |
CHEMBL548295 |
-6.621 |
-43.392 |
-52.329 |
|
9 |
CHEMBL144301 |
-6.615 |
-39.044 |
-53.993 |
|
10 |
CHEMBL1688937 |
-6.465 |
-36.62 |
-47.716 |
|
11 |
CHEMBL3126678 |
-6.283 |
-35.055 |
-40.894 |
|
12 |
CHEMBL231404 |
-6.231 |
-41.403 |
-57.535 |
|
13 |
CHEMBL3581077 |
-6.229 |
-35.622 |
-44.044 |
|
14 |
CHEMBL228562 |
-6.213 |
-38.21 |
-47.383 |
|
15 |
CHEMBL64002 |
-6.2 |
-54.248 |
-66.561 |
|
16 |
CHEMBL120659 |
-6.187 |
-39.807 |
-49.956 |
|
17 |
CHEMBL4081874 |
-5.959 |
-62.364 |
-89.45 |
|
18 |
CHEMBL206726 |
-5.858 |
-45.658 |
-62.59 |
|
19 |
CHEMBL543644 |
-5.753 |
-35.035 |
-43.973 |
|
20 |
CHEMBL362202 |
-5.735 |
-35.005 |
-47.442 |
|
21 |
CHEMBL543644 |
-5.734 |
-34.051 |
-39.443 |
|
22 |
CHEMBL181074 |
-5.663 |
-33.638 |
-40.732 |
|
23 |
CHEMBL1170193 |
-5.662 |
-30.393 |
-35.034 |
|
24 |
CHEMBL19844 |
-5.656 |
-28.712 |
-36.53 |
|
25 |
CHEMBL223910 |
-5.557 |
-41.095 |
-58.957 |
|
26 |
CHEMBL200715 |
-5.529 |
-30.428 |
-40.263 |
|
27 |
CHEMBL3966940 |
-5.507 |
-44.065 |
-65.28 |
|
28 |
CHEMBL2141241 |
-5.502 |
-30.585 |
-37.99 |
|
29 |
CHEMBL343412 |
-5.497 |
-25.703 |
-26.742 |
|
30 |
CHEMBL482098 |
-5.479 |
-44.162 |
-54.926 |
Table 3: Extra precision (XP) Glide docking results within the cavity 306 of RAGE
Binding modes of five hit molecules with the cavity 305
The careful analysis of Figure 6 suggests that all these compounds adopt almost similar binding modes with V-domain of RAGE by targeting the shallow binding pocket of RAGE that consists of various hydrophobic as well as hydrophilic amino acid residues. The five compounds obtained from different set of experiments exhibited interactions such as hydrogen bonding interactions, hydrophobic and polar interactions within the cavity 305 which are depicted in two and three dimensional pictures in Figure 6. Most potential binder of cavity 305, i.e. ChEMBL501494 exhibited XP dock score of -5.19 with the highest ΔG value (-65.80 kcal/mol) was considered as potent inhibitor of RAGE that binds through the cavity 305. The reason for its best binding free energy among the others is because of its number of interactions formed with the different amino acid residues. The molecule interacts with RAGE by making strong six hydrogen bonds. (Figures 6A and 6A’) The nitrogen atom present on terminal indole was found to be involved in hydrogen bonding with Glu50. The oxygen atom located in the amide linkage between two indole rings also involved in hydrogen bonding with Lys52 and Arg98 (Figure 6A’). Three additional hydrogen bonds were found with Lys110, Asn112 and Arg98. Various pi-cation interactions with the compound stabilize the molecule within the cavity 305 of RAGE. Second most efficient binder ChEMBL1075716 was also found to involved with various hydrogen and pi- cation interactions with RAGE 305 cavity (Figures 6B and 6B’). It was found that Lys52, Lys110 and Asn112 are commonly involved in these interactions between these two molecules (Figure 6A and 6B). Lys39 and Asn112 were found to be involved in hydrogen bonding interactions with carboxy oxygen atom available in closer proximity to the terminal indole moiety. Indole moiety is positioned and stabilized in the cavity formed by various hydrophobic amino acid residues such as Ala21, Met22 and Ala23. (Figure 6B’) Furthermore, compounds ChEMBL3908491 (Figures 6C and 6C’), ChEMBL4126863 (Figures 6D and 6D’), and ChEMBL3326332 (Figures 6E and 6E’) also found to be involved in various hydrogen bonding as well as cationic interactions with residues present inside the cavity 305. Most importantly, Lys110 and Asn112 are the residues that were found to be involved in all these potential binders of RAGE which means that these interactions are important in inhibiting the RAGE through cavity 305. Furthermore, pharmacophoric validation by overlaying the compound ChEMBL501494 on developed pharmacophore at cavity 305 was performed to check whether the discovered potent compound completely matches the developed features or not. It was clear from the Figure 9 that the discovered molecule ChEMBL501494 completely occupy the chemical features obtained inside the cavity 305 and could be responsible to block the bindings of various ligands that mainly bind through the same cavity. Interestingly, the terminal indole ring with the spacer containing two carbon chain along with amide linkage make the molecule perfect match for the acceptor (a), donor (b), aromatic ring (c), and negative ionic (d) features. The centered aromatic feature (e) was also found to overlap properly with the centrally present indole moiety that has also been involved in pi-cation interactions with Arg98 (Figures 6A’ and Figure 9A). The distance constraint between the feature (e) and (f) was observed 7.37Å and compound ChEMBL501494 also fulfilled the criteria having the spacer length of same size so that it can fully occupy the developed pharmacophoric features (Figure 9A).
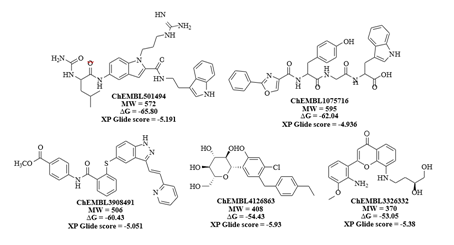
Figure 5: 2D structures, dock scores and ΔG values of identified lead molecules that can bind to the cavity 305 on V-domain of RAGE.
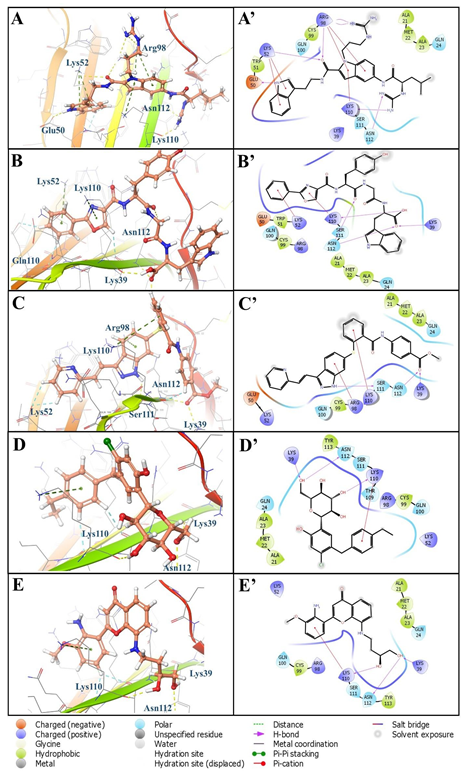
Figure 6: 3D and 2D binding interactions of hit compounds (A) ChEMBL501494, (B) ChEMBL1075716, (C) ChEMBL3908491, (D) ChEMBL4126863, (E) ChEMBL3326332 with binding site of RAGE cavity 305; A’, B’, C’, D’, and E’ are the 2D depiction of their respective poses.
Binding modes of five hit molecules with the cavity 306
Molecular docking studies analysis at cavity 306 was performed and five hit compounds were discovered with high XP dock scores with excellent binding free energies (ΔG in kcal/mol). (Figure 7) It has been found that compounds more effectively and more tightly bind to this cavity as compare to the compounds bind to the cavity 305 as confirmed from their XP dock scores and ΔG values. Most effective binder ChEMBL4081874 exhibited ΔG value of -68.53 kcal/mol with the XP dock score value of -5.95. Compound bind with the active site 306 with various electrostatic interactions such as hydrogen bonding, hydrophobic interactions, negative and positive charged interactions. (Figures 8A and 8A’) Importantly, the compound showed five hydrogen bonds with amino acid residues such as Glu50, Gln47, Pro66, and Val78 (Figure 8A). The right terminal thiophene ring was found to be completely occupied by the hydrophobic residues Val78, Leu79, Pro80, Phe85 and Pro87 (Figure 8A’). Negatively charged amino acid residue Glu50 helped to stabilize the molecule from another end (left) by offering two hydrogen bonds with terminal hydroxyl groups (Figure 8A’). Similarly, compound ChEMBL206726 also showed hydrogen bonding interactions with Gln47, Pro66, and Val78 (Figures 8B and 8B’). Nitrogen containing five membered rings of benzimidazole moiety showed pi-cation interaction with positively charged amino acid residue Arg77. Its ΔG value and XP dock score is provided in the figure 7. Third most effective compound ChEMBL3966940 was involved to form two hydrogen bonds with polar residue Gln47 and hydrophobic residue Val78 (Figures 8C and 8C’). Most of the interactions of this compound was found hydrophobic with residues such as Pro45, Pro46, Pro66, Val78, Leu79, Pro80, Phe85 and Pro87 (Figure 8C’). Similar interactions were found with compounds ChEMBL2179043 (Figures 8D and 8D’) and ChEMBL223910 (Figures 8E and 8E’). It has been observed that Gln47 and Val78 are commonly involved in all these compounds at cavity 306 which could be crucial for binding these compounds with the RAGE.
To further confirm the fitness of most effective binder of cavity 306, molecule ChEMBL4081874 was analyzed by incorporating directly into the developed pharmacophoric features associated to the cavity 306 (Figure 9B). Among the seven developed features, compound perfectly matched five features. Specifically, central oxepin carbonyl oxygen atom acts as hydrogen bond acceptor from residue Gln47 that matches the acceptor feature (a). (Figure 9B) Two donor features (b) and (c) were fulfilled by the presence of single secondary amine linker in between two phenyl rings that acts as hydrogen bond donor to hydrophobic residue Pro66. Fused oxepin benzene ring and terminal thiophene ring act as aromatic rings available in the closer proximity to the aromatic features (e and g). (Figure 9B) Overall, compound ChEMBL4081874 perfectly fit to the developed hypothesis of the pharmacophoric features at cavity 306 and exert its potent binding affinities as confirmed through molecular interactions, XP docking and ΔG scoring functions.
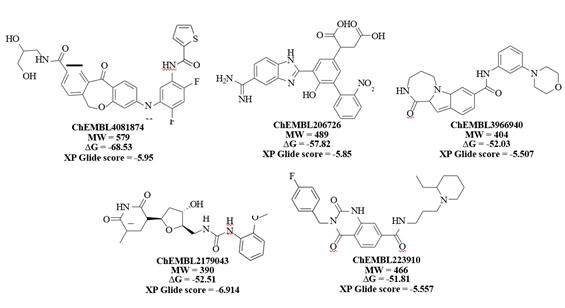
Figure 7: 2D structures, dock scores and ΔG values of identified lead molecules that can bind to the cavity 306 on V-domain of RAGE
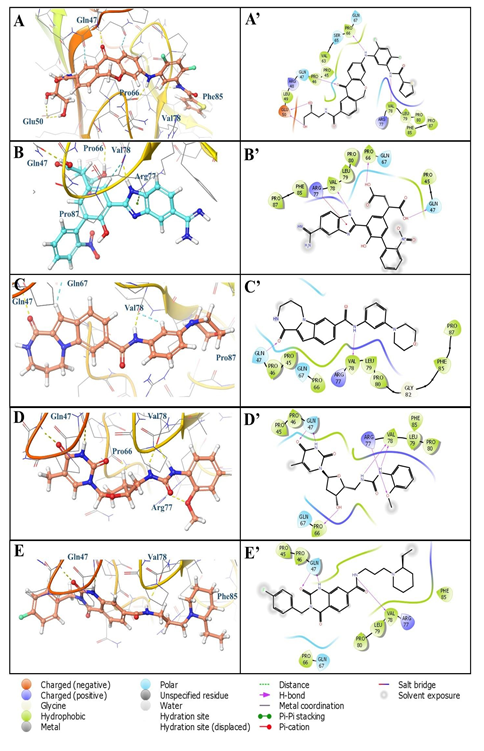
Figure 8: 3D and 2D binding interactions of hit compounds (A) ChEMBL4081874, (B) ChEMBL206726, (C) ChEMBL3966940, (D) ChEMBL2179043, (E) ChEMBL223910 with binding site of RAGE cavity 306; A’, B’, C’, D’, and E’ are the 2D depiction of their respective poses
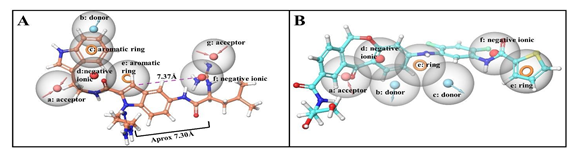
Figure 9: Overlay of compounds (A) ChEMBL501494 on developed features of cavity 305 and, (B) ChEMBL4081874 on developed features of cavity 306
Conclusion
In this study, phase screening of ChEMBL database was performed using two generated pharmacophores for two binding sites 305 and 306 on V-domain of RAGE. This process was followed by high-throughput virtual screening, extra precision docking studies, and binding free energy calculations to discover two compounds ChEMBL501494 and ChEMBL4081874 that passes all these steps. Free binding energy (MM-GBSA) calculations proved the stability of these two obtained compounds in their respective pocket of V-domain of RAGE justified with their lowest negative ΔG values. The compound ChEMBL501494 was found stabilized inside the 305cavity possessing significant interactions with Met22, Lys39, Arg98, and Lys110. While compound ChEMBL4081874 at cavity 306 interacted via hydrogen bonding, water bridges, hydrophobic and ionic interactions. Particularly, amino acid Gln47, Gln67 and Val78 were found to be the most interacting residues that helped to stabilize the molecule in cavity 306. Both the molecules were found to perfectly match with the developed pharmacophoric features. In conclusion, ChEMBL501494 and ChEMBL4081874 are considered as novel candidates for screening as inhibitors in RAGE associated pathways.
Authors’ Contributions
Concept and design: HS, DKA; Literature Search and Acquisition of findings: HS; Analysis and interpretation of the findings: HS, DKA; Drafting the article: HS; Revising and editing the manuscript: HS, DKA; Final approval of the article: HS, DKA.
Funding
The research work of DK Agrawal is supported by research grants R01 HL144125 and R01 HL147662 from the National Institutes of Health, USA. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Conflicts of Interest
The research work of DK Agrawal is supported by the NIH research grants. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants, or patents received or pending, or royalties.
References
- Neeper M, Schmidt AM, Brett J, et Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem 267 (1992): 14998-15004.
- Cheng C, Tsuneyama K, Kominami R, et al. Expression profiling of endogenous secretory receptor for advanced glycation end products in human organs. Mod Pathol 18 (2005): 1385-1396.
- Bierhaus A, Humpert PM, Morcos M, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med 83 (2005): 876-886.
- Hudson BI, Lippman Targeting RAGE Signaling in Inflammatory Disease. Annu Rev Med 69 (2018): 349-364.
- Brett J, Schmidt AM, Yan SD, et al. Survey of the distribution of a newly characterized receptor for advanced glycation end products in Am J Pathol 143 (1993): 1699-1712.
- Yonekura H, Yamamoto Y, Sakurai S, et al. Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury. Biochem J 370 (2003): 1097-1109.
- Chuah YK, Basir R, Talib H, et al. Receptor for advanced glycation end products and its involvement in inflammatory diseases. Int J Inflam 2013 (2013): 403460.
- Lin L, Park S, Lakatta EG. RAGE signaling in inflammation and arterial aging. Front Biosci (Landmark Ed) 14 (2009): 1403-1413.
- Fritz RAGE: a single receptor fits multiple ligands. Trends Biochem Sci 36 (2011): 625-632.
- Kierdorf K, Fritz RAGE regulation and signaling in inflammation and beyond. J Leukoc Biol 94 (2013): 55-68.
- Schmidt AM, Yan SD, Wautier JL, et al. Activation of receptor for advanced glycation end products: a mechanism for chronic vascular dysfunction in diabetic vasculopathy and atherosclerosis. Circ Res 84 (1999): 489-497.
- Basta G. Receptor for advanced glycation end products and atherosclerosis: From basic mechanisms to clinical implications. Atherosclerosis 196 (2008): 9-21.
- Riehl A, Nemeth J, Angel P, et al. The receptor RAGE: Bridging inflammation and cancer. Cell Commun Signal 7 (2009): 12.
- Nasser MW, Wani NA, Ahirwar DK, et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor Cancer Res 75 (2015): 974-985.
- Deane R, Singh I, Sagare AP, et al. A multimodal RAGE- specific inhibitor reduces amyloid beta-mediated brain disorder in a mouse model of Alzheimer disease. J Clin Invest 122 (2012): 1377-1392.
- Singh H, Rai V, Agrawal LPS and oxLDL-induced S100A12 and RAGE expression in carotid arteries of atherosclerotic Yucatan microswine. Mol Biol Rep 49 (2022): 8663-8672.
- Noothi S, Rai V, Radwan MM, et al. Oxidized low-density lipoproteins and lipopolysaccharides augment carotid artery plaque vulnerability in hypercholesterolemic Cardiol Cardiovasc Med 7 (2023): 273-294.
- Rai V, Le H, Agrawal Novel mediators regulating angiogenesis in diabetic foot ulcer healing. Can J Physiol Pharmacol 101 (2023):488-501.
- Kuan R, Nath S, Agrawal DK, et al. Response to acute hyperglycemia and high fructose in cultured tenocytes. J Biochem 174 (2023):71-80.
- Singh H, Agrawal DK. Therapeutic potential of targeting the HMGB1/RAGE axis in inflammatory Molecules 27 (2022): 7311.
- Rai V, Mathews G, Agrawal Translational and clinical significance of DAMPs, PAMPs, and PRRs in trauma-induced inflammation. Arch Clin Biomed Res 6 (2022): 673-685.
- Singh H, Agrawal Recent advances in the development of active hybrid molecules in the treatment of cardiovascular diseases. Bioorg Med Chem 62 (2022): 116706.
- Singh H, Rai V, Agrawal Discerning the promising binding sites of S100/calgranulins and their therapeutic potential in atherosclerosis. Expert Opinion on Therapeutic Patents 31 (2021): 1045-1057.
- Satish M, Saxena SK, Agrawal DK. Adipokine dysregulation and insulin resistance with atherosclerotic vascular disease: Metabolic syndrome or independent sequaelae? J Cardiovasc Translational Res 12 (2019): 415-424.
- Thankam FG, Dilisio MF, Dietz NE, et al. TREM-1, HMGB1 and RAGE in the shoulder tendon: Dual mechanisms for inflammation based on the coincidence of glenohumeral arthritis. PLoS One 11 (2016):
- UniProtKB - Q15109 (RAGE_HUMAN).
- Hori O, Brett J, Slattery T, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem 270 (1995): 25752-25761.
- Xue J, Rai V, Singer D, et al. Advanced glycation end product recognition by the receptor for AGEs. Structure 19 (2011): 722-732.
- Koch M, Chitayat S, Dattilo BM, et al. Structural basis for ligand recognition and activation of Structure 18 (2010): 1342-1352.
- Dattilo BM, Fritz G, Leclerc E, et al. The extracellular region of the receptor for advanced glycation end products is composed of two independent structural Biochemistry 46 (2007): 6957-6970.
- Sturchler E, Galichet A, Weibel M, et al. Site-specific blockade of RAGE-Vd prevents amyloid-beta oligomer J Neurosci 28 (2008): 5149-5158.
- Xue J, Ray R, Singer D, et al. The receptor for advanced glycation end products (RAGE) specifically recognizes methylglyoxal-derived AGEs. Biochemistry 53 (2014): 3327-3335.
- Dixon SL, Smondyrev AM, Rao SN. PHASE: A Novel Approach to Pharmacophore Modeling and 3D Database Searching. Chem Biol Drug Des 67 (2006): 370-372.
- Kozlyuk N, Gilston BA, Salay LE, et al. A fragment- based approach to discovery of Receptor for Advanced Glycation End products inhibitors. Proteins 89 (2021): 1399-13412.
- Jorgensen WL, Tirado-Rives J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci USA. 102 (2005): 6665-6670.
- Musyoka TM, Kanzi AM, Lobb KA, et al. Structure based docking and molecular dynamic studies of plasmodial cysteine proteases against a south African natural compound and its analogs. Sci Rep 6 (2016): 1-12.
- Project H. Specifications TATES T 2 – t s (2005): 8-9.
- Ligprep 2.3, Schrodinger. LLC, New York, NY (2009).
- Halgren TA, Murphy RB, Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening J Med Chem 47 (2004):1750.
- Friesner RA, Murphy RB, Repasky MP, et al. J Med Chem 49 (2006): 6177.
- Small-Molecule Drug Discovery Suite 2013-1: Glide, version 9, Schrödinger, LLC, New York (2013).
