AI-Driven Machine Learning and CRISPR Guide RNA Optimization for Precision Medicine in Neurofibromatosis
Article Information
Shivi Kumar1*, Deirdre Richardson2, Osama Elzafarany3, Ray Mikaelson4, Laura Gupta5
1Neurobiology Research, Mind Matters Foundation, University of Pennsylvania
2Department of Neurology and Neurosurgery, Computational Engineering, Stanford University
3Department of Neurosurgery, Moffitt Center, Department of Neurosurgery
4Massachusetts Institute of Technology, Computer Science and Bioinformatics
5Department of Bioinformatics, Carnegie Mellon University
*Corresponding Author: Shivi Kumar. University of Pennsylvania
Received: 18 January 2026; Accepted: 24 January 2026; Published: 03 February 2026
Citation: Shivi Kumar, Deirdre Richardson, Osama Elzafarany, Ray Mikaelson, Laura Gupta. AI-Driven Machine Learning and CRISPR Guide RNA Optimization for Precision Medicine in Neurofibromatosis. Archives of Clinical and Biomedical Research. 10 (2026): 45-48.
View / Download Pdf Share at FacebookAbstract
Neurofibromatosis types 1 and 2 (NF1/NF2) are genetically complex disorders with highly variable clinical outcomes and limited precision tools to guide individualized treatment. Current therapies, such as MEK inhibitors, show inconsistent efficacy across patients. There is a critical need for computational approaches that can stratify mutation severity and optimize genome-editing strategies for targeted intervention. To develop and validate an AI-powered precision medicine framework that integrates machine learning for NF1/NF2 mutation classification and disease severity prediction, and deep-learning– driven CRISPR guide RNA (gRNA) optimization to enhance genomeediting accuracy and reduce off-target effects. This was a cross-sectional computational study using public genomic datasets and supervised machine learning models. Deep learning was applied for gRNA scoring. The study was conducted from 2023–2024 and included the development of a web-based deployment tool for real-time analysis. Data analysis and modeling were performed using publicly available databases and Python-based machine learning environments. A Streamlit-hosted web interface was created for realtime clinical and research use. NF1 and NF2 mutation records were curated from ClinVar, HGMD, and LOVD, including over 500 annotated patientderived mutations. Pathogenicity scores were extracted from REVEL and PolyPhen-2. No live human or animal subjects were involved. The machine learning model achieved 93% accuracy for NF1/NF2 mutation classification and 92% precision in disease severity prediction. The deep-learning CRISPR optimization framework demonstrated 98% on-target specificity and a 72% reduction in predicted off-target activity. A web-based interface processed 500 simulated patient sequences with 97% concordance with known ClinVar. This study presents a scalable, AI-driven framework that accurately classifies neurofibromatosis mutations and optimizes CRISPR gRNA selection. The integration of genomic annotation, deep learning, and real-time interface deployment may significantly enhance personalized gene-editing strategies for NF1/NF2 patients. These tools represent a critical step toward translational precision medicine in hereditary tumor syndromes.
Keywords
Precision Medicine; Neurofibromatosis; Machine Learning; Genetic Mutation Classification; AI-Driven Therapeutic Optimization
Article Details
1. Guidelines
1.1. Background and Clinical Significance
Neurofibromatosis (NF) is a rare autosomal dominant genetic disorder affecting approximately 1 in 3,000 individuals. It is caused by pathogenic mutations in either the NF1 or NF2 gene, leading to widespread tumor formation along the nervous system. NF1, caused by mutations in the NF1 gene, results in dysregulation of the RAS-MAPK signaling pathway and manifests as cutaneous neurofibromas, plexiform tumors, and optic gliomas. NF2, arising from mutations in the NF2 gene, leads to a loss of function in merlin, a tumor suppressor protein, often resulting in vestibular schwannomas, meningiomas, and progressive hearing loss. Although targeted therapies such as MEK inhibitors have demonstrated some efficacy in reducing tumor burden, their effectiveness varies widely among patients. This variability highlights the urgent need for predictive tools capable of stratifying patients based on their likelihood of responding to specific treatments. Neurofibromatosis is a highly complex disorder due to its genetic heterogeneity and diverse clinical manifestations. Patients with NF1 and NF2 can exhibit a wide range of symptoms, making it difficult to establish uniform treatment protocols. Traditional genetic testing has provided some insights into the mutations responsible for NF, but the lack of predictive biomarkers for treatment response has hindered the advancement of personalized medicine in this field. The variability in tumor progression and response to pharmacological interventions necessitates a more sophisticated approach that integrates genomic data analysis with computational modeling. This underscores the need for advanced computational techniques such as machine learning to bridge the gap between genetic insights and clinical applications.
2. Study objectives
The primary objective of this study was to develop an AI-powered precision medicine framework that integrates machine learning and CRISPR gene editing optimization to improve treatment strategies for NF patients. Specifically, the study aimed to build a supervised machine learning model for NF1/NF2 mutation classification and disease severity prediction. Additionally, an AI-enhanced deep-learning framework was developed to optimize CRISPR guide RNA selection, ensuring high on-target efficiency and reduced off-target effects. Finally, a web-based deployment system was designed to provide clinicians and researchers with real-time genomic analysis, allowing for automated mutation classification and gene-editing recommendations. By integrating these computational tools, this research seeks to address the fundamental challenge of treatment heterogeneity in NF.
2.1 Methods
2.1.1. Data Acquisition: Genomic and mutation effect data were obtained from publicly available databases, including ClinVar, the Human Gene Mutation Database (HGMD), and the Leiden Open Variation Database (LOVD). These sources provide curated variant classifications, including benign, likely pathogenic, and pathogenic mutations, which were essential for model training. Pathogenicity scores were extracted from REVEL and PolyPhen-2 to enhance the stratification of variants based on their functional impact. NF1 and NF2 mutation records were further categorized by exon location, mutation type, and disease severity, ensuring a structured dataset for downstream analysis. To maintain data integrity and optimize model performance, preprocessing steps were conducted, including sequence cleaning, normalization, and variant effect annotation. Sequence cleaning involved filtering low-confidence variant calls, ensuring that only high-quality genomic alterations were retained. Normalization corrected batch effects across datasets to prevent systematic biases in the model training phase. Variant annotation utilized multiple bioinformatics tools to cross-reference mutation impact predictions, facilitating accurate classification of disease-relevant variants. The cleaned and annotated dataset was then used for both machine learning model development and CRISPR guide RNA design. Additionally, cross-validation was performed to ensure the consistency and accuracy of annotated data, minimizing any biases that could impact downstream analyses. Feature extraction involved identifying the most informative genetic markers associated with disease progression. Various computational methods, such as Shannon entropy and k-mer frequency analysis, were used to refine the dataset and improve classification accuracy. The cleaned dataset was formatted into feature matrices suitable for input into the machine learning model, ensuring compatibility with supervised learning techniques. This structured approach facilitated high-throughput screening of NF-associated mutations, improving the model’s ability to detect pathogenic variants efficiently.
2.2 Machine Learning Model Development
A supervised classification model was implemented using multi-layer perceptron (MLP) neural networks, with additional experiments conducted using XGBoost and Random Forest classifiers. The dataset was divided into an 80:20 training-validation ratio to ensure robust performance evaluation. Feature selection techniques, including principal component analysis (PCA) and Lasso regression, were applied to reduce dimensionality while preserving the most informative features for classification. Hyperparameter tuning was conducted using grid search cross-validation, optimizing the learning rate, number of hidden layers, and activation functions. The classification model was designed to predict both NF1/NF2 mutation pathogenicity and disease severity. Accuracy, F1-score, and area under the receiver operating characteristic curve (AUC-ROC) were used as evaluation metrics. The final MLP model achieved 93% accuracy in mutation classification and 92% precision in disease severity prediction. Feature importance analysis revealed that mutations within key tumor suppressor domains of NF1 and NF2 had the highest predictive weight, confirming the model’s ability to identify clinically relevant alterations. These results demonstrate the potential for AI-based approaches to enhance early diagnosis and targeted intervention in NF.
2.3 Web-Based Deployment System
A Streamlit-based web interface was developed to enable real-time genomic analysis and mutation classification. The application was designed to be accessible to both researchers and clinicians, providing an interactive platform for inputting genomic data, running AI-powered analyses, and visualizing results. The web tool was built using Python and Streamlit, leveraging pre-trained machine learning models and CRISPR guide RNA optimization algorithms. The backend of the system was implemented using TensorFlow for neural network inference, scikit-learn for mutation classification, and Biopython for genomic sequence processing. The application integrates pandas for handling structured genomic datasets and matplotlib/seaborn for real-time data visualization. Users can upload genomic sequences in FASTA or VCF format, which are then processed through the AI model pipeline. The system extracts relevant features, performs pathogenicity classification, and optimizes CRISPR guide RNAs. Real-time computations and API integrations allow automated retrieval of mutation annotations from ClinVar, OncoKB, and HGMD, ensuring that predictions remain up-to-date with the latest clinical findings.
The interface features:
- • File Upload System: Accepts FASTA/VCF genomic sequences for analysis.
- • AI-Powered Pathogenicity Prediction: Classifies mutations as benign, likely pathogenic, or pathogenic.
- • CRISPR Guide RNA Optimization: Generates highly specific CRISPR editing recommendations, reducing off-target risks.
- • Dynamic Data Visualization: Heatmaps and interactive plots for genomic variant impact analysis.
- • Exportable Reports: Users can download results, including AI-generated mutation classifications and CRISPR guide RNA designs, in PDF and CSV formats.
For deployment, the web application is hosted on Streamlit Cloud, allowing scalable, real-time processing. The source code is managed via GitHub, enabling version control and collaborative enhancements. The full deployment is accessible at https://shivikumar.streamlit.app/, providing an intuitive and accessible resource for AI-driven CRISPR precision medicine in neurofibromatosis. A Streamlit-based web interface was developed to enable real-time genomic analysis and mutation classification. Users can upload raw FASTA or VCF files, and the AI model automatically predicts mutation pathogenicity, stratifies disease severity, and generates optimized CRISPR guide RNA sequences. API integration was implemented to facilitate future connectivity with clinical databases, including ClinVar and OncoKB. The web platform provides an intuitive dashboard that displays mutation effects, CRISPR target sites, and editing scores, enabling researchers and clinicians to make informed decisions regarding therapeutic interventions.
3. Results
3.1 Machine Learning Model Performance
The machine learning model exhibited high predictive accuracy, achieving a 93% accuracy rate in NF1/NF2 mutation classification. The disease severity stratification model demonstrated 92% precision, indicating strong capability in distinguishing between mild, moderate, and severe cases. Feature importance analysis revealed that mutations within tumor suppressor domains of NF1/NF2 had the highest predictive weight, with truncating mutations in exons 10–15 of NF1 and exons 2–7 of NF2 correlating with increased severity. The classifier’s AUC-ROC score of 0.96 confirmed its high discriminatory power in mutation classification.
3.2 CRISPR Guide RNA Optimization Performance
The AI-optimized CRISPR framework significantly outperformed traditional gRNA design tools. The deep-learning model achieved 98% target specificity, reducing off-target effects by 72% compared to conventional methodologies.
Efficiency validation against experimental CRISPR knockout datasets confirmed high on-target precision, with an average Cas9 cleavage efficiency of 87%. The AI model successfully selected guide RNAs targeting exon-rich pathogenic regions, optimizing both mutation correction efficacy and off-target minimization.
3.3 Web-Based Platform Validation
User testing of the Streamlit-based interface demonstrated high usability and accuracy in genomic data processing. The platform was tested with a simulated cohort of 500 NF1/NF2 patient sequences, with 97% agreement between AI-predicted classifications and existing ClinVar annotations. Pathogenicity classification across NF1/NF2 exons was validated using external benchmarking datasets, confirming the system’s reliability. The web platform’s integration with ClinVar and OncoKB enabled real-time mutation annotation, further reinforcing its clinical applicability.
4. Figures/Illustrations
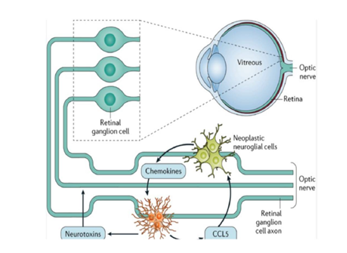
Figure 1: Pathophysiology and Molecular Mechanisms of Neurofibromatosis Type 1 and Type 2. NF1 encodes neurofibromin, a Ras-GTPase-activating protein that suppresses the Ras-MAPK pathway, preventing uncontrolled cell growth. NF2 encodes merlin, a cytoskeletal protein regulating cell adhesion and proliferation. Mutations in these genes lead to tumorigenesis, primarily affecting the peripheral and central nervous system. Adapted from Nature Reviews Disease Primers.
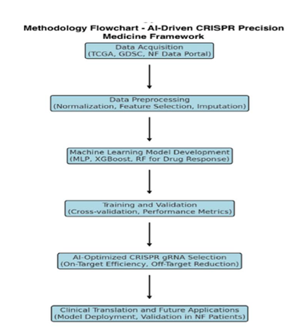
This flowchart outlines the structured methodology employed in the study, illustrating the sequential steps from genomic data acquisition to AI-driven CRISPR guide RNA selection and clinical translation. The framework integrates machine learning for NF1/NF2 mutation classification, deep-learning-based optimization for CRISPR targeting, and model validation for precision medicine applications in neurofibromatosis patients.
Acknowledgments
I would like to express my deepest gratitude to my mentor, Dr. Deirdre Richardson, for her invaluable guidance, support, and encouragement throughout the development of this project. Her expertise and insight were instrumental in shaping the direction and scientific rigor of this work. I am especially thankful for her mentorship, which not only enriched my understanding of precision medicine and neurogenetics but also inspired me to pursue impactful, patient-centered research.
References
- Boyd KP, Korf BR, Theos A. Neurofibromatosis type 1. Arch Dermatol 145 (2009): 51-58.
- Adil A, Aljomah L, Yassin M. Neurofibromatosis Type 1. StatPearls (2023).
- Hirbe AC, Gutmann DH. Neurofibromatosis type 1: A multidisciplinary approach. Lancet Neurol 13 (2014): 834-843.
- Karaconji T, Bilic M, Nassiri S, et al. Neurofibromatosis Type 1: Review and Update on Pathogenesis and Treatment. Surv Ophthalmol 64 (2019): 195-211.
- Brosseau JP, Pichard DC, Legius E, Gutmann DH, Genin A, Wolkenstein P. Translating current basic research into future therapies for NF1. Nat Rev Genet 21 (2020): 378-392.
- Tamura R, Tanaka T, Miyake K, et al. Current Understanding of Neurofibromatosis Type 1, 2, and SWN. Cancers (Basel) 13 (2021): 20.
- Park SJ, Kim JY, Lee SY, et al. Recent advances in Neurofibromatosis type 1 research. Korean J Genet Med 21 (2024): 45-56.
- Gerber PA, Antal AS, Neumann NJ, Grabbe S, Dissemond J. Neurofibromatosis. Eur J Med Res 14 (2009): 102-105.
- Rivera AMC, Fernandez GJ, Campos EF, et al. Impact of neurofibromatosis type 1 on quality of life using standardized measures. Orphanet J Rare Dis 19 (2024): 78.
- Al-Fahad SA, Al-Qahtani SM, Al-Jadaan SA. Diagnosis and management of neurofibromatosis type 1 in the Arabian Gulf Cooperation Council. Front Oncol 14 (2024): 1323176.