CDKs Functional Analysis in Low Proliferating Early-Stage Pancreatic Ductal Adenocarcinoma
Article Information
Shikai Zhu1,2,#, Huining Yang1,#, Lingling Liu3, Zhilin Jiang1, Juanjuan Ji1, Xiao Wang1, Lin Zhong1, Fulin Liu1, Xueliang Gao 3,4, Haizhen Wang3,4∗ and Yu Zhou1∗
1Sichuan Provincial Key Laboratory for Human Disease Gene Study, Department of Laboratory Medicine, Center for Medical Genetics, Sichuan Provincial People's Hospital, School of Medicine,University of Electronic Science and Technology of China, Chengdu, China
2Organ Transplant Center, Sichuan Provincial People's Hospital, School of Medicine,University of Electronic Science and Technology of China, Chengdu, China
3Department of Cell and Molecular Pharmacology & Experimental Therapeutics,
4Hollings Cancer Center, Medical University of South Carolina, Charleston, South Carolina, USA
*Corresponding author: Yu Zhou. Haizhen Wang. Sichuan Provincial Key Laboratory for Human Disease Gene Study, Department of Laboratory Medicine, Center for Medical Genetics, Sichuan Provincial People's Hospital, School of Medicine,University of Electronic Science and Technology of China, Chengdu, China. Department of Cell and Molecular Pharmacology & Experimental Therapeutics, Hollings Cancer Center, Medical University of South Carolina, Charleston, South Carolina, USA.
Received: 15 July 2023; Accepted: 24 July 2023; Published: 16 August 2023
Citation:
Shikai Zhu, Huining Yang, Lingling Liu, Zhilin Jiang, Juanjuan Ji, Xiao Wang, Lin Zhong, Fulin Liu, Xueliang Gao, Haizhen Wang and Yu Zhou. CDKs Functional Analysis in Low Proliferating Early-Stage Pancreatic Ductal Adenocarcinoma. Journal of Bioinformatics and Systems Biology. 6 (2023): 187-200.
View / Download Pdf Share at FacebookAbstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly devastating disease with a poor prognosis and growing incidence. In this study, we explored the potential roles of CDK1, CDK2, CDK4, and CDK6 in the progression of early-stage PDAC. Clinicopathologic and mRNA expression data and treatment information of 140 patients identified with stage I/II PDAC who underwent pancreaticoduodenectomy were obtained from the Cancer Genome Atlas data set. Our bioinformatic analysis showed that higher CDK1, CDK2, CDK4, or CDK6 expression was associated with a shorter median survival of the early-stage PDAC patients. Of note, in the low-proliferating pancreatic cancer group, CDKs expressions were significantly associated with proteins functioning in apoptosis, metastasis, immunity, or stemness. Among the low-proliferating PDAC, higher expression of CDK1 was associated with the shorter survival of patients, suggesting that CDK1 may regulate PDAC progression through cell cycleindependent mechanisms. Our experimental data showed that CDK1 knockdown/inhibition significantly suppressed the expression levels of AHR and POU5F1, two critical proteins functioning in cancer cell metastasis and stemness, in low-proliferating, but not in high-proliferating pancreatic cancer cells. In all, our study suggests that CDKs regulate PDAC progression not only through cell proliferation but also through apoptosis, metastasis, immunity, and stemness.
Keywords
Pancreatic ductal adenocarcinoma (PDAC), Cyclin-dependent kinase, Early stage, Survival, Metastasis, Apoptosis, Immunogenicity, Lowproliferating cancer cells.
Pancreatic ductal adenocarcinoma (PDAC) articles; Cyclin-dependent kinase articles; Early stage articles; Survival articles; Metastasis articles; Apoptosis articles; Immunogenicity articles; Low-proliferating cancer cells articles.
Pancreatic ductal adenocarcinoma (PDAC) articles Pancreatic ductal adenocarcinoma (PDAC) Research articles Pancreatic ductal adenocarcinoma (PDAC) review articles Pancreatic ductal adenocarcinoma (PDAC) PubMed articles Pancreatic ductal adenocarcinoma (PDAC) PubMed Central articles Pancreatic ductal adenocarcinoma (PDAC) 2023 articles Pancreatic ductal adenocarcinoma (PDAC) 2024 articles Pancreatic ductal adenocarcinoma (PDAC) Scopus articles Pancreatic ductal adenocarcinoma (PDAC) impact factor journals Pancreatic ductal adenocarcinoma (PDAC) Scopus journals Pancreatic ductal adenocarcinoma (PDAC) PubMed journals Pancreatic ductal adenocarcinoma (PDAC) medical journals Pancreatic ductal adenocarcinoma (PDAC) free journals Pancreatic ductal adenocarcinoma (PDAC) best journals Pancreatic ductal adenocarcinoma (PDAC) top journals Pancreatic ductal adenocarcinoma (PDAC) free medical journals Pancreatic ductal adenocarcinoma (PDAC) famous journals Pancreatic ductal adenocarcinoma (PDAC) Google Scholar indexed journals Cyclin-dependent kinase articles Cyclin-dependent kinase Research articles Cyclin-dependent kinase review articles Cyclin-dependent kinase PubMed articles Cyclin-dependent kinase PubMed Central articles Cyclin-dependent kinase 2023 articles Cyclin-dependent kinase 2024 articles Cyclin-dependent kinase Scopus articles Cyclin-dependent kinase impact factor journals Cyclin-dependent kinase Scopus journals Cyclin-dependent kinase PubMed journals Cyclin-dependent kinase medical journals Cyclin-dependent kinase free journals Cyclin-dependent kinase best journals Cyclin-dependent kinase top journals Cyclin-dependent kinase free medical journals Cyclin-dependent kinase famous journals Cyclin-dependent kinase Google Scholar indexed journals Early stage articles Early stage Research articles Early stage review articles Early stage PubMed articles Early stage PubMed Central articles Early stage 2023 articles Early stage 2024 articles Early stage Scopus articles Early stage impact factor journals Early stage Scopus journals Early stage PubMed journals Early stage medical journals Early stage free journals Early stage best journals Early stage top journals Early stage free medical journals Early stage famous journals Early stage Google Scholar indexed journals Survival articles Survival Research articles Survival review articles Survival PubMed articles Survival PubMed Central articles Survival 2023 articles Survival 2024 articles Survival Scopus articles Survival impact factor journals Survival Scopus journals Survival PubMed journals Survival medical journals Survival free journals Survival best journals Survival top journals Survival free medical journals Survival famous journals Survival Google Scholar indexed journals Metastasis articles Metastasis Research articles Metastasis review articles Metastasis PubMed articles Metastasis PubMed Central articles Metastasis 2023 articles Metastasis 2024 articles Metastasis Scopus articles Metastasis impact factor journals Metastasis Scopus journals Metastasis PubMed journals Metastasis medical journals Metastasis free journals Metastasis best journals Metastasis top journals Metastasis free medical journals Metastasis famous journals Metastasis Google Scholar indexed journals Apoptosis articles Apoptosis Research articles Apoptosis review articles Apoptosis PubMed articles Apoptosis PubMed Central articles Apoptosis 2023 articles Apoptosis 2024 articles Apoptosis Scopus articles Apoptosis impact factor journals Apoptosis Scopus journals Apoptosis PubMed journals Apoptosis medical journals Apoptosis free journals Apoptosis best journals Apoptosis top journals Apoptosis free medical journals Apoptosis famous journals Apoptosis Google Scholar indexed journals Immunogenicity articles Immunogenicity Research articles Immunogenicity review articles Immunogenicity PubMed articles Immunogenicity PubMed Central articles Immunogenicity 2023 articles Immunogenicity 2024 articles Immunogenicity Scopus articles Immunogenicity impact factor journals Immunogenicity Scopus journals Immunogenicity PubMed journals Immunogenicity medical journals Immunogenicity free journals Immunogenicity best journals Immunogenicity top journals Immunogenicity free medical journals Immunogenicity famous journals Immunogenicity Google Scholar indexed journals Lowproliferating cancer cells articles Lowproliferating cancer cells Research articles Lowproliferating cancer cells review articles Lowproliferating cancer cells PubMed articles Lowproliferating cancer cells PubMed Central articles Lowproliferating cancer cells 2023 articles Lowproliferating cancer cells 2024 articles Lowproliferating cancer cells Scopus articles Lowproliferating cancer cells impact factor journals Lowproliferating cancer cells Scopus journals Lowproliferating cancer cells PubMed journals Lowproliferating cancer cells medical journals Lowproliferating cancer cells free journals Lowproliferating cancer cells best journals Lowproliferating cancer cells top journals Lowproliferating cancer cells free medical journals Lowproliferating cancer cells famous journals Lowproliferating cancer cells Google Scholar indexed journals mRNA articles mRNA Research articles mRNA review articles mRNA PubMed articles mRNA PubMed Central articles mRNA 2023 articles mRNA 2024 articles mRNA Scopus articles mRNA impact factor journals mRNA Scopus journals mRNA PubMed journals mRNA medical journals mRNA free journals mRNA best journals mRNA top journals mRNA free medical journals mRNA famous journals mRNA Google Scholar indexed journals
Article Details
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most prevalent pancreatic neoplastic cancer, responsible for more than 90% of all malignancies in the pancreas. PDAC is the third and is projected to become the second leading cause of cancer deaths in the United States [1], and also the fourth most common cause of cancer-related deaths worldly [2]. Upon decades of effort, the 5-year survival rate of PDAC remains less than 8% [3-5]. PDAC can be divided into different stages, ranging from early - stage I- to late - stage IV - [6]. PDAC treatments are partially based on the stage of cancer [7,8]. When the disease is in an early stage (AJCC-UICC stages I and II), with no spreading to large blood vessels or distant organs, tumor surgical resection can be performed commonly [9]. However, pancreatic adenocarcinoma is usually diagnosed after it has progressed to an advanced stage, and palliative care as a treatment for symptoms is often the only option for patients [10]. In this study, we are particularly interested in the early-stage PDAC, which can be resected and usually has a favorable prognosis.
Cyclin-Dependent Kinases (CDKs) have been reportedly involved in pancreatic ductal adenocarcinoma development and progression [11]. CDKs are the families of protein kinases first discovered for their roles in regulating the cell cycle. CDKs are also involved in the regulation of transcription, mRNA processing, and differentiation of nerve cells [12]. CDK interacts with cyclin, a regulatory protein, to form the cyclin-CDK complex to phosphorylate its substrates on serine and/or threonine amino acid (s) to regulate the activities of downstream target proteins. Four major CDKs, i.e., CDK1, CDK2, CDK4, and CDK6, are involved in cell-cycle regulation. CDK4 and CDK6 are the G1 CDKs that are required to enter the cell cycle from the quiescent state. CDK2 is responsible for S- phase entry and promoting DNA replication, whereas CDK1 is the mitotic CDK that drives mitosis in M phase. CDKs are often elevated in PDAC and are associated with the unrestrained proliferation of tumor cells [11]. CDKs are considered potential therapeutic targets for anti-cancer medication, and several CDK inhibitors are undergoing clinical trials [13].
PDAC treatment options are very restricted and are highly dependent on the disease stage [14,15]. For medical decisions and individualized treatment modalities, comprehending the risk factors and long-term survival related to accurate staging in PDAC patients is crucial. Although CDKs dysregulation in PDAC has been reported, the critical role of CDKs in the early stage of PDAC is not well studied. To this end, we applied the TCGA database to explore survival and Cox proportional hazards analysis on CDK1, CDK2, CDK4, and CDK6 in stage I/II PDAC patients. We found that high expression of CDK1, CDK2, CDK4, or CDK6 was associated with shorter median survival of patients, and was also an independent hazardous risk for the early stage PDAC. Interestingly, in the low-proliferating pancreatic cancer group, CDKs were significantly associated with several apoptosis, metastasis, and immunity-related genes, listed as FAS, BIRC2, PTPN13, RELA, and TAP1, which were the independent hazardous risks for the early stage PDAC. Our finding indicates that CDKs may regulate apoptosis, metastasis, immunity, and stemness besides cancer proliferation in the early stage of low-proliferating PDAC.
2. Materials and Methods
2.1 Data source and patient information
TCGA Data on pancreatic cancer were downloaded on Oct 10, 2022. Clinical data, mRNA expression data, tumor methylation data, tumor DNA sequence, and tumor protein RPPA (Reverse Phase Protein Array, RPPA) data were all acquired from the TCGA dataset. In the current study, the clinical information and mRNA expression data were used in the following analysis. All patients underwent pathologic diagnosis by tissue biopsy, tumor resection, or cytology. As part of the TCGA network, the mRNA of pancreatic cancer specimens was sequenced with next-generation sequencing, usually on the Illumina platform with quality controls. The Cancer Genome Atlas (TCGA), a landmark cancer genomics program, molecularly characterized over 20,000 primary cancer and matched normal samples spanning 33 cancer types, has been well-described elsewhere[16]. TCGA project began in 2006 and has generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. The data has already improved our ability to diagnose, treat, and prevent cancer, and are publicly available for anyone in the research community to use.
2.2 Inclusion and exclusion criteria
Inclusion criteria were complete data available, demonstrated histology consistent with “pancreas-adenocarcinoma ductal type”, underwent pancreaticoduodenectomy, and American Joint Committee on Cancer (AJCC) stage I or II based on the 7th edition of the AJCC staging system. Patients with stage III or IV disease were excluded.
2.3 Expression and gene analyses
TCGA data portal (https://tcga-data.nci.nih.gov/) open access HTTP directory was the data source of the current study. RNASeqV2 normalized gene expression data, and all patient TCGA clinical data were downloaded from the data portal of the Genomic Data Commons (GDC). Genes with≥ 75th percentile log2 mRNA expression values for each gene were identified as having high expression. Low gene expression was based on values < 75th percentile log2 mRNA expression value for each gene. The relationship among CDK1, CDK2, CDK4, CDK6, and MKI67 and survival in patients with resected PDAC staged as I or II were specifically investigated. We also explored the relationship between CDKs and other genes, functioning in apoptosis, metastasis, and immunity, in the low-proliferating pancreatic cancer group (the expression of MKI67 mRNA was ≤ 25th percentile) and tried to find whether their expression was also an independent hazardous risk in the early stage PDAC.
2.4 Pancreatic cancer Cell culture, compound treatment, and Viral transduction/transfection
PANC-1, ASPC, CFPAC-1, and MiaPaca II cells were gifted from Dr. Denis Guttridge’s group at MUSC. ASPC cells were cultured in RPMI-1640 medium (Corning) supplemented with 10% FBS and 1% penicillin/streptomycin. CFPAC-1 cells were cultured in Iscove's Modified Dubecco's Medium (IMDM, Hyclone) supplemented with 10% FBS and 1% penicillin/streptomycin. MiaPaca II and PANC-1 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Corning) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco). The cells were maintained under standard culture conditions at 37°C in humidified air with 5% CO2. PANC-1 and MiaPaca II cells were treated with CDK1 inhibitors (10 uM CDK1-IN-2, MedChemExpress HY-112463; or 5 uM RO-3306, MedChemExpress HY12529) or vehicle for 24h or 48hrs in the presence/absence of 10%FBS. Lentiviral plasmids were packaged in HEK293T cells by co-transfecting expression plasmids together with the packaging plasmid (pCMV-Gag-Pol for retroviral infections, pCMV-delta8.9 for lentiviral infections) and the envelope plasmid (pCMV-VSV-G), using Lipofectamine 2000 transfection reagent (using the plasmid ratio of pCMV-VSV-G/pCMV-Gag-Pol/expression plasmid 3:5:10 for retroviral transductions, and the ratio of pCMV-VSV-G/pCMV-delta8.9 /expression plasmid 1:5:10 for lentiviral transductions). Virus-containing medium was collected two days after transfection, filtered, and used for infection in the presence of 10 µg/ml polybrene. After 24 h, the virus-containing medium was replaced by a fresh medium, and cells were selected for 2 days with puromycin (2 µg/ml). shRNAs against CDK1 (TRCN0000000583 and TRCN0000196602) in pLKO.1-puro vector were obtained from MilliporeSigma Mission shRNA.
2.5 Immunoblot and antibodies
Mammalian cells were lysed in RIPA buffer supplemented with protein protease and phosphatase inhibitors. Protein concentrations were measured with the microplate spectrophotometer Benchmark Plus using protein assay dye (Bio-Rad, 5000006). 6–10% SDS-PAGE was applied to separate cell lysate, and proteins were transferred to nitrocellulose membranes (0.45 µM). The membrane was blocked with 5% BSA in TBS for 30 min before incubating with primary antibody overnight at 4°C. After washing with TBS-Tween20 (0.1%) three times, the membrane was incubated with the secondary antibody at room temperature for an additional 1 hr. The signal was detected with a chemiluminescence imaging system with a CCD camera (LI-COR Biosciences, Odyssey). The following antibodies were used: anti-Ki67 antibodies (Proteintech, 28074-1-AP), anti- CDK1 antibodies (Proteintech, 19532-1-AP), anti-actin antibodies (Proteintech, 66009-1-lg), anti-AHR antibodies (Proteintech, 67785-1-Ig), and anti-OCT4/POU5F1 antibodies (Proteintech, 11263-1-AP). Western blot data are representative of at least three independent experiments.
2.6 Statistical analyses
Data analyses were performed with SPSS Statistics (IBM, Armonk, NY). Significance was set to p = 0.05. Differences in variables between the two groups were explored with Student t test or χ2 as appropriate. Overall survival was investigated with the Kaplan-Meier method utilizing the log-rank test by Prism-GraphPad. Correlation analysis was also explored by SPSS Statistics. Variables associated with survival were investigated with Cox proportional regression analyses with the proportional hazard ratio (HR).
3. Results
3.1 Stage I/II pancreatic ductal adenocarcinoma analysis in TCGA dataset
TCGA database was applied to explore the early-stage PDAC data set. 147 out of 178 tumor samples with pancreatic malignancies demonstrated histology consistent with “pancreas- adenocarcinoma ductal type”. Further, 7 samples were excluded from the 147 pancreas- adenocarcinoma ductal type samples due to the missing or mismatched stages information (stage III or IV diseases). With these inclusion and exclusion criteria, 140 samples were ultimately used for further analysis. The average age of these patients was 65 ± 11 years, 86.4% were white, and 45% were female. The median follow-up time was 225 days (range, 0-2182 days). Ten patients (7.1%) had a documented family history of pancreatic cancer. Eleven patients (7.8%) had a history of chronic pancreatitis. Only one patient received radiation treatment prior to resection. In this patient group, 13.6%, 57.1%, and 29.3% of tumors were neoplasm histologic grade I, II, and III/IV, respectively. The mean number of lymph nodes examined was 17.7 ± 8.5 per patient with an average of 3.1 ± 3.2 positive lymph nodes per patient. Overall, 113 patients (80.7%) received Whipple surgical procedure, 16 patients (11.4%) received a total/distal pancreatectomy surgical procedure, and 9 patients (6.4%) received other surgery or have no information of the surgical method. The detailed clinical information was shown in supplementary data 1.
3.2 High CDK1, CDK2, CDK4, and CDK6 expression is associated with poor survival in Stage I/II PDAC patients
The clinical and treatment characteristics for patients with tumors expressing high levels of CDK1, CDK2, CDK4, and CDK6 were similar to that of low levels (Table I). Log2mRNA expression value was used for CDK1, CDK2, CDK4, and CDK6 gene expression evaluation. The average expression of CDK1, CDK2, CDK4 and CDK6 in the top quartile (high expression group) was 30.9±9.3, 26.4±4.0, 64.6±18.7 and 34.6±11.1 whereas it was 12.7±5.5, 14.8±3.7, 36.4±8.4 and 15.3±5.1 in patients with low expression, respectively (P < 0.0001, Figure 1A-D).
Survival analysis showed that CDK1, CDK2, CDK4, and CDK6 expression were associated with patient survival. In brief, low CDK1 expression was associated with a median survival of 634 days, whereas high CDK1 expression was associated with a median survival of 277 days (Figure 1E; P=0.001). Low CDK2 expression was associated with a median survival of 607 days, whereas high CDK1 expression was associated with a median survival of 308 days (Figure 1F; P=0.0395). Low CDK4 expression was identified with a median survival of 627 days, whereas high CDK4 expression with a median survival of 460 days (Figure 1G; P=0.0355). Low CDK6 expression was associated with a median survival of 666 days, whereas High CDK6 expression was associated with a median survival of 460 days (Figure 1H; P=0.0198).
Table 1: Basic information of patient and treatment characteristics
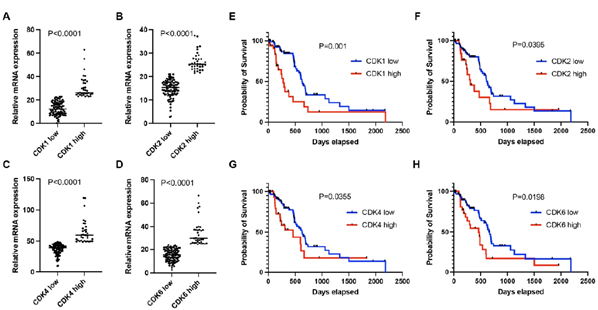
Figure 1: A-D. mRNA expression levels of CDK1 (A), CDK2 (B), CDK4 (C), and CDK6 (D) in the high-expression group were significantly enriched compared to the low-expression group in early-stage PDAC. E-H. Patients in the high-expression group had shorter median survival than the low-expression group as shown in (P<0.05). Patients were categorized into two groups according to the mRNA expression of individual CDKs. The high expression group had mRNA expression ≥75th percentile. The low expression group had mRNA expression <75th percentile.
3.3 Cox regression with TCGA data of early-stage PDAC
Of the 140 patients, the overall Kaplan-Meier median survival was 598 days. By Cox proportional hazards model analyses, radiotherapy, chemotherapy, number of lymph nodes, high CDK1, high CDK2, and high CDK6 expression were significantly associated with survival in stage I/II PDAC patients (Table II). Age, gender, family history of cancer, tumor grade, and low CDK4 expression were not statistically associated with survival (Table II).
Table 2: Characteristics associated with overall survival in early-stage PDAC.
|
Variable |
B |
SE |
P value |
HR |
|
age |
0.02 |
0.013 |
0.13 |
1.02 |
|
Gender |
-0.23 |
0.278 |
0.41 |
0.795 |
|
Family history of cancer |
-0.08 |
0.309 |
0.797 |
1.083 |
|
Number of lymph node |
0.094 |
0.036 |
0.01 |
1.098 |
|
Tumor Grade |
0.365 |
0.283 |
0.197 |
1.44 |
|
Radiotherapy |
1.231 |
0.409 |
0.003 |
3.424 |
|
Chemotherapy |
-1.186 |
0.279 |
<0.0001 |
0.305 |
|
High CDK1 expression |
0.921 |
0.289 |
0.001 |
2.512 |
|
High CDK2 expression |
0.622 |
0.307 |
0.043 |
1.862 |
|
High CDK4 expression |
0.279 |
0.384 |
0.468 |
1.322 |
|
High CDK6 expression |
0.658 |
0.288 |
0.022 |
1.932 |
3.4 Poor survival is associated with increased proliferation
Ki67 (gene name: MKI67) is an established marker for cancer cell proliferation. In this study, we analyzed Ki67 mRNA expression in PDAC. Log2 mRNA expression level of MKI67 was 32.5±13 in the high expression group (≥75th percentile) and 11.3 ± 4.9 in the low expression group (Figure 2A, P<0.0001). Patients with high MKI67 expression tumors had shorter median survival than those with low MKI67 expression tumors (473 days versus 627 days; P = 0.0454, Figure 2B). Patients with higher levels of CDK1, CDK2, CDK4 or CDK6, respectively, (log2 mRNA expression≥ 75th percentile of each gene) in the high MKI67 expression group were compared with patients in the low MKI67 expression group(log2 mRNA expression < 75th percentile of each gene; 29.3 versus 13.3, P <0.0001; 21.7 versus 16.4, P <0.0001; 50.8 versus 41.0, P= 0.0026; 23.3 versus 19.0, P=0.0442; respectively, Supplementary figure 1A-D). CDK1, CDK2, CDK4, and CDK6 are important cyclin-dependent kinases functioning in cell proliferation. We analyzed the correlation between the individual cyclin-dependent kinase and MKI67. As shown in figure 2C-F, CDK1 had a high correlation with MKI67 (r=0.7981, P<0.0001), while CDK2 had a median correlation with MKI67 (r=0.4214, P<0.0001). No correlation between CDK4/ CDK6 and MKI67 was found (r=0.2764, p=0.0009, for CDK4; and r=0.264, p=0.0016, for CDK6, respectively).
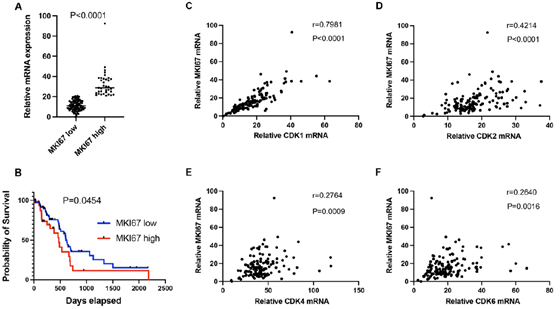
Figure 2: A. Log2 mRNA expression level of MKI67 in the high group (≥ 75th percentile) and low group (<75th percentile) in early-stage PDAC patients. B. Patients in the high MKI67 group (MKI67 high) had shorter median survival compared to the low MKI67 group (MKI67 low). Pearson's correlation analysis was performed to analyze the mRNA expression correlation between MKI67 and CDK1 (C), CDK2 (D), CDK4 (E), and CDK6 (F).
3.5 CDKs expression in low-proliferating pancreatic cancers
Besides the important roles in regulating cell proliferation, CDKs also carry out critical functions in apoptosis, metastasis, and immunity [17]. CDK1, CDK2, CDK4, CDK6, or MIK67 that ≥ 75th percentile log2 mRNA expression value for each gene were identified as having high expression. Low gene expression was based on values < 75th percentile log2 mRNA expression value for each gene. Interestingly, our analysis showed that CDKs were not only expressed in high proliferating (high Ki67), but also low proliferating (low Ki67) PDAC (Figure 3a). We found that in high CDK1 expressing pancreatic cancer, 31% of samples showed low Ki67 expression. In high CDK2-expressing pancreatic cancers, 49% of samples had low Ki67 expression. In high CDK4-expressing pancreatic cancers, 54% of samples showed low expression of Ki67. In high CDK6-expressing pancreatic cancers, 66% of samples displayed low expression of Ki67. This indicates CDK1/2/4/6 may have cell cycle-independent function in pancreatic cancers. Consistent with the well-studied functions of CDK1/2/4/6 in cell proliferation, we found that the majority showed low Ki67 expression in low CDK1/2/4/6 expression pancreatic cancers (Figure 3b).
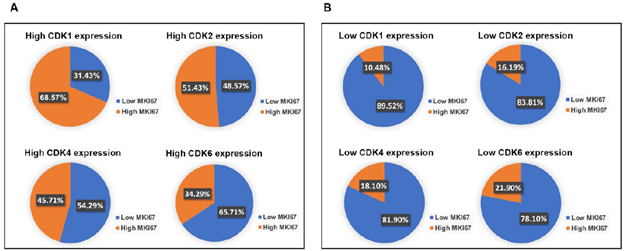
Figure 3: PDAC with high expression of CDKs had over 30% population with low MKI67 expression. A. In the high CDK1, CDK2, CDK4, or CDK6 expression group, approximately 31.43%, 48.57%, 54.29%, or 65.71% of the PDAC population showed low MKI67 expression, respectively. B. The majority of PDAC expressing low CDKs levels showed low MKI67 expression. In the low CDKs expression group, approximately 89.52%, 83.81%, 81.90%, or 78.10% of the PDAC population displayed high MKI67 expression, respectively.
3.6 The CDKs expression is associated with proteins functioning in apoptosis, metastasis, immunity, or stemness in the low-proliferating stage I/II PDAC group
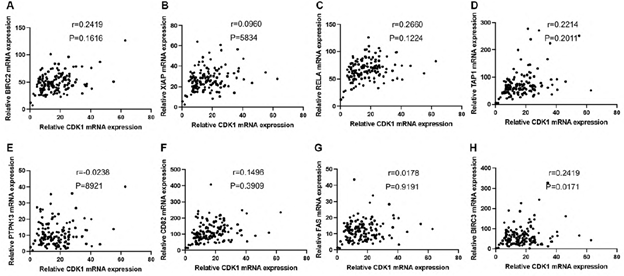
We defined PDAC with MKI67 expression <=25% as a low proliferating group. To explore the potential functions of CDKs in low-proliferating stage I/II PDAC, we analyzed the correlations between CDKs and several critical genes known to play significant roles in apoptosis/metastasis/immunity/stemness in PADC cells (supplementary data 2). BIRC2, BIRC3, PTPN13, and FAS regulate cancer survival [18-21]. Of note, the expressions of CDK2, CDK4, and CDK6 showed significant positive correlations with BIRC2 (R=0.75, P<0.05, R=0.64, P<0.0001, R=0.79, P<0.0001, respectively) (Figure 4A-C). Meanwhile, CDK6 expression correlated with BIRC3 (R=0.68, P<0.05) (Figure 4D); and CDK4 expression correlated with FAS (R=0.73, P<0.05) and PTPN13 (R=0.73, P<0.05)(Figure 4E&4F). AHR, CD82, and RELA function in cancer metastasis [22-24]. Our analysis showed that CDK2 expression correlated with CD82 (R=0.6, P<0.05) (Figure 4G). In addition, CDK2 and CDK6 were positively correlated with TAP1 (R=0.72, P<0.05, R=0.60, P<0.05, respectively) (Figure 4H, 4I), which functions in antigen presentation and immunogenicity. RELA has multiple functions in regulating cancer inflammation, apoptosis, and metastasis. The expression of CDK2, CDK4, and CDK6 was positively correlated with the RELA expression (R=0.75, P<0.05, R=0.64, P<0.05, R=0.60, P<0.05, respectively) (Figure 4J-4L). We also analyzed the correlations between CDKs and cancer stem cells related protein markers. We found that the expression of CDK2 (R=0.6735, p<0.0001) and CDK6 (R=0.6835, p=0,0015) significantly correlated with the expression level of CD44, a compelling marker for cancer stem cells (Figure 4M and 4O). All this information suggests CDK2, CDK4, and CDK6 may have survival/metastasis/immunity/stemness regulatory functions in low proliferating cells. Our analysis showed a correlation between CDK1 and AHR/POU5F1 (OCT4) was 0.5291/0.5336 (Figure 5E &5F). This suggests CDK1 may regulate cancer metastasis and stemness in low-proliferating PDAC. Of note is that the strong correlations between the CDKs expression and genes functioning in survival/metastasis/immunity/stemness did not exist in the high MKI67 expression group (Supplementary Figure 2). This suggests CDKs may regulate survival/metastasis/immunity/stemness in low-proliferating cancer cells.
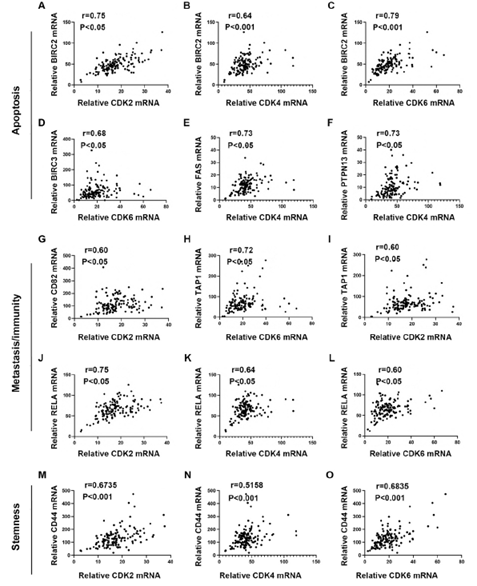
Figure 4: Pearson's correlation analysis showed the mRNA expression correlations between CDK2/4/6 and apoptosis/metastasis/immunogenicity/stemness-related genes in low-proliferating PDAC. A-F. The mRNA expression correlation between individual CDKs and protein functioning in apoptosis. CDK2 (A) and CDK6 (C) expressions showed strong (CDK2: r=0.75, p<0.05; CDK6: r=0.79, p<0.001) and CDK4 (B) showed a medium (r=0.64, p<0.001) correlations with BIRC2 expression. CDK6 (D) showed a medium correlation with BIRC3 (r=0.68, p<0.05). CDK4 showed a strong correlation with FAS (E) and PTPN13 (F) (FAS: r=0.73, p<0.05; PTPN13: r=0.73, p<0.05). The cutoff was set as r>0.6 and p value<0.05. G-L. The mRNA expression correlation between individual CDKs and protein functioning in metastasis or immunity. CDK2 expression showed a medium correlation with CD82 (r=0.60, p<0.05) (G). CDK6 showed a strong (r=0.72, p<0.05) (H), and CDK2 showed a medium (r=0.60, p<0.05) (I) correlation with TAP1. CDK2 showed a strong (r=0.75, p<0.05) (J), and CDK4 (K) and CDK6 (L) showed a medium (CDK4: r=0.64,
p<0.05; CDK6: r=0.60, p<0.05) correlation with RELA. The cutoff was set as r>0.6 and p value<0.05. M-O. The mRNA expression between individual CDKs and cancer stem cell markers. mRNA expression of CDK2 (M), CDK4 (N), and CDK6 (O) showed a medium correlation with CD44 (CDK2: r=0.6735, p<0.0001; CDK4: r=0.5158, p<0.0001; CDK6: r=0.6835, p=0.0011). The cutoff was set as r>0.5 and p value<0.05.
3.7 High CDK1 and low Ki67 expression Stage I/II PDAC patients display a shorter survival rate
To further analyze the role of CDKs in high-proliferating and low-proliferating cells, we investigated survival in patients by gene expression status. As expected, in the low CDK1/high MKI67 expression group, the median survival of PDAC patients was longer than in the high CDK1/high MKI67 group (532 versus 381 days, overall P=0.6507). Interestingly, the median survival of patients in the low CDK1/low MKI67 expression group was 627 days. In contrast, patients in the high CDK1/low MKI67 group demonstrated a median survival of 239 days(634 versus 239 days, overall P<0.0001) (Figure 5A). This further suggests CDK1 may have non-cell cycle-related functions to regulate low proliferating PDAC progression. There is no significant difference in the median survival of patients between low CDK2/low MKI67 and high CDK2/low MKI67 groups(627 versus 293 days, overall P=0.3954), and between low CDK2/high MKI67 and high CDK2/high MKI67 (532 versus 308 days, overall P=0.3050) (Figure 5B). There is also no significant difference in the median survival of patients between low CDK4/low MKI67 and high CDK4/low MKI67 groups(652 versus 598 days, overall P=0.1366), and between low CDK4/high MKI67 and high CDK4/high MKI67 (485 versus 308 days, overall P=0.4099) (Figure 5C). Of note, in the high CDK6/high MKI67 expression group, the median survival of PDAC patients was much shorter than the patient in the low CDK6/high MKI67 group (250 versus 684 days, overall P=0.0053)(Figure 5D). This suggests CDK6 may play a significant role in high-proliferating PDAC to promote PDAC progression.
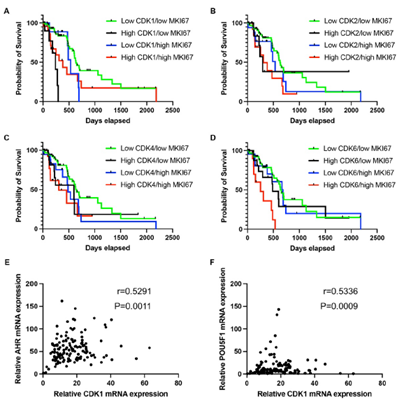
Figure 5: PDAC patients with high CDK1 and low ki67 expression showed reduced survival rates, and high CDK1 expression correlated with metastasis/stemness-related genes. A-D. The survival rate comparison in patients showing diverse mRNA expression levels of MKI67 and CDKs. Patients were grouped as low CDK/low MKI67 in which the mRNA expression of CDKs and MKI67 were <75th percentile; as high CDK/low MKI67 in which the mRNA expression of CDKs was ≥ 75th percentile and MKI67 was < 75th percentile; as low CDK/high MKI67 in which the mRNA expression of CDK expression < 75th percentile and MKI67 was ≥ 75th percentile; and as high CDK/high MKI67 in which the mRNA expression of both CDK and MKI67 were ≥ 75th percentile. E-F. mRNA expression correlation analysis was performed to analyze the association between CDK1 and proteins functioning in metastasis (AHR) and stemness (POU5F1). CDK1 mRNA expression showed a medium correlation with AHR (r=0.5291, p=0.0011) (E) and POU5F1 (r=0.5336, p=0.0009) (F). The cutoff was set as r>0.5 and p value<0.05.
3.8 CDK1 regulates the expressions of AHR and OCT4 under starvation conditions
CDK1 expression was positively associated with AHR and POU5F1/OCT4 expression in low-proliferating cells (low Ki67 expression) (Figure 5E&5F). We then applied both pharmacological and genetic approaches to test whether CDK1 regulated the expression of AHR and OCT4 in pancreatic cells. Four human pancreatic cancer cell lines were screened for the expression of CDK1 and Ki67. MiaPaca II cells expressed a relatively low level of CDK1, whereas PANC-1 displayed a relatively high level of CDK1 (Figure 6A). Two shRNA constructs were then applied to knock down CDK1 in pancreatic cancer cells. CDK1 knockdown in MiaPaca II cells and PANC-1 cells decreased the expressions of AHR and OCT4 (Figure 6B). CDK1 inhibitors were applied to test whether this regulation depended on the kinase activity of CDK1. Interestingly, we found that the expressions of AHR and OCT4 in pancreatic cells (MiaPaca II and PANC-1) were decreased when CDK1 was inhibited (CDK1-in-2) under starvation conditions, but not in the standard culture conditions (Figure 5C &5D). It is known that starvation slows down cell proliferation. This result suggests CDK1 may increase the expressions of AHR and OCT4 in low-proliferating cells to promote tumor progression.
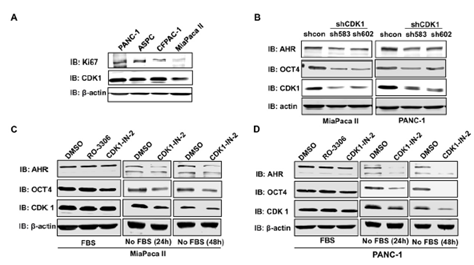
Figure 6: CDK1 depletion/inhibition downregulated the protein expressions of AHR and OCT4. A. Immunoblotting was performed to detect the protein expression levels of Ki67 and CDK1 in four pancreatic cancer cell lines. B. Two shRNA against CDK1 (sh583 and sh602) were applied to knock down CDK1 in MiaPaca II and PANC-1 pancreatic cell lines. Inhibitors (RO-2206, 5uM or CDK1-IN-1, 10uM) were applied to inhibit the CDK1 kinase activity in MiaPaca II cells (C) or PANC-1 (D) for 24 or 48 hr. The expression of Ki67, CDK1, AHR, and OCT4 was detected with immunoblotting using specific antibodies. b-actin is a loading control. In C and D, cells were cultured in the culture medium supplied with or without Fetal Bovine Serum (FBS).
4. Discussion
PDAC is a highly complex and malignant type of cancer in humans. Despite considerable improvements in surgical, radiation, and chemotherapeutic treatments, the mortality of PDAC didn’t decrease in decades. Thus, there is an urgent need to improve the understanding of the molecular mechanisms involved in PDAC pathogenesis. CDKs showed emerging attention in the pathogenesis and treatment of PDAC for its critical roles in cell cycle regulation and tumor progression [25] [26] [27]. However, the success of applying CDKs inhibitors, such as CDK4/6 inhibitors, for PDAC therapy is limited [28,29]. It is important to explore unknown mechanisms regulated by CDKs. In the current study, we first confirmed the association between the expression of CDKs and the survival in stage I/II of PDAC by the Kaplan-Meier method and Cox proportional hazards analysis. As expected, high CDK1, CDK2, CDK4, or CDK6 expression was associated with shorter median survival in the early stage of PDAC. MKI67 is a proliferative marker for cancers [30]. MKI67 increases expression in the S phase and is most highly expressed in M phase [31]. CDK4 and CDK6 are the G1 CDKs. CDK2 is responsible for S-phase entry, whereas CDK1 is the mitotic CDK that drives mitosis in M phase [32]. Our correlation analysis showed CDK1 expression being strongest associated with MKI67 in stage I/II PDAC among four CDKs is in alignment with previous observations.
Interestingly, in the low-proliferating early-stage PDAC, high expression of CDK1 is associated with shorter survival of the patients, which indicates CDK1 may have cell cycle-independent functions in regulating PDAC progression. Importantly, we found that CDK1 regulates the expressions of POU5F1 and AHR. This regulation is more significant in low-proliferating cells than in high-proliferating cells. POU5F1 is a marker for stemness and AHR functions in apoptosis. This suggests CDK1 may regulate cancer cell stemness and metastasis in low-proliferating cells to promote the progression of stage I/II PDAC, and targeting CDK1 may be a promising therapeutic strategy for low-proliferating PDAC patients. Interestingly, CDK2, CDK4, and CDK6 showed significant correlations with the expressions of proteins functioning in apoptosis, immunogenicity, and metastasis in low proliferating PDAC. This information also suggests that CDK2/4/6 may regulate PDAC progression through those cell cycle-independent functions. As those correlations only existed in low-proliferating cells, CDKs may be occupied for cell cycle function in high-proliferating cells, and they may be released for other functions when cells are in low proliferating status. This study has clinical significance as it may explain why inhibiting cell proliferation alone is ineffective for PDAC therapy. Importantly, our analysis showed that in 25% low MKI67 PDAC, low MKI67 expression is not associated with longer survival (P=0.7896). This suggests that in low-proliferating PDAC, proliferation is not the leading cause of PDAC progression.
CDK6 is first investigated as a product of a close analogous gene of CDK4 [33]. Even though CDK6 and CDK4 share 71% amino acid identity and exert redundant functions in cell cycle regulation, their roles differ in several aspects. For example, CDK6 has unique roles in cell development [34], DNA protection [35], metabolic homeostasis [36], centrosome stability [37] as well as in cancer cell survival [38], infiltration [39], immune evasion [40], and T-cell activation [41]. In this study, we found that both high expressions of CDK6 and CDK4 were associated with poor survival in stage I/II PDAC.
Inhibition of CDK4/6 with palbociclib significantly induces apoptosis in pancreatic tumor overexpressing Rb and also enhances the apoptotic effect of chemotherapeutic gemcitabine in patient- derived xenografts in mice [42]. CDK4/CDK6 inhibitor has been made into phase III clinical studies and is even FDA-approved for certain advanced breast cancers. However, clinical trials using CDK4/6 inhibitors are unsuccessful in PDAC [29,43]. Interestingly, in high-proliferating PDAC, only high expression of CDK6, but not other CDKs, correlates with shorter survival of PDAC patients. And in low-proliferating PDAC, CDK1 may be released for different functions (such as metastasis and stemness) to promote PDAC progression. This suggests we may target CDK6 in high proliferating PDAC and follow with CDK1 inhibition/depletion after the proliferation is inhibited.
Our analysis showed that expressions of CDKs are significantly associated with expression levels of proteins functioning in cell apoptosis/metastasis/immunity/stemness. Future studies may focus on whether and how CDKs regulate these proteins’ expressions. Of note, CDKs may regulate protein expressions through kinase-independent functions [44].It is important to know whether the regulation of CDKs in cell apoptosis/metastasis/immunity/stemness depends on their kinase activities. Pancreatic cancer is recognized as having a complex multistep etiology with the interaction between genetic susceptibility and environmental toxins. The most consistently mutated genes in PDAC are KRAS, CDKN2A, TP53, and SMAD4/DPC4 [45]. Tobacco smoking, alcohol, and obesity are known modifiable risk factors. New onset of diabetes (NoD) in subjects >50 years has also been documented as a high-risk factor in sporadic PDAC [46]. The current study used Cox proportional hazards regression to evaluate hazard ratios (HRs) in early-stage PDAC. Within the factors we explored, radiotherapy, chemotherapy, CDK1 expression, CDK2 expression, and CDK6 expression are associated with overall survival in stage I/II PDAC.
5. Conclusion
In summary, this study suggests CDKs may function in cell apoptosis, metastasis, immunity, and stemness besides cell proliferation in low-proliferating early-stage PDAC. As current clinical trials using CDKs inhibitors are not successful [47], developing selective and efficient CDK degraders, such as CDK1 degrader, might be promising for the early stage PDAC therapy.
Author Contributions:
Y.Z. and H.W. conceived the idea. S.Z., X.G. wrote the manuscript. H.Y. and L.L. helped with data curation, analysis, and plotting of the data. J.J. and Z.J. assisted with manuscript review and graphical representation. X. W., L. Z. and F. L. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding:
This work was supported by the National Natural Science Foundation of China (no. 81970825 to YZ), Department of Science and Technology of Sichuan Province (no. 2022JDJQ0062 to YZ, no. 2019YJ0592 to SZ), Chengdu Science and Technology Department (no. 2018-YF05-01080-SN to YZ), Sichuan returned oversea talent funding, and the National University Basic funding (ZYGX2021J034). This work was also supported by the NIH (R37CA251165 to HW) and Melanoma Research Alliance Young Investigator Award (821901 to HW).
Conflict of Interest:
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 66 (2016): 7-30.
- Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers 2 (2016): 16022.
- Strobel O, Neoptolemos J, Jager D, et al. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol 16 (2019) 11-26.
- Stark AP, Sacks GD, Rochefort MM, et al. Long-term survival in patients with pancreatic ductal adenocarcinoma. Surgery 159 (2016): 1520-1527.
- Yao W, Maitra A, Ying H. Recent insights into the biology of pancreatic cancer. EBioMedicine 53 (2020): 102655.
- Bond-Smith G, Banga N, Hammond TM, et al. Pancreatic adenocarcinoma. BMJ 344 (2012): 2476.
- National Cancer Institute. National Institutes of Health (2014).
- Singh RR, O'Reilly EM. New Treatment Strategies for Metastatic Pancreatic Ductal Adenocarcinoma. Drugs 80 (2020): 647-669.
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 371 (2014): 1039-1049.
- Buanes TA. Pancreatic cancer-improved care achievable. World J Gastroenterol 20 (2014): 10405-10418.
- Garcia-Reyes B, Kretz AL, Ruff JP, et al. The Emerging Role of Cyclin- Dependent Kinases (CDKs) in Pancreatic Ductal Adenocarcinoma. Int J Mol Sci 19 (2018).
- Ghelli Luserna di Rora A, Martinelli G, Simonetti, G. The balance between mitotic death and mitotic slippage in acute leukemia: a new therapeutic window? J Hematol Oncol 12 (2019): 123
- Law ME, Corsino PE, Narayan S, et al. Cyclin-Dependent Kinase Inhibitors as Anticancer Therapeutics. Mol Pharmacol 88 (2015): 846-852.
- Adamska A, Domenichini A, Falasca M. Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. Int J Mol Sci 18 (2017)
- Katz MH, Pisters PW, Evans DB, et al. Borderline resectable pancreatic cancer: the importance of this emerging stage of disease. J Am Coll Surg 206 (2008): 833-846.
- Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45 (2013): 1113- 1120.
- Ghafouri-Fard S, Khoshbakht T, Hussen BM, et al. A review on the role of cyclin dependent kinases in cancers. Cancer Cell Int 22 (2022): 325.
- Yamato A, Soda M, Ueno T, et al. Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor-kappaB-activating potential. Cancer Sci 106 (2015): 1137-1142.
- Dromard M, Bompard G, Glondu-Lassis M, et al. The putative tumor suppressor gene PTPN13/PTPL1 induces apoptosis through insulin receptor substrate-1 dephosphorylation. Cancer Res 67 (2007): 6806-6813.
- Bamberg A, Redente EF, Groshong SD, et al. Protein Tyrosine Phosphatase-N13 Promotes Myofibroblast Resistance to Apoptosis in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 198 (2018): 914-927.
- Nagata S. Fas ligand-induced apoptosis. Annu Rev Genet 33 (1999): 29-55.
- Nothdurft S, Thumser-Henner C, Breitenbucher F, et al. Functional screening identifies aryl hydrocarbon receptor as suppressor of lung cancer metastasis. Oncogenesis 102 (2020).
- Viera M, Yip GWC, Shen HM, et al. Targeting CD82/KAI1 for Precision Therapeutics in Surmounting Metastatic Potential in Breast Cancer. Cancers (Basel) 13 (2021).
- Chen T, Li J, Xu M, et al. PKCepsilon phosphorylates MIIP and promotes colorectal cancer metastasis through inhibition of RelA deacetylation. Nat Commun 8 (2017): 939-949.
- Diril MK, Ratnacaram CK, Padmakumar VC, et al. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci U S A 109 (2012): 3826-3831.
- Guo J, Kleeff J, Li J, et al. Expression and functional significance of CDC25B in human pancreatic ductal adenocarcinoma. Oncogene 21 (2004): 71-81.
- Calbo J, Serna C, Garriga J, et al. The fate of pancreatic tumor cell lines following p16 overexpression depends on the modulation of CDK2 activity. Cell Death Differ 11 (2004): 1055-1065.
- Dickson I. Pancreatic cancer: Tailored therapy for pancreatic tumour subtype. Nat Rev Gastroenterol Hepatol 15 (2018): 5.
- Goodwin CM, Waters AM, Klomp JE, et al. Combination Therapies with CDK4/6 Inhibitors to Treat KRAS-Mutant Pancreatic Cancer. Cancer Res 83 (2023): 141-157.
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol 182 (2000): 311-322.
- Darzynkiewicz Z, Zhao H, Zhang S, et al. Initiation and termination of DNA replication during S phase in relation to cyclins D1, E and A, p21WAF1, Cdt1 and the p12 subunit of DNA polymerase delta revealed in individual cells by cytometry. Oncotarget 6 (2015): 11735-11750.
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9 (2009): 153-166.
- Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol 14 (1994): 2077-2086.
- Grossel MJ, Hinds PW. From cell cycle to differentiation: an expanding role for cdk6. Cell Cycle 6 (2006): 266-270.
- Nagasawa M, Gelfand EW, Lucas JJ. Accumulation of high levels of the p53 and p130 growth-suppressing proteins in cell lines stably over-expressing cyclin- dependent kinase 6 (cdk6). Oncogene 20 (2001): 2889-2899.
- Zanuy M, Ramos-MontoyaA, Villacanas O, et al. Cyclin-dependent kinases 4 and 6 control tumor progression and direct glucose oxidation in the pentose cycle. Metabolomics 8 (2012): 454-464.
- Hussain MS, Baig SM, Neumann S, et al. CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Hum Mol Genet 22 (2013): 5199-5214.
- Wang H, Nicolay BN, Chick JM, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546 (2017): 426-430.
- Gao X, Qin S, Wu Y, et al. Nuclear PFKP promotes CXCR4-dependent infiltration by T cell acute lymphoblastic leukemia. J Clin Invest 131 (2021).
- Zhang J, Bu X, Wang H, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553 (2018): 91-95.
- Gao X, Wu Y, Chick JM, et al. Targeting protein tyrosine phosphatases for CDK6-induced immunotherapy resistance. Cell Rep 23 (2023): 112314.
- Chou A, Froio D, Nagrial AM, et al. Tailored first-line and second-line CDK4- targeting treatment combinations in mouse models of pancreatic cancer. Gut 67 (2018): 2142-2155.
- Digiacomo G, Volta F, Garajova I, et al. Biological Hallmarks and New Therapeutic Approaches for the Treatment of PDAC. Life (Basel) 11 (2021).
- Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol 17 (2016): 280-292.
- Miller MS, Allen P, Brentnall TA, et al. Pancreatic Cancer Chemoprevention Translational Workshop: Meeting Report. Pancreas 45 (2016): 1080-1091.
- Kenner BJ, Chari ST, Maitra A, et al. Early Detection of Pancreatic Cancer-a Defined Future Using Lessons From Other Cancers: A White Paper. Pancreas 45 (2016): 1073- 1079.
- Silvis MR, Silva D, Rohweder R, et al. MYC-mediated resistance to trametinib and HCQ in PDAC is overcome by CDK4/6 and lysosomal inhibition. J Exp Med 220 (2023).
Supplementary Tables:
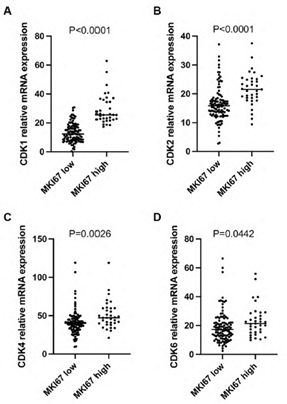
Supplementary Figure 1: The mRNA expression level of CDK1 (A), CDK2 (B), CDK4 (C), and CDK6 (D) was significantly higher in the MKI67 high expression group than the MKI67 low expression group in early-stage PDAC patients. Patients were categorized as a high expression group in which MKI67 mRNA expression was ≥75th percentile, and as a low expression group in which MKI67 mRNA expression was <75th percentile.
Supplementary Figure 2: Correlation analysis was performed to examine the mRNA expression between CDK1 and apoptosis, immunity, and metastasis-associated genes. mRNA expression of CDK1 showed no correlation with BIRC2 (A), XIAP (B), RELA (C), TAP1 (D), PTPN13 (E), CD82 (F), FAS (G), and BIRC3 (H). The cutoff was set as r>0.6 and p value<0.05.
