Bioinformatics Analysis and Identification of the Expression and Mechanism of Immune-Related Gene MAPT in Hepatocellular Carcinoma
Article Information
Qi-ming Gong1#, Duan-kai Chen2#, Fa-hui Liu2#, Yuan-xun Gong2, Yan Jiang3*
1Department of Nephrology, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise , China
2Hepatobiliary Diseases Clinical Medicine Research Center of Guangxi, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise , China
3Institutes of Life Science, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise , China
# - These authors have equally contributed to this work.
*Corresponding author: Yan Jiang, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise, China
Received: 12 October 2021; Accepted: 21 October 2021; Published: 29 October 2021
Citation: Qi-ming Gong, Duan-kai Chen, Fa-hui Liu, Yuan-xun Gong, Yan Jiang. Bioinformatics Analysis and Identification of the Expression and Mechanism of Immune-Related Gene MAPT in Hepatocellular Carcinoma. Journal of Bioinformatics and Systems Biology 4 (2021): 140-157.
View / Download Pdf Share at FacebookAbstract
MAPT (Microtubule Associated Protein Tau) is a Protein Coding gene. Aberrant expression of MAPT has been reported to be associated with several types of tumors, such as gastric, breast, and colorectal cancer, and so on. However, the role of MAPT in HCC (Hepatic carcinoma) is still poorly understood. In our research, MAPT-related data mining and analyzing were used publicly-available data from TCGA, GEO, Oncomine, and HPA databases. The survival curve was shown and analyzed by using Kaplan Meier Plotter. Gene Set Enrichment Analysis (GSEA), TIMER, STRING, and R package were used to explore the function and potential mechanism of MAPT in HCC. The results show that MAPT is overexpressed in HCC samples and is correlated with worse prognosis of patients with HCC. Bioinformatics analysis showed that MAPT contributed to the development of tumors through a variety of mechanisms. In conclusion, the Up-regulation of MAPT was significantly linked to poor prognosis in HCC patients, and it could be a new therapeutic target for HCC therapy.
Keywords
MAPT; Hepatocellular carcinoma; Prognosis; Bioinformatics analysis
MAPT articles; Hepatocellular carcinoma articles; Prognosis articles; Bioinformatics analysis articles
MAPT articles MAPT Research articles MAPT review articles MAPT PubMed articles MAPT PubMed Central articles MAPT 2023 articles MAPT 2024 articles MAPT Scopus articles MAPT impact factor journals MAPT Scopus journals MAPT PubMed journals MAPT medical journals MAPT free journals MAPT best journals MAPT top journals MAPT free medical journals MAPT famous journals MAPT Google Scholar indexed journals Hepatocellular carcinoma articles Hepatocellular carcinoma Research articles Hepatocellular carcinoma review articles Hepatocellular carcinoma PubMed articles Hepatocellular carcinoma PubMed Central articles Hepatocellular carcinoma 2023 articles Hepatocellular carcinoma 2024 articles Hepatocellular carcinoma Scopus articles Hepatocellular carcinoma impact factor journals Hepatocellular carcinoma Scopus journals Hepatocellular carcinoma PubMed journals Hepatocellular carcinoma medical journals Hepatocellular carcinoma free journals Hepatocellular carcinoma best journals Hepatocellular carcinoma top journals Hepatocellular carcinoma free medical journals Hepatocellular carcinoma famous journals Hepatocellular carcinoma Google Scholar indexed journals Prognosis articles Prognosis Research articles Prognosis review articles Prognosis PubMed articles Prognosis PubMed Central articles Prognosis 2023 articles Prognosis 2024 articles Prognosis Scopus articles Prognosis impact factor journals Prognosis Scopus journals Prognosis PubMed journals Prognosis medical journals Prognosis free journals Prognosis best journals Prognosis top journals Prognosis free medical journals Prognosis famous journals Prognosis Google Scholar indexed journals Bioinformatics analysis articles Bioinformatics analysis Research articles Bioinformatics analysis review articles Bioinformatics analysis PubMed articles Bioinformatics analysis PubMed Central articles Bioinformatics analysis 2023 articles Bioinformatics analysis 2024 articles Bioinformatics analysis Scopus articles Bioinformatics analysis impact factor journals Bioinformatics analysis Scopus journals Bioinformatics analysis PubMed journals Bioinformatics analysis medical journals Bioinformatics analysis free journals Bioinformatics analysis best journals Bioinformatics analysis top journals Bioinformatics analysis free medical journals Bioinformatics analysis famous journals Bioinformatics analysis Google Scholar indexed journals MAPT articles MAPT Research articles MAPT review articles MAPT PubMed articles MAPT PubMed Central articles MAPT 2023 articles MAPT 2024 articles MAPT Scopus articles MAPT impact factor journals MAPT Scopus journals MAPT PubMed journals MAPT medical journals MAPT free journals MAPT best journals MAPT top journals MAPT free medical journals MAPT famous journals MAPT Google Scholar indexed journals AMELY articles AMELY Research articles AMELY review articles AMELY PubMed articles AMELY PubMed Central articles AMELY 2023 articles AMELY 2024 articles AMELY Scopus articles AMELY impact factor journals AMELY Scopus journals AMELY PubMed journals AMELY medical journals AMELY free journals AMELY best journals AMELY top journals AMELY free medical journals AMELY famous journals AMELY Google Scholar indexed journals Gene expression data articles Gene expression data Research articles Gene expression data review articles Gene expression data PubMed articles Gene expression data PubMed Central articles Gene expression data 2023 articles Gene expression data 2024 articles Gene expression data Scopus articles Gene expression data impact factor journals Gene expression data Scopus journals Gene expression data PubMed journals Gene expression data medical journals Gene expression data free journals Gene expression data best journals Gene expression data top journals Gene expression data free medical journals Gene expression data famous journals Gene expression data Google Scholar indexed journals Protein-Protein interaction articles Protein-Protein interaction Research articles Protein-Protein interaction review articles Protein-Protein interaction PubMed articles Protein-Protein interaction PubMed Central articles Protein-Protein interaction 2023 articles Protein-Protein interaction 2024 articles Protein-Protein interaction Scopus articles Protein-Protein interaction impact factor journals Protein-Protein interaction Scopus journals Protein-Protein interaction PubMed journals Protein-Protein interaction medical journals Protein-Protein interaction free journals Protein-Protein interaction best journals Protein-Protein interaction top journals Protein-Protein interaction free medical journals Protein-Protein interaction famous journals Protein-Protein interaction Google Scholar indexed journals Immune infiltration analysis articles Immune infiltration analysis Research articles Immune infiltration analysis review articles Immune infiltration analysis PubMed articles Immune infiltration analysis PubMed Central articles Immune infiltration analysis 2023 articles Immune infiltration analysis 2024 articles Immune infiltration analysis Scopus articles Immune infiltration analysis impact factor journals Immune infiltration analysis Scopus journals Immune infiltration analysis PubMed journals Immune infiltration analysis medical journals Immune infiltration analysis free journals Immune infiltration analysis best journals Immune infiltration analysis top journals Immune infiltration analysis free medical journals Immune infiltration analysis famous journals Immune infiltration analysis Google Scholar indexed journals
Article Details
1. Introduction
Hepatocellular carcinoma (HCC) is the most common malignancy with a high mortality rate in many parts of the world [1]. During the last few decades, significant advances have been achieved in understanding the epidemiology, risk factors, and molecular mechanisms in HCC [2]. Nevertheless, the annual mortality rate associated with HCC has increased substantially over the past two decades [3]. Determining accurate prognostic biomarkers and revealing therapeutic targets for HCC is therefore urgent. The microtubule-associated protein tau (MAPT) gene is located on chromosome 17 (17q21) and encodes for the protein Tau [4]. The MAPT gene is of central importance for several neurodegenerative diseases [5]. Interestingly, growing numbers of studies in recent years have been conducted on the function of MAPT in tumors; its expression and role in different tumors have been found to be changed. Previous research demonstrated that higher serum levels of MAPT were independently related to poor prognosis in patients with metastatic breast cancer [6]. Conversely, initial analyses, reported by Saif Zaman et al., suggested that a higher expression of MAPT associated with better overall survival in neuroblastoma [7].
However, MAPT in HCC has been rarely discussed. Zhang developed and validated a prognostic nomogram to predict the overall survival of HCC patients, in which MAPT acts as a significant variable [8]. Bioinformatics analysis by Qi revealed that MAPT serves as an upstream regulator of AMELY in HCC [9]. Despite this, the expression level, clinicopathological significance, and the biological functions and associated mechanisms of MAPT in HCC had not reported to date. In this study, GEO, Oncomine, HPA, and TCGA databases were used to clarify the potential connection between the expression of MAPT and prognosis of HCC patients, as well as the association between MAPT and clinical parameters. Furthermore, the correlation between the expression of MAPT and prognosis of HCC patients was performed by using survival analysis based on Kaplan Meier Plotter. Finally, initial studies of the mechanisms of MAPT in HCC were carried out by a series of online databases and bioinformatics software packages, hoping to provide useful insights into the current research in HCC.
2. Methods
2.1 Data resource and description
Gene expression data (HTSeq-counts) of 424 HCC samples, including 374 primary tumors and 50 solid tissue normal samples, were downloaded from TCGA (https://cancergenome.nih.gov/). Gene expression analysis was performed by R(v3.6.2) using the limma [10] packages to look for the differentially expressed of MAPT between tumor and non-tumor tissue samples. A meta-analysis of MAPT expression in 5 analyses was shown by using Oncomine (https://www.oncomine.org) [11]. Three expression microarray series GSE25097, GSE45436, GSE55092, including HCC tumor and non-tumor tissue samples were obtained from the Gene Expression Omnibus dataset (GEO, https://www.ncbi.nlm.nih.gov/geo/). The Human Protein Atlas database (HPA, https://www.proteinatlas.org/) [12] was used to identify the difference of MAPT protein expression between HCC tissues and adjacent normal samples. A total of 2498 immune?related genes were downloaded from the ImmPort database (http://www.immport.org/), and 500 most differential survival genes in HCC were obtained from Gene Expression Profiling Interactive Analysis (GEPIA) online database (http://gepia.cancer-pku.cn/) [13]. Additionally, survival curves of MAPT in HCC patients in different subgroups were constructed by using the Kaplan Meier plotter (http://kmplot. com/analysis/) [14].
2.2 Gene set enrichment analysis
Gene expression data were divided into a high and a low group based on the median level of MAPT expression to elucidate the significant function and pathway of MAPT in HCC prognosis by using GSEA software (https://software.broadinstitute.org/gsea/) [15]. A function or pathway term with P-value <0.05. FDR<0.05 was considered statistically significant.
2.3 Protein interaction network of MAPT and GO enrichment analyses
We perform a Protein-Protein interaction network of MAPT by using the STRING database (https://string-db.org/). Gene Ontology (GO) enrichment analyses were used to investigate gene sets related to MAPT by using the R package "cluster profiler" [16]. In our research. GO terms were visualized by R package "GO plot" [17], and terms with FDR < 0.05 were considered statistically significant.
2.4 Analysis of gene expression and tumor-infiltrating immune cells
Immune infiltration analysis of HCC was performed by TIMER2.0 (http://timer.cistrome.org/) [18, 19] to explore the correlation between MAPT and the infiltration levels of immune cells. The association between MAPT and six types of immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) was analyzed by the Spearman's correlation, and Wilcoxon rank-sum test was used to clarify the association of infiltration of immune cells with the different expression groups of MAPT.
3. Results
3.1 Identification of differentially expressed genes
By comparing transcriptome in HCC from TCGA, in total, 2068 differentially expressed genes were identified by limma, including 1991 up-regulated genes and 77 down-regulated genes (DEGs) (Figure 1A). To confirm immune?related genes associated with HCC, we selected all 116 immune?related genes from the DEGs that were considered statistically significant (Figure 1B).
3.2 Prognosis-associated and immune-related genes
The overlap between the 116 immune?related genes and 500 most differential survival genes in HCC contained three genes (BIRC5?MAPT?SPP1) as shown in the Venn diagram (Figure 1C), Three genes are highly expressed in HCC, and GEPIA shows that their expression has a significant relationship with the prognosis of patients. Among the three genes mentioned above, we selected MAPT, still poorly understood in HCC, to assess its diagnostic and prognostic value and the mechanisms it may involve in HCC.
3.3 MAPT expression comparison
A meta-analysis of MAPT expression in 5 analyses with verge values was determined as p-Value 0.05, fold change >1, and the top 10% gene rank in the Oncomine database was performed in our research. The findings have shown that the expression levels of MAPT mRNA were significantly higher in human HCC samples than those in the non-tumor samples (Figure 2A). Analysis of different databases in GEO revealed that MAPT mRNA was significantly upregulated in HCC in GSE25097, GSE45436, GSE55092, compared with that in normal liver tissues (P < 0.001, Figure 2B-2D). The details of the GEO series used in this analysis can be seen in Table 1. Further analysis shows that the protein expression of MAPT was significantly elevated in HCC samples compared with normal tissues in the HPA database (Figure 3).
3.4 Association between the expression of MAPT and the clinicopathology variables
In our study, gender, age, histologic grade, TNM stage, race, and other a total of 21 factors in clinical data were tested in this study to identify the relationship between MAPT and clinicopathological characteristics (Table 2). The findings indicated that MAPT overexpression was significantly associated with new tumor events after initial treatment (P <0.001, Table 2). However, there is no correlation between MAPT expression and gender, age, histologic grade, TNM stage, or other clinicpatho-logic characteristics. A univariate analysis revealed that MAPT expression levels, pathologic stage, and TNM stage correlated with the prognosis of patients (P < 0.05). To further explore factors associated with patients' survival, a multivariate Cox regression analysis was performed on eight variables. The results showed that MAPT expression levels (P < 0.001), histologic grade (P = 0.018) and stage T (P = 0.001) and stage M (P = 0.047) were independent prognostic factors associated with overall survival (OS) in HCC patients (Table 3).
3.5 The relationship between MAPT expression and prognosis in HCC patients
We analyzed the overall survival (OS), disease-free survival (DSS), progression-free survival (PFS), and relapse-free survival (RFS) rates to evaluate the prognostic value of MAPT expression in HCC. As shown in figure 4, HCC patients with high MAPT expression had shorter OS (P= 0.00013, Figure 4A), RFS (P = 0.0019, Figure 4B), DSS (P = 0.0014, Figure 4C) and PFS (P = 0.0029, Figure 4D) than those with low MAPT expression. Also, subgroup survival analysis in different populations was performed in our research. The high expression level of MAPT has significantly affected the worse OS in HCC patients with male sex, but not associated with poor prognoses in female patients (Figure 5) and MAPT overexpression was only significantly associated with worse OS in HCC patients without vascular invasion (Figure 6). In addition to this, the high expression level of MAPT was significantly correlated with poor prognosis in other groupings.
3.6 Association of MAPT expression with immune infiltration
The correlation between the expression level of MAPT and the immune infiltration in the tumor microenvironment was analyzed by Spearman correlation and then generated by TIMER. The plot shows that the MAPT was positively correlated with macrophages, Conversely, no associations were observed between MAPT and infiltration of B cells, CD4+ T cells, CD8+ T cells, neutrophils, and dendritic cells (Figure 7).
3.7 PPI and GO enrichment
Protein-protein interaction network of MAPT revealed that ten genes, including CDK5R1, APP, CASP3, CDK5, GSK3B, CAMK2A, BRSK2, MAPK2, MAPK8, and TUBA1B, were interacted with MAPT (Figure 8A). Peptidyl−serine phosphorylation, peptidyl−serine modification, regulation of autophagy, neuron death, etc. were the most enriched GO biological process of MAPT interactive genes. Additionally, protein serine/threonine kinase activity, tau−protein kinase activity, tau protein binding, protease binding, etc. was the most enriched GO molecular functions of MAPT interactive genes (Figure 8B).
3.8 Result of the GSEA tool reporting the most significant KEGG pathways
Subsequent single-gene GSEA analysis confirmed high expression of MAPT was linked to the Wnt signaling pathway, the Notch signaling pathway, the VEGF signaling pathway, pathways in cancer, and promoted the development of bladder cancer, small cell lung cancer, thyroid cancer and so on. The results confirmed that MAPT was tightly associated with tumors (Figure 8C).
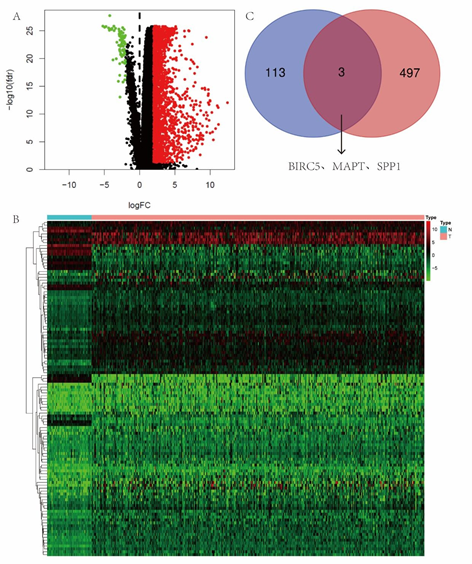
Figure 1: (A) 1991 up-regulated genes and 77 down-regulated genes were identified in HCC. (B) 116 immune?related genes from the DEGs that were considered statistically significant. (C) The overlap between the 116 immune?related genes and 500 most differential survival genes in HCC contained 3 genes.
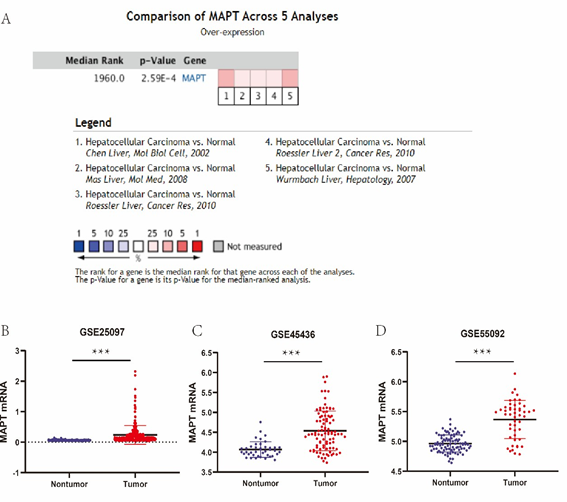
Figure 2: (A) Oncomine analysis of MAPT expression in HCC. (B) MAPT mRNA expression levels between tumor and nontumor tissues in HCC patients in GEO database series.
|
GEO series |
Contributor(s) |
Tumo r |
Nontumor |
Platform |
|
GSE25097 |
Zhang C, 2010 |
268 |
243 |
Rosetta/Merck Human RSTA Affymetrix 1.0 microarray, Custom CDF |
|
GSE45436 |
Hsieh J, 2013 |
93 |
42 |
Affymetrix Human Genome U133 Plus 2.0 Array |
|
GSE55092 |
Melis M et al, 2014 |
49 |
91 |
Affymetrix Human Genome U133 Plus 2.0 Array |
Table 1: Details of GEO series included in this analysis.
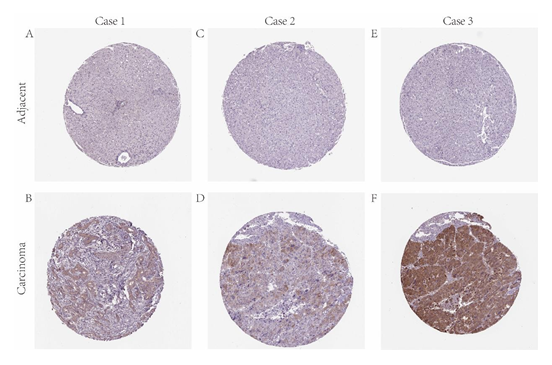
Figure 3: IHC staining of MAPT protein in HCC (HPA database). Representative images of IHC staining of MAPT expression in HCC samples and matched adjacent normal tissues (A-F).
|
Variables |
MAPT expression level |
P value |
|
|
Low (n = 185) |
High (n = 185) |
||
|
Age |
0.754 |
||
|
<60 |
83 |
86 |
|
|
≥60 |
102 |
99 |
|
|
Gender |
0.74 |
||
|
Male |
123 |
123 |
|
|
Female |
59 |
62 |
|
|
Pathologic stage |
0.797 |
||
|
? |
88 |
79 |
|
|
? |
43 |
46 |
|
|
? |
52 |
57 |
|
|
? |
2 |
3 |
|
|
Histologic grade |
0.166 |
||
|
G1 |
28 |
27 |
|
|
G2 |
98 |
83 |
|
|
G3 |
56 |
66 |
|
|
G4 |
3 |
9 |
|
|
T classification |
0.704 |
||
|
T1 |
94 |
87 |
|
|
T2 |
47 |
48 |
|
|
T3 |
38 |
42 |
|
|
T4 |
5 |
8 |
|
|
TX |
1 |
0 |
|
|
N classification |
0.582 |
||
|
N0 |
127 |
126 |
|
|
N1 |
3 |
1 |
|
|
NX |
55 |
58 |
|
|
M classification |
0.973 |
||
|
M0 |
134 |
134 |
|
|
M1 |
2 |
2 |
|
|
MX |
51 |
49 |
|
|
New Event |
0.795 |
||
|
No occurrent |
97 |
97 |
|
|
Extrahepatic Recurrence |
14 |
17 |
|
|
Intrahepatic Recurrence |
42 |
36 |
|
|
Locoregional Recurrence |
27 |
32 |
|
|
New Primary Tumor |
5 |
3 |
|
|
New tumor event after initial treatment |
<0.001 |
||
|
Yes |
39 |
56 |
|
|
No |
73 |
101 |
|
|
NA |
73 |
28 |
|
|
Relative family cancer history |
0.366 |
||
|
Yes |
62 |
50 |
|
|
No |
100 |
107 |
|
|
NA |
23 |
28 |
|
|
Vascular tumor cell type |
0.633 |
||
|
Macro |
9 |
7 |
|
|
Micro |
42 |
50 |
|
|
None |
108 |
98 |
|
|
NA |
26 |
30 |
|
|
Cancer Status |
0.546 |
||
|
With tumor |
58 |
51 |
|
|
Tumor free |
112 |
122 |
|
|
NA |
15 |
12 |
|
|
Hepatic inflammation |
0.156 |
||
|
None |
63 |
54 |
|
|
Mild |
49 |
50 |
|
|
Severe |
12 |
5 |
|
|
NA |
61 |
76 |
|
|
Albumin result specified value |
0.481 |
||
|
<3.5 |
38 |
31 |
|
|
3.5~5.1 |
109 |
108 |
|
|
>5.1 |
6 |
4 |
|
|
NA |
32 |
42 |
|
|
Child-pugh classification |
0.062 |
||
|
A |
106 |
110 |
|
|
B |
16 |
5 |
|
|
C |
1 |
0 |
|
|
NA |
62 |
70 |
|
|
Ishak fibrosis status |
0.384 |
||
|
No Fibrosis |
44 |
30 |
|
|
Portal Fibrosis |
16 |
15 |
|
|
Fibrous Speta |
11 |
17 |
|
|
Nodular Formation and Incomplete Cirrhosis |
5 |
4 |
|
|
Established Cirrhosis |
36 |
33 |
|
|
NA |
73 |
86 |
|
|
Race |
0.279 |
||
|
American Indian or Alaska Native |
2 |
0 |
|
|
Asian |
70 |
87 |
|
|
Black or African American |
9 |
8 |
|
|
White |
98 |
86 |
|
|
NA |
6 |
4 |
|
|
Prothrombin time result value |
0.65 |
||
|
<11 |
134 |
126 |
|
|
11~13 |
15 |
13 |
|
|
>13 |
2 |
3 |
|
|
NA |
34 |
43 |
|
|
Platelet result count |
0.475 |
||
|
<100 |
11 |
6 |
|
|
100~300 |
103 |
109 |
|
|
>300 |
40 |
34 |
|
|
NA |
31 |
36 |
|
|
Creatinine value in mg/dl |
0.297 |
||
|
<0.5 |
2 |
3 |
|
|
0.5~1.2 |
131 |
118 |
|
|
>1.2 |
23 |
21 |
|
|
NA |
29 |
43 |
|
|
Residual tumor |
0.124 |
||
|
R0 |
164 |
159 |
|
|
R1 |
11 |
6 |
|
|
R2 |
1 |
0 |
|
|
RX |
6 |
16 |
|
|
NA |
3 |
4 |
|
Table 2: Clinicopathological characteristics of HCC patients between MAPT high and low groups.
|
Patients |
||||||
|
Univariate analysis |
Multivariate analysis |
|||||
|
Variable |
HR |
95% CI |
P value |
HR |
95% CI |
P value |
|
Age |
1.213 |
0.855-1.7 21 |
0.278 |
1.143 |
0.790-1.6 52 |
0.478 |
|
Gender |
1.226 |
0.861-1.7 47 |
0.259 |
1.086 |
0.757-1.5 58 |
0.655 |
|
Histologic grade |
1.122 |
0.890-1.4 15 |
0.330 |
1.347 |
1.052-1.7 25 |
0.018 |
|
Pathologic stage |
1.283 |
1.170-1.4 07 |
< 0.001 |
1.026 |
0.861-1.2 22 |
0.777 |
|
Stage T |
1.245 |
1.159-1.3 38 |
< 0.001 |
1.234 |
1.088-1.4 00 |
0.001 |
|
Stage N |
1.222 |
1.016-1.4 69 |
0.033 |
1.123 |
0.872-1.4 47 |
0.370 |
|
Stage M |
1.273 |
1.058-1.5 31 |
0.010 |
1.293 |
1.004-1.6 65 |
0.047 |
|
MAPT |
1.497 |
1.223-1.8 33 |
< 0.001 |
1.542 |
1.233-1.9 29 |
< 0.001 |
Table 3: Univariate and multivariate Cox regression analyses of overall survival in 370 HCC patients form TCGA.
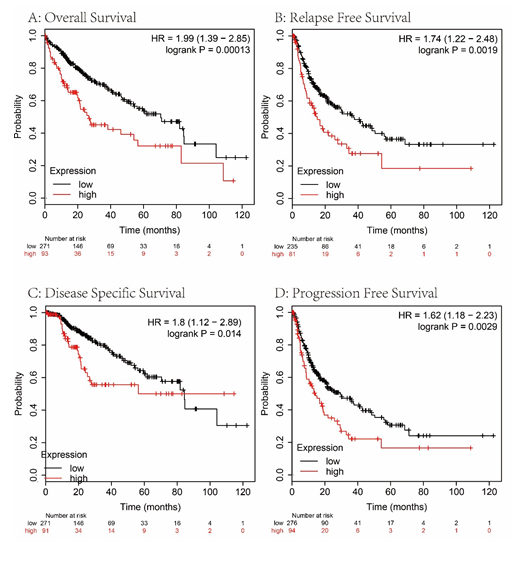
Figure 4: The effect of low vs. high MAPT expression in patients with HCC on OS, RFS, DSS and PFS time (A-D).
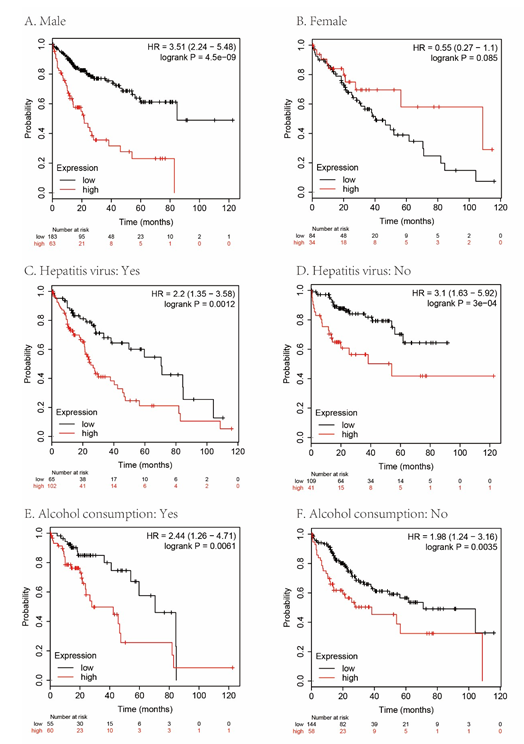
Figure 5: Subgroup analyses of overall survival comparison in different population [gender (A, B), gender hepatitis virus (C, D) and alcohol consumption (E, F)] with MAPT median cutoffs in HCC patients.
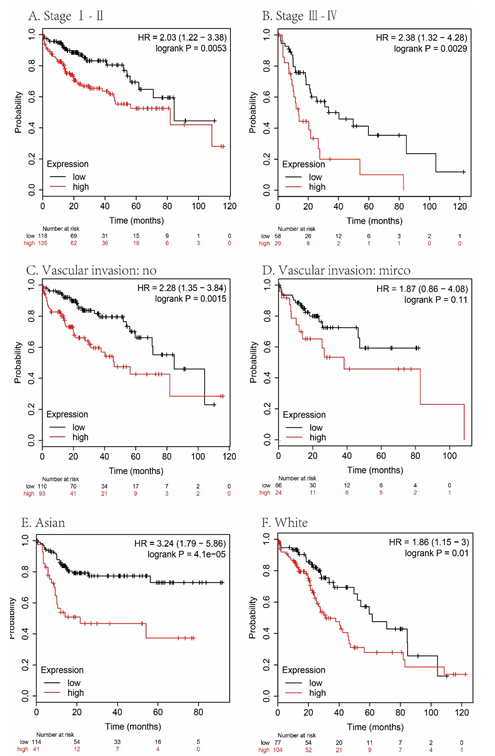
Figure 6: Subgroup analyses of overall survival comparison in different population [stage (A, B), vascular invasion (C, D) and race (E, F)] with MAPT median cutoffs in HCC patients.
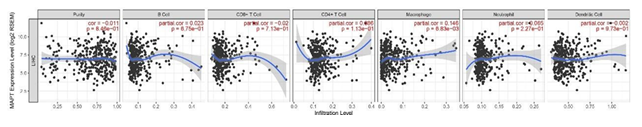
Figure 7: Association of MAPT expression with immune infiltration in HCC.
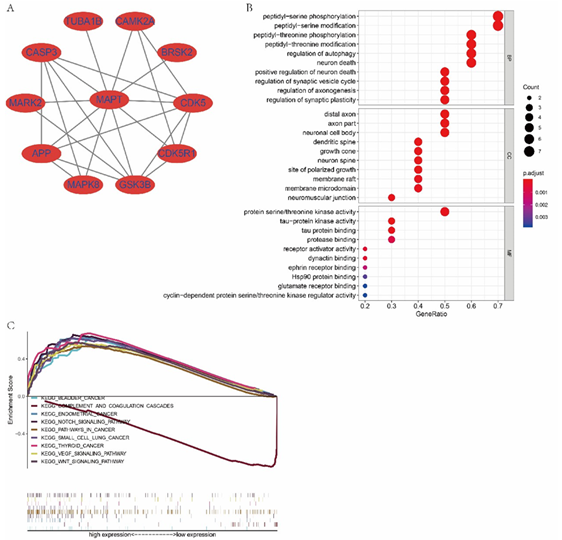
Figure 8: (A) Protein-protein interaction of MAPT using String analysis. (B) GO enrichment of its interactive genes. (C) Representative signal pathways of MAPT single-gene GSEA analysis.
4. Discussion
Previous studies about MAPT related diseases mainly focused on neurological disorders such as Dementia, Alzheimer's Disease, Parkinson's Disease, and so on [20-23]. However, recent studies have drawn much attention to its role in the tumor over the past few years. Analysis by Ricardo Gargini et al. suggests that MAPT is aberrantly expressed in a number of tumor types and shows a general association with prognosis [24], although its exact molecular mechanism and function in cancer have not yet been elucidated. Besides, many studies showed that the aberrant expression of MAPT in many cancers, such as neuroblastoma, breast, and prostate cancer [6, 7, 25]. Nonetheless, the survey about MAPT in HCC was still lacking. In light of the previous studies mentioned above, we undertook this research to investigate the potential value of MAPT in HCC. In this research, we found that overexpression of MAPT is significantly in HCC, and the high expression levels of MAPT in HCC were significantly associated with worse OS. This conclusion was in general agreement with that in breast cancer. Regretfully, the serum MAPT levels in HCC patients were not collected in the present study. In addition, MAPT expression was associated with recurrence after initial treatment. It indicates that HCC patients with high MAPT expression are more likely to relapse. Furthermore, a weak positive correlation was observed between MAPT and macrophages in HCC. Liver macrophages play a significant role in acute and chronic hepatic diseases, and they orchestrate inflammation, fibrosis, tumor progression, and angiogenesis, as well as tissue repair and tumor surveillance [26]. The result suggests that there are subtle correlations between MAPT and HCC.
Functionally, among gastric cancer and epithelial ovarian cancer patients, lower expression of MAPT is correlated with increased susceptibility to paclitaxel therapy [27, 28]. This suggests that MAPT levels might be a marker for predicting the sensitivity of tumors to chemotherapy. More than that, MAPT promotes microtubule assembly and stability [29]. However, MAPT was aberrantly expressed in various tumors. Due to the different microtubule composition of different cell types, a tentative inference on the result is that MAPT-induced microtubule dynamics changes may have different effects on cell behavior, depending on the origin of the cell tissue [25]. A complete description of a protein's function requires an understanding of all the chaperones to which it specifically binds. KEGG and GO enrichment analysis was performed on the genes in the protein-protein interaction network, and the results demonstrated that MAPT might participate in several tumor-associated pathways and oxidation-reduction processes in our research. GO enrichment analysis of genes that show MAPT is mainly involved in biological processes such as peptidyl−threonine phosphorylation and peptidyl−serine phosphorylation. Its molecular function is primarily related to the regulation of serine-threonine protein kinase activity. Serine-threonine protein kinase involves a variety of proteins and plays a wide range of molecular features in many tumors [30, 31, 32]. The above results show that MAPT may function as an upstream regulatory factor to participate in the regulation of serine-threonine protein kinase activity and participate in both tumor initiation and progression of human cancers. Unfortunately, no experimental study has been carried out to find the potential oncogenic mechanisms of MAPT in HCC development.
Accordingly, further experiments are required to demonstrate the specific mechanism of its cancer-promoting effect. Taken together, this study provides the first detailed analysis of evaluating the expression level of human MAPT protein and mRNA in HCC. The expression of MAPT was significantly overexpressed compared with that in non-tumor liver tissues in HCC based on TCGA, Oncomine, HPA, and GEO databases. MAPT was remarkably overexpressed in HCC tissues and was inversely associated with OS and RFS of HCC patients. Furthermore, our initial analyses also show the signaling pathways that MAPT may be involved in, and the molecular functions, biological processes that MAPT may participate in. We will inform the design of future experimental studies in light of our results. In summary, MAPT may be useful as a predictive diagnostic factor in HCC and as a possible therapeutic target.
Acknowledgments
This study was supported in part by Guangxi Clinical Medical Research Center of Hepatobiliary Diseas es (No: AD17129025), Special Funding for Guangxi Special Experts (No: GRCT[2019]13), Guangxi Medical High-level Leading Talents Training “139” Project (No: GWKJ[2018] 22.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: a cancer journal for clinicians 68 (2018): 394-424.
- Yang J D, Hainaut P, Gores G J, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management[J]. Nature Reviews Gastroenterology & Hepato-logy (2019): 1-16.
- Castelli G, Pelosi E, Testa U. Liver cancer: molecular characterization, clonal evolution and cancer stem cells[J]. Cancers 9 (2017): 127.
- Butler V J, Salazar D A, Soriano-Castell D, et al. Tau/MAPT disease-associated variant A152T alters tau function and toxicity via impaired retrograde axonal transport[J]. Human molecular genetics 28 (2018): 1498-1514.
- Strang K H, Golde T E, Giasson B I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration[J]. Laboratory Investiga-tion (2019): 1.
- Darlix A, Hirtz C, Thezenas S, et al. The prognostic value of the Tau protein serum level in metastatic breast cancer patients and its correlation with brain metastases[J]. BMC cancer 19 (2019): 110-110.
- Zaman S, Chobrutskiy B I, Blanck G. MAPT (Tau) expression is a biomarker for an increased rate of survival in pediatric neuroblastoma[J]. Cell Cycle 17 (2018): 2474-2483.
- Zhang Z, Li J, He T, et al. Two predictive precision medicine tools for hepatocellular carcinoma[J]. Cancer Cell International 19 (2019): 1-19.
- Qi L, Wang L, Huang J, et al. Activated amelogenin Y-linked (AMELY) regulation and angiogenesis in human hepatocellular carcinoma by biocomputation[J]. Oncology letters 5 (2013): 1075-1079.
- Ritchie M E, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic acids research 43 (2015): e47-e47.
- Rhodes D R, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform[J]. Neoplasia 6 (2004): 1-6.
- Uhlén M, Björling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics[J]. Molecular & cellular proteomics 4 (2005): 1920-1932.
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses[J]. Nucleic acids research 45 (2017): W98-W102.
- Menyhárt O, Nagy Á, Gy?rffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepato-cellular carcinoma[J]. Royal Society open science 5 (2018): 181006.
- Subramanian A, Tamayo P, Mootha V K, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles[J]. Proceedings of the National Academy of Sciences 102 (2005): 15545-15550.
- Yu G, Wang L G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. Omics: a journal of integrative biology 16 (2012): 284-287.
- Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis[J]. Bioinformatics 31 (2015): 2912-2914.
- Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells[J]. Cancer research 77 (2017): e108-e110.
- Li B, Severson E, Pignon J C, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy[J]. Genome biology 17 (2016): 174.
- Laws S M, Friedrich P, Diehl-Schmid J, et al. Genetic analysis of MAPT haplotype diversity in frontotemporal dementia[J]. Neurobiology of aging 29 (2008): 1276-1278.
- Balaji V, Kaniyappan S, Mandelkow E, et al. Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems[J]. Autophagy 14 (2018): 2139-2154.
- Li J, Ruskey J A, Arnulf I, et al. Full sequencing and haplotype analysis of MAPT in Parkinson's disease and rapid eye movement sleep behavior disorder[J]. Movement Disorders 33 (2018): 1016-1020.
- Zhang C C, Xing A, Tan M S, et al. The role of MAPT in neurodegenerative diseases: genetics, mechanisms and therapy[J]. Molecular neurobiology 53 (2016): 4893-4904.
- Gargini R, Segura-Collar B, Sánchez-Gómez P. Novel Functions of the Neurodegenerative-Related Gene Tau in Cancer[J]. Frontiers in aging neuroscience 11 (2019): 231-231.
- Schroeder C, Grell J, Hube-Magg C, et al. Aberrant expression of the microtubule-associated protein tau is an independent prognostic feature in prostate cancer[J]. BMC cancer 19 (2019): 193-193.
- Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease[J]. Nature reviews. Immunology 17 (2017): 306-321.
- He W, Zhang D, Jiang J, et al. The relationships between the chemosensitivity of human gastric cancer to paclitaxel and the expressions of class III β-tubulin, MAPT, and survivin[J]. Medical Oncology 5 (2014): 1-7.
- Smoter M, Bodnar L, Grala B, et al. Tau protein as a potential predictive marker in epithelial ovarian cancer patients treated with paclitaxel/platinum first-line chemotherapy[J]. Journal of Experimental & Clinical Cancer Research 32 (2013): 25.
- Kellogg E H, Hejab N M A, Poepsel S, et al. Near-atomic model of microtubule-tau inter-actions[J]. Science 360 (2018): 1242-1246.
- Roskoski Jr R. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs[J]. Pharmacological Research 139 (2019): 471-488.
- Liu Y Y, Tanikawa C, Ueda K, et al. INKA2, a novel p53 target that interacts with the serine/threonine kinase PAK4[J]. International journal of oncology 54 (2019): 1907-1920.
- Roskoski Jr R. Targeting oncogenic Raf protein-serine/threonine kinases in human cancers[J]. Pharmacological Research 135 (2018): 239-258.
