Artificial Intelligence-Nanomedicine Interface: Today’s Theory Tomorrow's Technology
Article Information
Sarah I Mazi1#, Abdulahi M Hassen2#, Fahad A Alrashed1, Fatiha M. Benslimane3, Nura A Mohamed3*
1Department of Cardiac Sciences, College of Medicine, King Saud University, P.O. Box 7805, Riyadh 11472, Saudi Arabia.
2Computer Science, College of Engineering, Qatar University, Doha P.O. Box 2713, Qatar.
3Biomedical Research Center, Qatar University, Doha P.O. Box 2713, Qatar.
*Corresponding author: Nura A Mohamed. Biomedical Research Center, Qatar University, Doha P.O. Box 2713, Qatar.
Received: 27 September 2023; Accepted: 03 October 2023; Published: 12 December 2023
Citation: Sarah I Mazi, Abdulahi M Hassen, Fahad A Alrashed, Fatiha M. Benslimane, Nura A Mohamed. Artificial Intelligence-Nanomedicine Interface: Today’s Theory Tomorrow's Technology. Archives of Clinical and Biomedical Research. 7 (2023): 635-641.
View / Download Pdf Share at FacebookAbstract
Substantial strides were made in the nano-therapeutic and nanodiagnostic fields. The clinical deployment of the investigational nanomedicine improved treatment outcomes significantly. Currently, there is an urge to develop a single nanoparticle that can serve as both detection and treatment tools, in addition to the possibility of tailoring it to integrate more than one therapeutic agent. Furthermore, scientists are interested in developing functionalized-nanoparticles that can be activated only at the desired tissues and organs or be taken up by specific cells. Finding nanoparticles with these unique properties is insufficient as concerns about nailing the appropriate doses and accumulation, tolerance, timedependent, and patient-dependent issues persist. Such concerns necessitate establishing better data mining tools as available data is scattered. By integrating artificial intelligence (AI) with nanomedicine, information platforms for this very promising field can be created. Furthermore, through the use of AI-nanomedicine-interface, combinatorial-nanotherapy can be optimized and made more sustainable, moving it one step closer to clinical application. This article, to this end, will focus on implementing AI to improve the sustainability of nanotherapeutics and nanodiagnostics as well as highlighting the possible concerns that AI can address to ensure a smooth translation of the developed nanotherapeutics and nanodiagnostics from bench to clinical use.
Keywords
Artificial Intelligence (AI); Nanomedicine; Data Mining; Sustainability
Article Details
1. Introduction
The nanomedicine field offers various attractive opportunities. The precise engineering of nanomaterials with desirable physiochemical properties while controlling their behavior in the human body is an example. Such control can be gained through chemically engineering nanomaterials with tunable parameters to tailor their half-life in the body; designing nanomaterials with different porosities to control the release kinetics (e.g., release and clearance profiles) of the incorporated pharmaceuticals. Which, by default, will affect tissue selectivity and their uptake/accumulation in cells, tissues, and organs, as well as the local and systemic toxicity. It is nearly impossible to control these parameters when dealing with small-sized drugs due to the lack of a quantitative understanding of the structure–activity relationships (QSAR) [1]. In contrast, the same parameters are achievable for the larger nanoformulated drugs, such as the advanced drug de-livery systems (DDS), due to the possibility of controlling their dimensions, porosity, surface charge and chemistry, and physicochemical properties precisely. Furthermore, DDS can protect the loaded therapeutic agents from environmental factors, enzymes’ early degradation, and immune system clearance. They can also interact with the loaded drugs (synergistic action), providing passive targeting and being traceable/imageable to localize nanoparticles accumulations sites. A key feature of DDS is that they can be functionalized to activate and release the loaded therapeutic agents under certain circumstances. However, the diversity in the human tissues, organs, and cells requires using both small and large nanoformulations as tools for drug delivery and disease detection, which is why nanomedicine cannot only focus on DDS. Despite the many advances made in the nanomedicine field, it is still associated with poor clinical translation. This is attributed to the lack of a quantitative and/or qualitative understanding of the exact link between the nanomaterials and the biological outcomes [2,3]. Notwithstanding, nanomaterials heuristic and pseudo-rational design practices were commonly used. Unsurprisingly this practice did not lead to many significant outcomes, and neither did it help enhance the clinical translation of the nanomaterials. Thus, a different approach is being considered now in the nanomedicine field to better understand the relationships between nanomaterials and physicochemical and biological outcomes. Such approaches include implementing artificial intelligence (AI) in nanomedicine [1]. Early research on AI dates back to the 1950s[4], when machines were recognized as trainable tools that could be used in setting up problem-solving strategies. Since then, AI has been used by scientists in the nanomedicine field to determine and analyze the nanoformulation’s pharmacokinetics [4], pharmacodynamics [5], and image analysis using SEM and TEM [6] [25]. Moreover, AI was used to rationalize the selection and the design of the nanoparticles as well as the other chemical modifications that could be used to achieve controlled release, adjust the drug’s pharmacokinetics, provide targeting delivery, and avoid local and systemic toxicity. In addition, AI-nanomedicine interface is preferred because it can provide a principally different vision that could be used in generating hypothesis, designing experimental meth-odologies, planning evaluation procedures, and analyzing generated data [2,3].
1.1 AI use in the Nanomedicine field
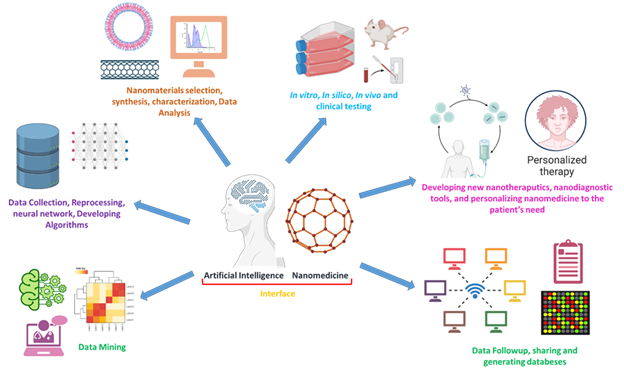
In addition to the use of AI in determining drug targeting and ratiometric delivery, AI-nanomedicine interface can help indemnify the dynamic patient response to different treatments and provide better accountability for the treatments’ dynamic modulation as current conventional approaches do not provide the required accountability in clinics[7]. Harnessing AI can agnostically optimize nanomedicine-based treatment strategies in a mechanism-independent manner. It correlates inputs, including drug selection, dose adjustment, stimuli (responsive), and frequency determination, with the outputs, including drug efficacy and safety, as well as targeting and localization effects. This indicates that AI can be regarded as a powerful tool that could be used to rationally optimize the combination of nanotherapy. Such data can be used by both individuals and populations to overcome the many challenges that confront the nano-medicine field as it uses different algorithms (e.g., deep learning algorithms)[8-10]. Such algorithms can optimize drug delivery, efficacy, and synergy and guide the nanotherapy’s clinical use. AI-based optimization was recently suggested to develop the advances in combinatorial nanomedicine further. Such interfacing can help execute substantial improvements in the treatment outcomes simply because nanomedicine-based combination alone is subjected to the same constraints that unmodified combination therapies face. At the same time, AI can overcome these constrains [11]. Figure 1 summarizes the different ways AI can be used to revolutionize and improve the nanomedicine field.
1.2 AI and the Drug selection paradigm
During the conventional screening strategies, drugs with no apparent treatment efficacy are eliminated. However, a virtually insurmountable brute force interrogation is used in AI where the drug and dose space is prohibitively ample. As such, even if the drug alone has no efficacy, the same drug might have or mediate a beneficial effect when delivered in combination with other drugs (right drugs), and the appropriate doses or when it interacts with the nanocarrier is used in delivering it. Noteworthy, AI methodologies can be used in predicting the drug combinations’ efficacy based on the drug synergy with the nanocarrier. However, without an AI interface, it is very difficult to identify the potential of combination therapy [7,12-14]. An example is using an AI platform named Quadratic Phenotypic Optimization Platform (QPOP). QPOP is a multi-parametric global optimization approach that can overcome the different hurdles facing the drug development process, resulting in finding both efficacious and safer nanotherapies, which will redefine the drug development roadmap and pave the way for a smooth clinical translation. Furthermore, in this platform, large pools of drugs for a desired disease are used to design a novel combination therapy, then computational results are used in designing patient-specific regimens.
2. AI implementation in the nanomedicine-based combination therapy design
Besides the nanomedicine-based combination therapy design, AI will play a major role in determining the synergistic effect between the selected drugs and their interaction with the used nanocarrier. Moreover, AI can help optimize the route of administration and possible ways of activating unmodified drug combinations [15-22]. In doing so, AI uses different big data-driven approaches, where the outcome of the patient’s electronic medical records, their genetic and broader–omics profiling, and other information are employed in the drug selection process. Collectively these strategies represent a crucial first step towards establishing valuable nanomaterials-drugs databases with the required in-formation to refine the nanotherapeutic regimen (Dose)-design process, alternatively improving the efficacy and safety of the broader patient population. However, even after regimens selection, identifying synergies and patient-specific responses are key challenges that must be addressed. Interestingly, AI uses a powerful technology platform, the so-called CURATE, which comprises dosing strategies such as parabolic personalized dosing (PPD) [7]. Furthermore, CURATE.AI works by mapping the relationship between the input (e.g., intervention) and the output (e.g., individual’s phenotypic result), making it exclusively based or personalized to that individual’s data. Some studies showed that using CURATE.AI can significantly shorten the treatment time, enhance treatment efficacy, improve patient outcomes, reduce the care cost, and implement a broad clinical use for this application. Most importantly, using CURATE.AI in nanomedicine and drug development programs can predict and show the inter-patient variability that could occur in response to drug interactions, which can establish stronger grounds for AI-based future improvements in response rates [23]192.
2.1 Data mining and acquisition
Collecting adequate material-related data is required to access the aforementioned biological responses. Since many open materials science databases exist, these data can be extracted from published articles using data mining tools and/or other screening techniques. However, the lack of comprehensive nanomedicine databases creates the need for screening studies based on comprehensive AI and machine learning (ML)-assisted research to curate and reproduce experimental and theoretical data. Examples of partially or fully automated AI and ML-assessed data mining data approaches include HTSy and HTSc. Proper data mining is a powerful approach that uses existing data in testing hy-potheses through data extraction, cleaning, and representing materials digitally, as well as data augmentation, clus-tering, and visualization, throughout patterns uncovering, detecting data anomalies and the data’s statistically signif-icant structures. In doing so, Natural language processing (NLP) is required to extract data from research articles [24,25]. The lack of sufficient data is one of the main obstacles NLP faces in nanomedicine. Hence, several data augmentation approaches to achieve optimal performance exists, such as generative adversarial networks (GANs) [26], TL [27], molecular rotation [28], tautomerism structural properties [29], start token replacement [30], descriptor noising[31], and physical domain knowledge [32]…etc. Nevertheless, these data augmentation techniques suggest some assumptions whose validity strongly depends on the form of the user data and the relationships between the dependent and independent variables. Following mining, data goes through several preprocessing stages that include data cleaning, missing data handling, normalization, data integration, data reduction, transformation, and noise identification are performed [33]. Nanomedicine-related data is usually sparse and is only found in high-dimensional parameter space. Kim et al. [34] proposed method to generate sufficient amount of data while minimizing the number of parameters used. This is achieved by applying a variational autoencoder (VAE) to learn a lower dimensional representation of the data through a neural network (NN) learning. Furthermore, using the data augmentation technique for the NLP was suggested to overcome the data sparsity and scarcity problems. Moreover, some platforms are known to exist in full-cycle data mining as they have large data set retrieval and preprocessing stages enabling the prediction of the ML models con-struction [35]. Unfortunately, that is not the case in the nanomedicine field, indicating the need for high-throughput experimental data acquisition [36,37]. Limited nanomedicine data often accumulate manually, which is why HTSc/HTSy automatization via decision-making platforms development is highly desired for the identification of novel nanoformulations[38]. In doing so, AI and ML algorithms can help in determining nanoparticles composition[39], enhancing synthesis procedures[40], self-assembly [41], tuning nanoparticle’s size, shape and providing surface functionalization options[5], which intern gives more realistic idea about the nanoparticle’s safety [42], pharmaco-kinetics [43], tissue/organ specificity [44], and most importantly the therapeutic action [41].
2.2 Determining nanomaterials-body interaction by AI
Despite the rapid development in the nanomedicine field, little is known about the fundamental nanoparticles inter-actions with biological systems, and even less is known about designing nanoparticles with desired biological effects. Data mining and computer simulation can be employed to determine essential design parameters. Thus, nanoinfor-matics was largely recognized as a rational approach to employ weight-of-the-evidence strategies to ensure safe na-noparticle development. Nanoinformatics enable data collection, representation, sharing, collaboration, data sharing, and the semantic (meaningful) search and integration of data using AI and ML. This approach led to the establishment of methods that can predict different nanoparticles that could be used in the nanomedicine field. Moreover, a set of issues were addressed in this context using data mining and ML techniques. Such issues include functional and structural properties, cytotoxicity[45], nanoparticle’s size and shape, nanoparticle’s surface charge and composition, nanoparticle’s adherence and cellular uptake[46], polydispersity, and molecular loading and release, which could then be used for medical purposes[47]. Furthermore, nanomaterials interaction with the different cells, tissues, and organs can be determined using QSAR-based studies. Therefore to achieve a QSAR-based study, the first step is to identify and reference different molecules, followed by the identification of the explanatory variable, and finally, defining the quantitative character-istics of the nanoparticles and the model selection for the in vitro and in silicio experimental designs to pave the way for the in vivo treatments[48]. The lack of common reporting standards and non-uniformity of the information reported are significant barriers to data sharing and re-use.
2.3 Data Sharing in Nanomedicine
Currently, there is a lack of nanomedicine data reporting standards in addition to the un-uniformity of the reported data making it less useful for sharing and re-using. Besides, most of the available data on nanomedicine is found in journal articles in the form of texts which is inherently difficult to process. The situation is further exacerbated by the lack of terminology standards and the substantial gaps between the nanomedicine’s chemical, physical, and biological data as a result of the inadequate nanomaterials characterization. In addition to the absence of the minimum information standards needed for reporting data generated in nanomedicine-specific journal articles and datasets, such standards include data quality, completeness, and reliability. Unfortunately, one of the main struggles of the nanomedicine field is irreproducibility, as a result of the lack of standardized protocols for nanoparticle preparation and characterization, data exchange and transfer of nanoparticle synthesis, chemical composition and characterization, safe handling, and toxicity. In the nanoinformatic field, there is a lack of sufficient raw data to be compared with analyzed data, which is an essential step in normalizing data from different sources to maintain consistency [49] through meta-analysis. The FDA first recognized model-informed drug development (MID) in 2017. It refers to the application of various quantitative models acquired from preclinical and clinical data to ease early decision-making and increase the chances for clinical approval [50,51]. Another evolving area in nanomedicine-AI is the systems thinking and quality-by-design (QbD) strategies that were used in producing “computational pharmaceutics” and the multi-scale modeling techniques, which together can alter the pharmaceutical sciences through the virtual constructing process and reducing the experiments N-number that is needed in the optimization process [52-54].
3. Conclusion
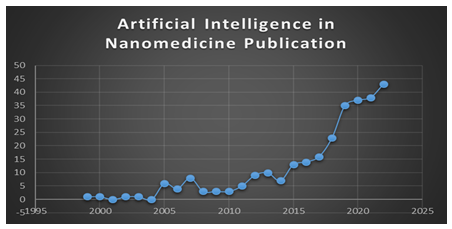
In the last 23 years, there has been an increase in AI-related nanomedicine research indicating the benefits that com-putational aids can offer to the drug development field, with a PubMed return of 281 results for the words AI and nanomedicine in December 2022 (Figure 2). Interfacing AI with nanomedicine can overcome several issues that face the development of the nanomedicine field, such as the nanoparticle’s selection, synthesis and characterization, chemical functionalization, size/shape adjustment, cellular adherence and uptake, tissue and organ accumulation, cytotoxicity, targeted delivery, pharmacokinetics and pharmacodynamics of the loaded molecule, synergistic effects, patient’s selectivity, and personalized medicine. Furthermore, such an interface can be used for the in vitro, in silicio, and in vivo model selection and the prediction of the success rates. However, an adequate amount of data is not always available in nanomedicine. Also, the current data is sparse and also found in high-dimensional parameters. In addition to the complex nature of the nanomaterials indicating the need for using specific algorithms, NLP to better use these data in overcoming the issues that face the development of the nanomedicine field and providing better answers. Therefore, It is becoming increasingly evident that the use of AI in nanomedicine, coupled with the development of advanced computational models, can greatly enhance our understanding of smart nanotherapeutics. This can lead to significant advancements in the clinical development and translation of these therapies.
Authors' contributions
Conceptualization, S.I.M, A.M.H and N.A.M; data curation, S.I.M, A.M.H and N.A.M; writing—original draft prepa-ration, S.I.M, A.M.H and F.A.A; writing—article and editing, N.A.M, and F.M.B; funding acquisition, N.A.M. All authors have read and agreed to the published version of the manuscript.
Funding Sources
NAM is a recipient of the Early Career Research Award (ECRA; ECRA03-006-3-004) from the Qatar National Research Fund (QNRF), and the L’Oréal-UNESCO for Women in Science Middle East Regional Young Talents Award 2021. In addition, the authors extend their appreciation to the Deputyship for Research & Innovation, “Ministry of Education“ in Saudi Arabia for funding this research workthrough the project no.( IFKSURG-2-861)".
Conflicts of Interest
The authors declare no conflict of interest
References
- Serov N, Vinogradov V. Artificial intelligence to bring nanomedicine to life. Adv Drug Deliv Rev 184 (2022): 114194.
- Chan WCW. Nanomedicine 2.0. Acc Chem Res 50 (2017): 627-632.
- Park K. The beginning of the end of the nanomedicine hype. J Control Release 305 (2019): 221-222.
- Mast MP, Modh H, Champanhac C, et al. Nano-medicine at the crossroads - A quick guide for IVIVC. Adv Drug Deliv Rev 179 (2021): 113829.
- Yamankurt G, Berns EJ, Xue A, et al. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat Biomed Eng 3 (2019): 318-327.
- Lee B, Yoon S, Lee, et al. Statistical Characterization of the Morphologies of Nanoparticles through Machine Learning Based Electron Microscopy Image Analysis. ACS Nano 14 (2020): 17125-17133.
- Zarrinpar A, Lee DK, Silva A, et al. Indi-vidualizing liver transplant immunosuppression using a phenotypic personalized medicine platform. Sci Transl Med 8 (2016): 333-349,
- Harrison C. University of California's first CRISPR patent win. Nat Biotechnol 36 (2018): 673.
- Prevedello LM, Erdal BS, Ryu, JL, et al. Automated Critical Test Findings Identification and Online Notification System Using Artificial Intelligence in Imaging. Radiology 285 (2017): 923-931.
- Dilsizian SE, Siegel EL, Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr Cardiol Rep 16 (2014): 441
- von Maltzahn G, Park JH, Lin KY, et al. Nanoparticles that communicate in vivo to amplify tumour targeting. Nat Mater 10 (2011): 545-552.
- Ho D, Wang CH, Chow EK. Nanodiamonds: The intersection of nanotechnology, drug development, and personalized medicine. Sci Adv 1 (2015): e1500439.
- Zimmer A, Tendler A, Katzir I, et al. Prediction of drug cocktail effects when the number of measurements is limited. PLoS Biol 15 (2017): e2002518.
- Ho D, Wang P, Kee T. Artificial intelligence in nanomedicine. Nanoscale Horiz 4 (2019): 365-377.
- Goldberg HS, Paterno MD, Grundmeier RW, et al. Use of a remote clinical decision support service for a multicenter trial to implement prediction rules for children with minor blunt head trauma. Int J Med Inform 87 (2016): 101-110.
- Kawamoto K, Houlihan CA, Balas EA, et al. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ 330 (2005): 765.
- Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism 69S (2017): S36-S40.
- Williams AM, Liu Y, Regner KR, et al. Artificial intelligence, physiological genomics, and precision medicine. Physiol Genomics 50 (2018): 237-243.
- Zellweger MJ, Tsirkin A, Vasilchenko V, et al. A new non-invasive diagnostic tool in coronary artery disease: artificial intelligence as an essential element of predictive, preventive, and personalized medicine. EPMA J 9 (2018): 235-247.
- Guglielmo A, Staropoli N, Giancotti M, et al. Personalized medicine in colorectal cancer diagnosis and treatment: a systematic review of health economic evaluations. Cost Eff Resour Alloc 16 (2018): 2.
- Warnken ZN, Smyth HDC, Davis DA, et al. Personalized Medicine in Nasal Deliv-ery: The Use of Patient-Specific Administration Parameters To Improve Nasal Drug Targeting Using 3D-Printed Nasal Replica Casts. Mol Pharm 15 (2018): 1392-1402
- Shinko D, Diakos CI, Clarke SJ, et al. Cancer-Related Systemic Inflammation: The Challenges and Therapeutic Opportunities for Personalized Medicine. Clin Pharmacol Ther 102 (2017): 599-610.
- Pantuck A, LDK, Kee T, et al. Modulating BET bromodomain inhibitor ZEN-3694 and enzalutamide combination dosing in a metastatic prostate cancer patient using CURATE.AI, an artificial intelligence platform. Adv Ther 1 (2018): 1800104.
- EA Olivetti JMC, E Kim, O Kononova, et al. Data-driven materials research enabled by natural language processing and information extraction. Appl Phys Rev 7 (2020)
- Lewinski NA, McInnes BT. Using natural language processing techniques to inform research on nanotechnology. Beilstein J Nanotechnol 6 (2015): 1439-1449.
- M Schwarzer BR, Y Ruan Z Song, DY Lee, et al. Learning to fail: Predicting fracture evolution in brittle material models using recurrent graph convolutional neural networks. Comput Mater Sci 162 (2019): 322-332.
- Ma XW, C Liu, X Ban, et al. Data augmentation in microscopic images for material data mining, Npj Comput Mater 6 (2020).
- Kim J, Kim Y, Lee EK, et al. Rotational Variance-Based Data Augmentation in 3D Graph Convolutional Network. Chem Asian J 16 (2021): 2610-2613.
- N Ulrich KUG, A Ebert. Exploring the octanol–water partition coefficient dataset using deep learning techniques and data augmentation. Commun Chem. 4 (2021).
- Grisoni F, Moret, M, Lingwood R, et al. Bidirectional Molecule Generation with Recurrent Neural Networks. J Chem Inf Model 60 (2020): 1175-1183.
- Cortes-Ciriano I, Bender A. Improved Chemical Structure-Activity Modeling Through Data Augmentation. J Chem Inf Model 55 (2015): 2682-2692.
- F Oviedo ZR, S Sun, C Settens, et al. Buonassisi. Fast and interpretable classification of small X-ray diffraction datasets using data augmentation and deep neural networks, Npj Comput Mater 5 (2019).
- S García SRG, J Luengo, JM Benítez, et al. Big data preprocessing: methods and prospects. Big Data Anal 1 (2016).
- E Kim KH, S Jegelka, E Olivetti. Virtual screening of inorganic materials synthesis parameters with deep learning, Npj Comput Mater 3 (2017).
- L Ward AD, A Faghaninia, NER Zimmermann, et al. Jain. Matminer: An open source toolkit for materials data mining, Comput Mater Sci 152 (2018).
- R Banerjee AP, Bo Wang C, Knobler H.et al. Yaghi. High-Throughput Synthesis of Zeolitic Imidazolate Frameworks and Application to CO 2 Capture. Sci 319 (2018): 939–943.
- K Gao, Xu T Zarganes-Tzitzikas, Li. Gao M, et al. Dömling. Nanoscale, au-tomated, high throughput synthesis and screening for the accelerated discovery of protein modifiers. RSC Med Chem 12 (2021): 809-818.
- S Sun NTPH, ZD Ren F, Oviedo AM, et al. Accelerated Development of Perovskite-Inspired Materials via High-Throughput Synthesis and Machine-Learning Diagnosis. Joule 3 (2019): 1437-1451.
- Reker D, Rybakova Y, Kirtane AR, et al. Computationally guided high-throughput design of self-assembling drug nanoparticles. Nat Nanotechnol 16 (2021): 725-733.
- BM Castro ME, JJ Ong, T Pollard Z, et al. Machine learning applied to over 900 3D printed drug delivery systems. J Control Release (2021).
- Long AW, Ferguson AL. Nonlinear machine learning of patchy colloid self-assembly pathways and mechanisms. J Phys Chem B 118 (2014): 4228-4244.
- Mohammadinejad R, Moosavi MA, Tavakol S, et al. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 15 (2019): 4-33.
- Mullis AS, Broderick SR, Binnebose AM, et al. Data Analytics Approach for Rational Design of Nanomedicines with Programmable Drug Release. Mol Pharm 16 (2019): 1917-1928.
- Boso DP, Lee SY, Ferrari M, et al. Optimizing particle size for targeting diseased microvasculature: from experiments to artificial neural networks. Int J Nanomedicine 6 (2011): 1517-1526.
- Singh AV, Ansari MHD, Rosenkranz D, et al. Artificial Intelligence and Machine Learning in Computational Nanotoxicology: Unlocking and Empowering Nanomedicine. Adv Healthc Mater 9 (2020): e1901862.
- Dimitri A, M T. The use of data mining and machine learning in nanomedicine: a survey 4 (2018).
- Jones DE, Ghandehari H, Facelli JC. A review of the applications of data mining and machine learning for the prediction of biomedical properties of nanoparticles. Comput Methods Programs Biomed 132 (2016): 93-103.
- Liu R, RR, Cohen Y. Quantitative Structure-Activity-Relationships for Cellular Uptake of Nanoparticles. Comb Chem High Throughput Screen 18 (2015): 365-375.
- Thomas DG, Klaessig F, Harper SL, et al. Informatics and standards for nanomedicine technology. Wiley Interdiscip Rev Nanomed Nanobiotechnol 3 (2011): 511-532.
- Wang Y, Zhu H, Madabushi R, et al. Model-Informed Drug Development: Current US Regula-tory Practice and Future Considerations. Clin Pharmacol Ther 105 (2019): 899-911.
- Us-Fda. Prescription Drug User Fee Act (PDUFA) IV Commitment Letter. PDUFA reauthorization performance goals and procedures fiscal years 2018 through 2022.
- Madabushi R, Wang Y, Zineh I. A Holistic and Integrative Approach for Advancing Model-Informed Drug Development. CPT Pharmacometrics Syst Pharmacol 8 (2019): 9-11.
- Sanadgol N, Wackerlig J. Developments of Smart Drug-Delivery Systems Based on Magnetic Molecularly Imprinted Polymers for Targeted Cancer Therapy: A Short Review. Pharmaceutics 12 (2020): 831.
- Wang W, Ye Z, Gao H, et al. Computational pharmaceutics - A new paradigm of drug delivery. J Control Release 338 (2021): 119-136