Bioinformatics Analysis of the Novel Conserved Micropeptides Encoded by the Plants of Family Brassicaceae
Article Information
Sergey Y Morozov1,2*, Dmitriy Y Ryazantsev3, Tatiana N Erokhina3
1Department of Virology, Biological Faculty, Lomonosov Moscow State University, Moscow, Russia
2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia
3Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Science, Moscow, Russia
*Corresponding Authors: Sergey Morozov, A. N. Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992, Moscow, Russia
Received: 25 September 2019; Accepted: 09 October 2019; Published: 15 October 2019
Citation: Sergey Y Morozov, Dmitriy Y Ryazantsev, Tatiana N Erokhina. Bioinformatics Analysis of the Novel Conserved Micropeptides Encoded by the Plants of Family Brassicaceae. Journal of Bioinformatics and Systems Biology 2 (2019): 066-077.
View / Download Pdf Share at FacebookAbstract
Background: The new class of plant small peptide regulators was recently shown to be encoded by primiRNA transcripts which can be transported to cytoplasm in unprocessed mRNA-like form. Striking similarities in general phenotypic activities between human miR200a/b and plant miR156a suggested us that a comparison with the coding potential between the corresponding pri-miRNAs could identify parallels in the encoded miPEPs.
Method: The study aimed to explore the protein coding ability of the pri-miR156a in Brassicaceae plants using bioinformatics analysis of the available proteomes and translatomes reported for Arabidopsis thaliana. Also, the physicochemical parameters of miPEP-156a were examined.
Results: Our analysis showed that predicted miPEP-156a micropeptide is evolutionarily conserved in plant family Brassicaceae. We propose that functional properties of miPEP-156a can be affected by posttranslational modifications.
Conclusion: Despite the well-known fact that primiRNAs are acting as non-protein-coding RNAs, the published data suggest that, in the plant genomes, some pri-miRNAs can also be found in polysomes, and the expression of these miRNA precursors may results in formation of micropeptides which may be involved in regulation of gene expression.
Keywords
microRNA, Pri-miRNA, Plant genome, Long non-coding RNA, Micropeptide, miPEP
microRNA articles, Pri-miRNA articles, Plant genome articles, Long non-coding RNA articles, Micropeptide articles, miPEP articles
microRNA articles microRNA Research articles microRNA review articles microRNA PubMed articles microRNA PubMed Central articles microRNA 2023 articles microRNA 2024 articles microRNA Scopus articles microRNA impact factor journals microRNA Scopus journals microRNA PubMed journals microRNA medical journals microRNA free journals microRNA best journals microRNA top journals microRNA free medical journals microRNA famous journals microRNA Google Scholar indexed journals Pri-miRNA articles Pri-miRNA Research articles Pri-miRNA review articles Pri-miRNA PubMed articles Pri-miRNA PubMed Central articles Pri-miRNA 2023 articles Pri-miRNA 2024 articles Pri-miRNA Scopus articles Pri-miRNA impact factor journals Pri-miRNA Scopus journals Pri-miRNA PubMed journals Pri-miRNA medical journals Pri-miRNA free journals Pri-miRNA best journals Pri-miRNA top journals Pri-miRNA free medical journals Pri-miRNA famous journals Pri-miRNA Google Scholar indexed journals Plant genome articles Plant genome Research articles Plant genome review articles Plant genome PubMed articles Plant genome PubMed Central articles Plant genome 2023 articles Plant genome 2024 articles Plant genome Scopus articles Plant genome impact factor journals Plant genome Scopus journals Plant genome PubMed journals Plant genome medical journals Plant genome free journals Plant genome best journals Plant genome top journals Plant genome free medical journals Plant genome famous journals Plant genome Google Scholar indexed journals Long non-coding RNA articles Long non-coding RNA Research articles Long non-coding RNA review articles Long non-coding RNA PubMed articles Long non-coding RNA PubMed Central articles Long non-coding RNA 2023 articles Long non-coding RNA 2024 articles Long non-coding RNA Scopus articles Long non-coding RNA impact factor journals Long non-coding RNA Scopus journals Long non-coding RNA PubMed journals Long non-coding RNA medical journals Long non-coding RNA free journals Long non-coding RNA best journals Long non-coding RNA top journals Long non-coding RNA free medical journals Long non-coding RNA famous journals Long non-coding RNA Google Scholar indexed journals Micropeptide articles Micropeptide Research articles Micropeptide review articles Micropeptide PubMed articles Micropeptide PubMed Central articles Micropeptide 2023 articles Micropeptide 2024 articles Micropeptide Scopus articles Micropeptide impact factor journals Micropeptide Scopus journals Micropeptide PubMed journals Micropeptide medical journals Micropeptide free journals Micropeptide best journals Micropeptide top journals Micropeptide free medical journals Micropeptide famous journals Micropeptide Google Scholar indexed journals miPEP articles miPEP Research articles miPEP review articles miPEP PubMed articles miPEP PubMed Central articles miPEP 2023 articles miPEP 2024 articles miPEP Scopus articles miPEP impact factor journals miPEP Scopus journals miPEP PubMed journals miPEP medical journals miPEP free journals miPEP best journals miPEP top journals miPEP free medical journals miPEP famous journals miPEP Google Scholar indexed journals DCL enzyme articles DCL enzyme Research articles DCL enzyme review articles DCL enzyme PubMed articles DCL enzyme PubMed Central articles DCL enzyme 2023 articles DCL enzyme 2024 articles DCL enzyme Scopus articles DCL enzyme impact factor journals DCL enzyme Scopus journals DCL enzyme PubMed journals DCL enzyme medical journals DCL enzyme free journals DCL enzyme best journals DCL enzyme top journals DCL enzyme free medical journals DCL enzyme famous journals DCL enzyme Google Scholar indexed journals genome sequences articles genome sequences Research articles genome sequences review articles genome sequences PubMed articles genome sequences PubMed Central articles genome sequences 2023 articles genome sequences 2024 articles genome sequences Scopus articles genome sequences impact factor journals genome sequences Scopus journals genome sequences PubMed journals genome sequences medical journals genome sequences free journals genome sequences best journals genome sequences top journals genome sequences free medical journals genome sequences famous journals genome sequences Google Scholar indexed journals bioinformatics BLAST articles bioinformatics BLAST Research articles bioinformatics BLAST review articles bioinformatics BLAST PubMed articles bioinformatics BLAST PubMed Central articles bioinformatics BLAST 2023 articles bioinformatics BLAST 2024 articles bioinformatics BLAST Scopus articles bioinformatics BLAST impact factor journals bioinformatics BLAST Scopus journals bioinformatics BLAST PubMed journals bioinformatics BLAST medical journals bioinformatics BLAST free journals bioinformatics BLAST best journals bioinformatics BLAST top journals bioinformatics BLAST free medical journals bioinformatics BLAST famous journals bioinformatics BLAST Google Scholar indexed journals ORFs articles ORFs Research articles ORFs review articles ORFs PubMed articles ORFs PubMed Central articles ORFs 2023 articles ORFs 2024 articles ORFs Scopus articles ORFs impact factor journals ORFs Scopus journals ORFs PubMed journals ORFs medical journals ORFs free journals ORFs best journals ORFs top journals ORFs free medical journals ORFs famous journals ORFs Google Scholar indexed journals
Article Details
1. Introduction
Previously, non-coding RNAs (ncRNAs), including micro-RNAs (miRNAs) and other long non-coding RNAs (lncRNAs), have been generally considered unable to encode proteins both in plants and animals [1-9]. The peptide encoded by lncRNA first attracted the attention of a group of scientists in the study of plant lncRNA in legumes [7, 10]. It was discovered that the gene, called an early nodulin 40 (Enod40), previously annotated as transcribed with the formation of lncRNA encodes actually two short peptides (with a length of 12 and 24 amino acid residues) in plants, where they participate in the organogenesis of the root nodules [10, 11]. Since then, many studies have been conducted to identify potential candidates among lncRNAs that can encode functional peptides (in a number of papers they are called microproteins, or sPEPs) [4, 5, 12]. Recent advances in bioinformatics, proteomics, and transcriptomics have shown that traditional computational algorithms used in the search for translatable open reading frames (ORFs) may have had omissions, as many modern studies have already identified hundreds of previously incorrectly annotated ORFs that have the potential to encode peptides. Researchers now consider the peptides encoded by lncRNAs as a new functional type because of their role in many biological processes [2-9, 12].
The first identification of microproteins in animals is related to studies of lncRNAs in Drosophila. It turned out that four peptides, encoded by a number of long non-coding RNAs, have a length of 11 to 32 residues and are necessary for embryonic development of flies [13, 14]. Since then, several microproteins have been functionally characterized, which may act, for example, as signals promoting cell migration and differentiation of human cells [5, 7]. It has recently been found that a group of such peptides plays an important role in calcium homeostasis, and thus affects regular muscle contractions [5, 7]. Another peptide has recently been identified and called a ”minion".The functional characteristic of this peptide [15] showed that the “minion” controls cell fusion and formation of muscles [16]. The functionality of microproteins has also been shown in the process of oncogenesis. For example, a small peptide that is encoded by lncRNA HOXB-AS3 inhibits oncogenesis by regulating alternative splicing and metabolic reprogramming of colon cancer cells [5-7, 17].
Since the first microproteins were functionally characterized in plants, analyses using high-throughput sequencing revealed a large number of ”translatable" long non-coding RNAs in various organisms [11, 12, 18]. These RNAs have been found to be involved in various biological processes, including plant growth and development, as well as response to environmental stresses [18]. It was shown that a peptide with a length of 36 residues, which is encoded by the gene POLARIS (PLS) in Arabidopsis affect root growth and microanatomy of the leaf blade (reviewed in [7]). In addition, two more microproteins, ROT18/ DLV1 and KOD, were characterized in Arabidopsis and found to participate in the processes of organogenesis and regulation of programmed cell death [5, 7, 19]. Two corn microproteins, Zm401p10 and Zm908p11, have also been recently identified and were shown to be involved in pollen development [12, 20]. Thus, the characteristics of microproteins indicate their functional diversity - from the effect on the morphogenesis of leaves and roots, pollen development to the programmable cell death.
Micro-RNAs derived from primary miRNAs (pri-miRNA) play a crucial role in posttranscription gene regulation by inhibiting translation or directing degradation of mRNA targets [21]. Currently, pri-miRNAs are regarded as specialized subclass of lncRNAs [3]. Indeed, pri-miRNAs like lncRNAs contain no long ORFs. However, obvious mark for pri-miRNAs is the hairpin region corresponding to pre-miRNA which is precursor for micro-RNAs [3] (Figure 1). Some miRNA genes are transcribed as lncRNAs by DNA-dependent RNA polymerase II. These lncRNAs were shown to contain “cap”-structure and poly(A)-tail. Most important, such lncRNAs include specific imperfect hairpin structures which are processed inside nucleus by DCL enzyme complexes giving rise to mature short double-stranded miRNA molecules with a length of 21-24 residues.
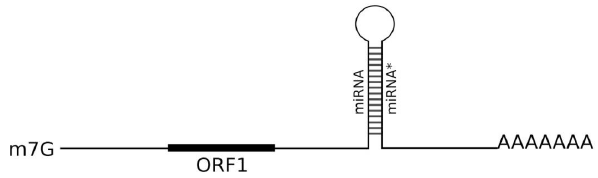
Figure 1: Schematic representation of pri-miRNA encoding miPEP microprotein upstream of the pre-miRNA stem-loop structure. Cap-structure (7-methyl guanosine [m7G]) and poly-A tract are shown.
One or sometimes both strands of such dsRNAs may function as molecular anchors in cytoplasmic AGO enzyme complexes through the base pairing with complementary sequences in the target RNAs, mediating the polynucleotide chain splitting or inhibition of their translation [22].
The new class of plant small peptide regulators was recently shown to be encoded by pri-miRNA transcripts which can be transported to cytoplasm in unprocessed mRNA-like form. Studies on Arabidopsis thaliana and Medicago truncatula have shown that some pri-miRNAs contain in their 5'-end part functional ORFs encoding peptides, the so-called miPEPs (Figure 1) [1, 2, 23]. Evidences obtained by in vivo overexpression of the corresponding ORFs or external spraying of plants with synthetic peptides, show that microproteins miPEP165a from Arabidopsis and miPEP171b of Medicago are able to activate the transcription of their own pri-miRNA messengers. Thus, a positive feedback loop is formed and resulted in increase the level of miRNA biogenesis. Treatment of M. truncatula plants with synthetic peptide miPEP171b increases endogenous expression of miR171b, which leads to a decrease in the density of the lateral roots. This effect of miPEP171b was specific because the peptide did not affect the expression of other miRNAs. Treatment of A. thaliana seedlings with miPEP165a also led to the specifically increased accumulation of miR165a [2, 23]. Recently, miPEP172c has been shown to control nodulation in soybean [5, 24]. It is known that several miRNAs regulate various stages of the process of nodule formation [25]. Soybean pri-miR172c was shown to stimulate nodulation by reducing the activity of factor nnc1 [26]. Even watering soybean plants with a solution containing a synthetic peptide miPEP172c, led to an increased number of nodules. This enhanced nodule formation is also correlated with increased pri-miR172c expression [24].
Occurrence of miPEP micropeptides is not unique for plants. Recently, it was shown that human miPEP-200a and miPEP-200b are encoded by the 5’-terminal upstream regions in pri-miRNAs of miR-200a and miR-200b, respectively [6, 27]. Importantly, miR-200a and miR-200b function per se as tumor suppressors inhibiting epithelial-mesenchymal transition in human hepatocellular carcinoma cell line [28]. However, miPEP-200a and miPEP-200b also affect the epithelial to mesenchymal transition in the case of prostate cancer cells [6, 27] suggesting common general functional pathways for the pri-miRNA-encoded peptide and small RNA.
Recently, we drew attention to the interesting fact that chemically synthesized miR156a encoded by plants of genus Brassica represses the epithelial–mesenchymal transition of human nasopharyngeal cancer cells and can be regarded as the main medicinal anticancer substance in broccoli assuming the ability of plant miRNAs to pass through the gastrointestinal tract of mammals [29]. Striking similarities in general phenotypic activities between human miR200a/b and plant miR156a suggested us that a comparison with the coding potential between the corresponding pri-miRNAs could identify parallels in the encoded miPEPs. We show here that more than 20 genomes of genus Brassica encode miPEPs of 33 amino acids with ORFs positioned in the 5’-proximal regions of pri-miR156a. Although these plant micropeptides demonstrate no sequence similarity to miPEPs-200a/b, their striking homology throughout plant species of whole family Brassicaceae suggests functional significance of miPEP-156a.
2. Materials and Methods
Sequences for comparative analysis were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/). The nucleic acid sequences and deduced amino acid sequences were analyzed and assembled using the NCBI. BLAST searches were carried out using the NCBI server with all available databases. An ORF search in plant genomic and transcriptomic sequences was performed with the ORF Finder on ExPasy platform (http://web.expasy.org). The secondary and tertiary structures of the proteins were predicted with the I-TASSER tool. In the absence of structural homologues, threading protein structure prediction approach was used for miPEP-156a. Threading is a fold recognition method to predict 3D structure of proteins. I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER) was used for three dimensional structure prediction. I-TASSER is an iterative threading program that builds 3D structure by using the hierarchical method. The miPEP-156a amino acid sequence was used to determine the polarity, accessibility, bulkiness and refractivity by using Protscale Server on ExPasy platform (http://web.expasy.org/protscale/). Prediction of glycosylation sites were determined through NetOGlyc and NetNGlyc web server (http://www.cbs.dtu.dk/services). Netphos 3.1 server (http://www.cbs.dtu.dk/services/NetPhos) determined the phosphorylation sites for each Thr, Ser and Tyr residues with 0.5 cut off threshold value. Protparam Server (http://web.expasy.org/protparam/) estimated half-life, molecular weight, and amino acid composition of miPEP-156a.
3. Results and Discussion
3.1 The coding potential of the miR156a transcript precursors (pri-miRNAs) in plants of genus Brassica
The availability of the B. napus (assembly Bra_napus_v2.0, release 2014/05/05), B. rapa (assembly Brapa_1.0, release 2018/05/26) and B. oleracea (assembly BOL, release 2018/05/26)genome sequences (http://www.ncbi.nlm.nih.gov/) has allowed us to perform comparative analysis of pri-miRNA in Brassica. Importantly, Brassica napus (rapeseed) is allopolyploid species evolved from diploid species B. oleracea and B. rapa [30]. So, to search for pri-miR156a-like sequences in the genomes and RNA transcripts inside genus Brassica, we performed bioinformatics BLAST analysis of the nucleotide sequences in the databases available at NCBI and used as a query B. rapa pre-miR156a sequence which is the only sequence of miR156a precursors available for genus Brassica at www.mirbase.org (bra-MIR156a MI0030547). Using this approach we first revealed several precursor RNA transcripts for pri-miR156a in B. napus, B. rapa and B. oleracea (Table 1). In the case of B. rapa and B. oleracea, pri-miR156a contained highly conserved ORFs of 32 and 34 codons (including termination codon), respectively. These ORFs were found 40-90 nucleotides from the 5’ end of sequence reads and located 342 and 52 nucleotides upstream of miRNA sequence in pre-miR156a of B. oleracea and B. rapa, respectively (Table 1). Allopolyploid Brassica napuscontained both types of transcripts originated from parental species (Table 1).
The conserved short ORFs were revealed also in the genomic sequences of the Brassica species mentioned above (Table 1). Moreover, analysis of genomic sequences allowed us to reveal conserved ORFs coding for miPEP-156a in two more species, Brassica cretica and Brassica juncea (Table 1).
3.2 The coding potential of the miR156a transcript precursors in plants of family Brassicaceae
Comparative analysis of amino acid sequences of miPEP-156a in genus Brassica (Figure 2) showed that a similar peptide was predicted in a computer analysis performed previously in a pioneering study of miPEPs in Arabidopsis thaliana (see Table 2, Extended data, in reference [23]). To investigate whether the predicted miPEP-156a micropeptides are conserved not only in A. thaliana but also in other plants of family Brassicaceae, we performed BLASTN and TBLASTN analyses of databases presented in NCBI. Unexpectedly, we found that miPEP-156a micropeptides are conserved at least in 11 tribes of family Brassicaceae (Table 1). These tribes include Brassiceae, Camelineae, Arabideae, Boechereae, Cardamineae, Conringieae, Euclidieae, Eutremeae, Schizopetaleae, Sisymbrieae, Thlaspideae (Table 1, Figure 2). No protein sequences similar to predicted miPEP-156a were revealed to be encoded in plant families other than Brassicaceae.
3.3 Putative deviations in the expression modes of the miPEP-156a ORFs
Comparison of the available genomic and transcriptomic nucleotide sequences for themiPEP-156a ORFs showed that in most cases (including tribe Brassiceae) the coding sequences of miPEPs are identical ingenomic loci and transcribed RNAs. However, we revealed two peculiar cases where post-transcriptional splicing may change the sequence of the encoded miPEP. First, splicing of the Camelina sativa pri-miR156a results in reducing the length of predicted miPEP-156a by 6 amino acids and changing the primary structure of the micropeptide C-terminal half (Table 1). Second, spliced transcript of the Capsella rubella pri-miR156a encodes predicted miPEP-156a which is shorter from the C terminus by 34 amino acids in comparison with variant of the micropeptide encoded by genomic DNA (Table 1). We also revealed an additional deviation in the expression mode of some miPEPs. It was found that in three plant species miPEP-156a ORFs use non-canonical translation initiation codons instead of usual AUG triplet (see below). Currently, it is well known that eukaryotic ribosomes have an ability to start translation from the rare initiation sites representing, particularly, some codons for Leu, ACG codon for Thr, GUG codon for Val and all codons for Ile [31]. Such non-canonical start codons are often found as initiators for short upstream open reading frames (uORFs) in the 5' untranslated terminal regions of eukaryotic mRNAs [32, 33]. In the case of miPEP-156a ORFs, non-canonical potential translation initiation codons of Ile, Leu and Val were revealed instead of AUG in Eutrema yunnanense, Leavenworthia alabamica and Euclidium syriacum (Table 1 and Figure 2). Interestingly, unlike E. yunnanense miPEP-156a ORF, which uses non-canonical AUA potential initiator, closely related plant E. heterophyllum encodes miPEP-156a ORF with normal AUG start codon (Table 1).
3.4 Peculiarities of the primary structure of miPEP-156a
Sequence alignment of predicted miPEP-156a micropeptides was obtained using complete miPEP-156a ORF sequences from Brassicaceae retrieved from NCBI DataBank (Table 1). We conducted multiple alignments using Clustal W algorithms in MEGA 6.06 software [34]. Based on this alignment, family-wide conserved residues were identified (Figure 2). It was revealed that miPEP-156a microproteins from genus Brassica contain all most conserved residues and can be regarded as a representative protein family member. It should be noted that several microproteins, for example Barbarea vulgaris miPEP-156a (Figure 2), show much less similarity to micropeptides from genus Brassica than most other members of peptide family.
3.4.1 Amino acid sequence parameters of miPEP-156a:
The physical parameters of Brassica rapa miPEP-156a microprotein were predicted using Protparam server (https://web.expasy.org/protparam/) (Table 2). The physico-chemical properties were determined by Protscale server (https://web.expasy.org/protscale/). The hydrophobicity prediction values according to Hopp and Woods were between −1.5 and 2.0. These data revealed no highly hydrophobic region and significantly hydrophilic C-terminal half of predicted miPEP-156a. It is known that the protein bulkiness may affect the local structure of a protein. We revealed that the bulkiness values of miPEP-156a range from 10.5 to 19.5. The dipole-dipole intermolecular interactions between the positively and negatively charged particles depicted polarity as predicted through Zimmerman score. The predicted score for predicted miPEP-156a was found between 1.2 and 34. These data showed that micropeptide possesses significant internal polarity.
3.4.2 Possible protein modification sites in the miPEP-156a:
We also attempted to predict the protein modification sites that could potentially modify the structure miPEP-156a using http://www.cbs.dtu.dk/ services [35]. According to Netphos 3.1 server predictions, we revealed the presence of two potential Ser and Thr phosphorylation sites (Ser-4 and Thr-24). Moreover, potential O-(alpha)-GlcNAc glycosylation site was found in the N-terminal region of the micropeptide at Ser-4 (Figure 2).
3.5 Peculiarities of the secondary and tertiary structures of the predicted miPEP-156a
Due to the absence of suitable experimental structural models, traditional sequence similarity modeling cannot be used. The secondary structure and three-dimensional models of the predicted miPEP-156a were predicted in silico using protein structure prediction method I-TASSER (Iterative Threading ASSEmbly Refinement). The secondary structure of micropeptide showed that the sequence contains mainly alpha helices (residues 7-12 and 17-28), as well as coils (residues 13-16 and 29-33) and extended strand (residues 1-6) (Figure 2). In order to get the spatial structure of predicted miPEP-156a, an ab-initio approach was used. By using this approach several models were generated through I-Tasser for B. rapa and A. thaliana. All models suggested that miPEP-156a is indeed alpha-helical protein with two ordered domains (Figure 3).
|
Plant species |
Tribe |
Genome vs RNA |
Number of codons/ distance between stop codon and mature miR156 sequence |
Sequence source (accession) |
|
|
Brassica rapa |
Brassiceae |
Genome |
34 codons/348 nts |
OVXL02000005 |
|
|
Brassica napus |
Brassiceae |
Genome |
34 codons/340 nts |
JMKK02008479 |
|
|
Brassica oleracea |
Brassiceae |
Genome |
34 codons/351 nts |
JJMF01000005 |
|
|
Brassica juncea |
Brassiceae |
Genome |
34 codons/351 nts |
LFQT01009712 |
|
|
Brassica cretica |
Brassiceae |
Genome |
34 codons/441 nts |
QGKX01337315 |
|
|
Arabidopsis thaliana |
Camelineae |
Genome |
34 codons/338 nts |
OMOK01000005 |
|
|
Arabidopsis lyrata |
Camelineae |
Genome |
33 codons/341 nts |
ADBK01000693 |
|
|
Arabidopsis halleri |
Camelineae |
Genome |
34 codons/423 nts |
RCNM01027982 |
|
|
Camelina sativa |
Camelineae |
Genome |
34 codons/356 nts |
JFZQ01000450 |
|
|
Capsella rubella |
Camelineae |
Genome |
71 codons/224 nts |
ANNY01001269 |
|
|
Arabis nordmanniana |
Arabideae |
Genome |
37 codons/377 nts |
LNCG01256614 |
|
|
Arabis montbretiana |
Arabideae |
Genome |
27 codons/357 nts |
LNCH01011198 |
|
|
Boechera stricta |
Boechereae |
Genome |
34 codons/331 nts |
MLHT01001679 |
|
|
Barbarea vulgaris |
Cardamineae |
Genome |
31 codons/335 nts |
LXTM01000340 |
|
|
Conringia planisiliqua |
Conringieae |
Genome |
34 codons/368 nts |
FNXX01000016 |
|
|
Euclidium syriacum |
Euclidieae |
Genome |
>41 codons*/344 nts |
FPAK01000008 |
|
|
Eutrema heterophyllum |
Eutremeae |
Genome |
34 codons/331 nts |
PKMM01027123 |
|
|
Eutrema yunnanense |
Eutremeae |
Genome |
>38 codons*/338 nts |
PKML01043689 |
|
|
Sisymbrium irio |
Sisymbrieae |
Genome |
34 codons/340 nts |
ASZH01009632 |
|
|
Thlaspi arvense |
Thlaspideae |
Genome |
28 codons/375 nts |
AZNP01000437 |
|
|
Caulanthus amplexicaulis |
Schizopetaleae |
RNA |
33 codons/31 nts |
GGBZ01008949 |
|
|
Brassica napus |
Brassiceae |
RNA |
34 codons/52 nts |
XR_001274070 |
|
|
Brassica oleracea |
Brassiceae |
RNA |
34 codons/351 nts |
XR_001263889 |
|
|
Brassica napus |
Brassiceae |
RNA |
34 codons/340 nts |
XR_001278112 |
|
|
Camelina sativa |
Camelineae |
RNA |
28 codons/356 nts |
XR_002035989 |
|
|
Capsella rubella |
Camelineae |
RNA |
37 codons/45 nts |
XR_002834317 |
|
|
Arabidopsis lyrata |
Camelineae |
RNA |
33 codons/341 nts |
XR_002332546 |
*-indicates that miPEP-156a ORF does not contain canonical initiation AUG codon. Upper part of table contains genome-derived sequence data, whereas lower part represents RNA-derived data.
Table 1: List of the miPEP-156a ORFs in genomic and transcribed sequences of Brassicaceae.

Figure 2: Multiple sequence alignment of representative sequences of miPEP-156a micropeptides from Brassicaceae plants. Alignment was generated at MEGA 6.06 software. Amino acid sites that are different from B. rapa sequence are in yellow; micropeptides, which ORFs start with non-canonical translation start, are marked by blue starting residues and by asterisks. Two residues in B. rapa sequence (Ser and Thr), which are proposed to be modified postranslationally, are in italic (see section 3.4.2.). Potential O-(alpha)-GlcNAc glycosylation site (Ser) is underlined (see section 3.4.2.).The proposed secondary structure of the predicted B. rapa miPEP-156a (according to I-TASSER, see section 3.4.2.) is shown above the corresponding sequence.

Table 2: Predicted physical parameters of Brassica rapa miPEP-156a according Protparam.
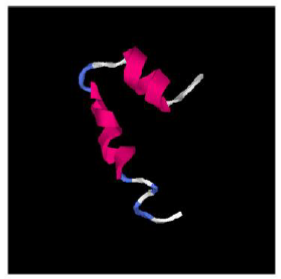
Figure 3: The B. rapa miPEP-156a spatial model based on the I-TASSER top ranked model (by consensus score). The N-terminal peptide part is in the top part of the figure.
Moreover, using http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=HOMOMER we predicted that miPEP-156a can form tetramers (Figure 4).
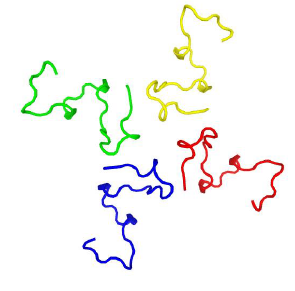
Figure 4: The B. rapa miPEP-156a oligomeric spatial model based on http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=HOMOMER top ranked model (by consensus score). The N-terminal peptide part is in the top part of the figure.
4. Conclusions
In the current study the occurrence and structural characterization of predicted miPEP-156a in the family Brassicaceae has been undertaken using bioinformatic approaches. Our analysis showed that this micropeptide is evolutionarily conserved in a particular plant family. We propose that functional properties of miPEP-156a can be affected by post-translational modifications. Particularly, it was predicted that phosphorylation and glycosylation which are the most common types of post-translational modifications of proteins are predicted for micropeptide with significant confidence [36, 37].
The peptides predicted in our study may affect some steps in the plant development as it was shown for miPEP165a from Arabidopsis and miPEP171b of Medicago [23, 24]. These peptides regulate the expression of their corresponding miRNAs and potentiate the activity of target genes involved in organ and tissue development. Finally, to get more direct evidence for coding the miPEP-156a in Brassicaceae plants, we performed analysis of the available proteomes and translatomes reported for Arabidopsis thaliana. Although proteomic search of miPEP-156a was failed, large-scale transcriptomic studies have indicated that miPEP-156a-specific transcripts show obvious positive expression profiles in seedlings. We performed BLAST analysis of Sequence Read Archive (SRA), which is the NCBI database collecting sequence data obtained by the use of next generation sequence (NGS) technology, using the A. thalianamiPEP-156a genome coding region as a query. Ribosome-associated miPEP-156a RNAs were found for polysomes isolated by conventional biochemical methods (NCBI accessions SRX345247, SRX1756766, SRX1756767, SRX1808281, SRX1808312) as well as by immunoprecipitation of an epitope-tagged ribosomal protein L18 [38] (NCBI accessions SRX3204187, SRX3204194, SRX3204195, SRX3204199). Thus we achieved first direct evidence that pri-miR156 RNAs undergo translation at least in seedlings of A. thaliana.
Co-authors Contributions
S.Y.M.: proposed the research problem, wrote, revised and submitted the manuscript; T.N.E.: run statistical analysis of results, and wrote the manuscript; D.Y.R.: conducted computer analysis and wrote the first draft of the manuscript.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Acknowledgements
The work of Sergey Morozov, Tatiana Erokhina and Dmitriy Ryazantsev was supported by the Russian Foundation for Basic Reasearch (grant 19-04-00174).. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Hellens RP, Brown CM, Chisnall MAW, et al. The Emerging World of Small ORFs. Trends Plant Sci 21 (2016): 317-328.
- Couzigou JM, Lauressergues D, Bécard G, et al. miRNA-encoded peptides (miPEPs): A new tool to analyze the roles of miRNAs in plant biology. RNA Biol. 12 (2015): 1178-1180.
- Li LJ, Leng RX, Fan YG, et al. Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res 361 (2017): 1-8.
- Li Q, Ahsan MA, Chen H, et al. Discovering Putative Peptides Encoded from Noncoding RNAs in Ribosome Profiling Data of Arabidopsis thaliana. ACS Synth Biol 7 (2018): 655-663.
- Matsumoto A, Nakayama KI. Hidden Peptides Encoded by Putative Noncoding RNAs. Cell Struct Funct 43 (2018): 75-83.
- Zhu S, Wang J, He Y, et al. Peptides/Proteins Encoded by Non-coding RNA: A Novel Resource Bank for Drug Targets and Biomarkers. Front. Pharmacol 9 (2018): 1295.
- Yeasmin F, Yada T, Akimitsu N. Micropeptides Encoded in Transcripts Previously Identified as Long Noncoding RNAs: A New Chapter in Transcriptomics and Proteomics. Front Genet 9 (2018): 144.
- Ruiz-Orera J, Albà MM. Translation of Small Open Reading Frames: Roles in Regulation And Evolutionary Innovation. Trends Genet 35 (2019): 186-198.
- Choi SW, Kim HW, Nam JW. The small peptide world in long noncoding RNAs. Brief Bioinform (2018).
- Rohrig H, Schmidt J, Miklashevichs E, et al. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci U.S.A 99 (2002): 1915-1920.
- Kereszt A, Mergaert P, Montiel J, et al. Impact of Plant Peptides On Symbiotic Nodule Development and Functioning. Front Plant Sci 9 (2018): 1026.
- Sousa ME, Farkas MH. Micropeptide. PLoS Genet 14 (2018): 1007764.
- Galindo MI, Pueyo JI, Fouix S, et al. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5 (2007): 106.
- Kondo T, Hashimoto Y, Kato K, et al. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9 (2007): 660-665.
- Nelson BR, Makarewich CA, Anderson DM, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351 (2016): 271-275.
- Zhang Q, Vashisht AA, Rourke JO, et al. The microprotein Minion controls cell fusion and muscle formation. Nat Commun 8 (2017): 15664.
- Huang JZ, Chen M, Chen D, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 68 (2017): 171-184.
- Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet 30 (2014): 439-452.
- Guo P, Yoshimura A, Ishikawa N, et al. Comparative analysis of the RTFL peptide family on the control of plant organogenesis. J Plant Res 128 (2015): 497-510.
- Dong X, Wang D, Liu P, et al. Zm908p11, encoded by a short open reading frame (sORF) gene, functions in pollen tube growth as a profilin ligand in maize. J Exp Bot 64 (2013): 2359-2372.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2004): 281-297.
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25 (2013): 2383-2399.
- Lauressergues D, Couzigou JM, Clemente HS, et al. Primary transcripts of microRNAs encode regulatory peptides. Nature 520 (2015): 90-93.
- Couzigou JM, André O, Guillotin B, et al. Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean. New Phytol 211 (2016): 379-381.
- Couzigou JM, Combier JP. Plant microRNAs: key regulators of root architecture and biotic interactions,. New Phytol 212 (2016): 22-35.
- Wang L, Liu L, Ma Y, et al. Transcriptome profiling analysis characterized the gene expression patterns responded to combined drought and heat stresses in soybean. Comput Biol Chem 77 (2018): 413-429.
- Fang J, Morsalin S, Rao VN, et al. Decoding of non-coding DNA and non-coding RNA: pri-micro RNA-encoded novel peptides regulate migration of cancer cells. J Pharm Sci Pharm 3 (2017): 23-27.
- Zhong C, Li MY, Chen ZY, et al. Micro RNA-200a inhibits epithelial-mesenchymal transition in human hepatocellular carcinoma cell line. Int J Clin. Exp Pathol 8 (2015): 9922-9931.
- Tian Y, Cai L, Tu Y, et al. miR156a mimic represses the epithelial-mesenchymal transition of human nasopharyngeal cancer cells by targeting junctional adhesion molecule A. PLoS One 11 (2016): e0157686.
- Bin Z, Qi P, Dongao H, et al. Transcriptional Aneuploidy Responses of Brassicarapa-oleraceaMonosomic Alien Addition Lines (MAALs) Derived From Natural Allopolyploid B. napus, Front Genet 10 (2019): 67.
- Diaz de Arce AJ, Noderer WL, Wang CL. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46 (2018): 985-994.
- Ivanov IP, Wei J, Caster SZ, et al. Translation Initiation from Conserved Non-AUG Codons Provides Additional Layers of Regulation and Coding Capacity. M Bio 8 (2017): 844-917.
- Jankowsky E, Guenther UP. A helicase links upstream ORFs and RNA structure. Curr Genet 65 (2019): 453-456.
- Tamura K, Stecher G, Peterson D, et al. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30 (2013): 2725-2729.
- Blom N, Sicheritz-Ponten T, Gupta R, et al. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4 (2004): 1633-1649.
- Koh GCKW, Porras P, Aranda B, et al. Analyzing protein-protein interaction networks. Journal of Proteome Research 11 (2012): 2014-2031.
- Yu CS, Cheng CW, Su WC, et al. CELLO2GO: a web server for protein subcellular localization prediction with functional gene ontology annotation. PLOS ONE 9 (2014).
- Lin SY, Chen PW, Chuang MH, et al. Profiling of translatomes of in vivo-grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell 26 (2014): 602-618.
