Multi-Omics Analyses Revealed Transcriptional Regulators associated with Immune Checkpoint Inhibitor Treatment in Advanced Bladder Cancer
Article Information
Feng Xu1#, Zuheng Wang2#, Dian Fu1#, Xiuquan Shi3#, Jie Huang2, Yuhao Chen1, Jianping Da1, Tingling Zhang1, Jingping Ge1, Xiaofeng Xu1*, Wen Cheng1,2 *
1Department of Urology, Nanjing Jinling Hospital, Nanjing University School of Medicine, Nanjing 210002, China
2Jinling Clinical Medical College, Nanjing Medical University, Nanjing, Jiangsu, 210002, China
3Medical School of Nanjing University, China
#Equal contribution to this work.
*Corresponding authors: Wen Cheng, Department of Urology, Nanjing Jinling Hospital, Nanjing University School of Medicine, Nanjing 210002, China.
Xiaofeng Xu, Department of Urology, Nanjing Jinling Hospital, Nanjing University School of Medicine, Nanjing 210002, China.
Received: 20 January 2023; Accepted: 26 January 2023; Published: 23 February 2023
Citation: Feng Xu, Zuheng Wang, Dian Fu, Xiuquan Shi, Jie Huang, Yuhao Chen, Jianping Da, Tingling Zhang, Jingping Ge, Xiaofeng Xu, Wen Cheng. Multi-Omics Analyses Revealed Transcriptional Regulators associated with Immune Checkpoint Inhibitor Treatment in Advanced Bladder Cancer. Journal of Biotechnology and Biomedicine 6 (2023): 49-66.
View / Download Pdf Share at FacebookAbstract
Abstract Background: Urothelial Bladder Cancer (UBC) is one of the most lethal cancers worldwide, the 5-year survival rate remains poor with platinum-based chemotherapy regimens as the standard of cancer treatment protocol. Recent FDA approval of a programmed death ligand-1 (PD-L1) inhibitor, atezolizumab, in advanced UBC patients is changing the therapeutic landscape. Although the response to anti-PD-L1 is correlated to PD-L1 expression and tumor mutation burden, the molecule determinants of responsiveness or non-responsiveness to Immune Checkpoint Inhibitor (ICI) is largely unknown.
Methods: R package maftools was used for genomic characterization and differential mutational analysis. EdgeR and DysRegSig algorithm were used for differential gene expression and dysregulator analysis. ConcensusTME algorithm was used for deconvolution of cell types within tumor microenvironment from bulk RNAseq data.
Result: A published immunotherapy cohort with whole exome sequencing, RNAseq and clinic outcome data for 29 metastatic urothelial cancer patients was used, paralleled with The Cancer Genome Altas (TCGA) Bladder Cancer cohort, GSE78220 cohort and MSKCC-bladder cancer cohort. Genomic mutational profiling, mutational signature, a panel genes in antigen presentation and interferon signaling in bladder cancer were delineated with potential correlation with Durable Clinic Benefit (DCB) or non-DCB of PD-L1 inhibitor treatment. Characterized immune-responsive or resistant associated genes showed differentially expressed between DCB group and non-DCB group. Furthermore, transcriptional signature and transcriptional regulators between DCB and non- DCB were identified from transcriptomic data.
Conclusion: Our exploratory analyses provide multidimensional view of complexity of molecular determinants of immune responsiveness and suggest the influences of transcriptional reprogram in i
Keywords
Antigen-Processing Machinery; Atezolizumab; Checkpoint Blockade; PD-1; PDL1; Urothelial Bladder Cancer
Antigen-Processing Machinery articles Antigen-Processing Machinery Research articles Antigen-Processing Machinery review articles Antigen-Processing Machinery PubMed articles Antigen-Processing Machinery PubMed Central articles Antigen-Processing Machinery 2023 articles Antigen-Processing Machinery 2024 articles Antigen-Processing Machinery Scopus articles Antigen-Processing Machinery impact factor journals Antigen-Processing Machinery Scopus journals Antigen-Processing Machinery PubMed journals Antigen-Processing Machinery medical journals Antigen-Processing Machinery free journals Antigen-Processing Machinery best journals Antigen-Processing Machinery top journals Antigen-Processing Machinery free medical journals Antigen-Processing Machinery famous journals Antigen-Processing Machinery Google Scholar indexed journals Atezolizumab articles Atezolizumab Research articles Atezolizumab review articles Atezolizumab PubMed articles Atezolizumab PubMed Central articles Atezolizumab 2023 articles Atezolizumab 2024 articles Atezolizumab Scopus articles Atezolizumab impact factor journals Atezolizumab Scopus journals Atezolizumab PubMed journals Atezolizumab medical journals Atezolizumab free journals Atezolizumab best journals Atezolizumab top journals Atezolizumab free medical journals Atezolizumab famous journals Atezolizumab Google Scholar indexed journals Checkpoint Blockade articles Checkpoint Blockade Research articles Checkpoint Blockade review articles Checkpoint Blockade PubMed articles Checkpoint Blockade PubMed Central articles Checkpoint Blockade 2023 articles Checkpoint Blockade 2024 articles Checkpoint Blockade Scopus articles Checkpoint Blockade impact factor journals Checkpoint Blockade Scopus journals Checkpoint Blockade PubMed journals Checkpoint Blockade medical journals Checkpoint Blockade free journals Checkpoint Blockade best journals Checkpoint Blockade top journals Checkpoint Blockade free medical journals Checkpoint Blockade famous journals Checkpoint Blockade Google Scholar indexed journals PD-1 articles PD-1 Research articles PD-1 review articles PD-1 PubMed articles PD-1 PubMed Central articles PD-1 2023 articles PD-1 2024 articles PD-1 Scopus articles PD-1 impact factor journals PD-1 Scopus journals PD-1 PubMed journals PD-1 medical journals PD-1 free journals PD-1 best journals PD-1 top journals PD-1 free medical journals PD-1 famous journals PD-1 Google Scholar indexed journals PDL1 articles PDL1 Research articles PDL1 review articles PDL1 PubMed articles PDL1 PubMed Central articles PDL1 2023 articles PDL1 2024 articles PDL1 Scopus articles PDL1 impact factor journals PDL1 Scopus journals PDL1 PubMed journals PDL1 medical journals PDL1 free journals PDL1 best journals PDL1 top journals PDL1 free medical journals PDL1 famous journals PDL1 Google Scholar indexed journals Urothelial Bladder Cancer articles Urothelial Bladder Cancer Research articles Urothelial Bladder Cancer review articles Urothelial Bladder Cancer PubMed articles Urothelial Bladder Cancer PubMed Central articles Urothelial Bladder Cancer 2023 articles Urothelial Bladder Cancer 2024 articles Urothelial Bladder Cancer Scopus articles Urothelial Bladder Cancer impact factor journals Urothelial Bladder Cancer Scopus journals Urothelial Bladder Cancer PubMed journals Urothelial Bladder Cancer medical journals Urothelial Bladder Cancer free journals Urothelial Bladder Cancer best journals Urothelial Bladder Cancer top journals Urothelial Bladder Cancer free medical journals Urothelial Bladder Cancer famous journals Urothelial Bladder Cancer Google Scholar indexed journals COSMIC database articles COSMIC database Research articles COSMIC database review articles COSMIC database PubMed articles COSMIC database PubMed Central articles COSMIC database 2023 articles COSMIC database 2024 articles COSMIC database Scopus articles COSMIC database impact factor journals COSMIC database Scopus journals COSMIC database PubMed journals COSMIC database medical journals COSMIC database free journals COSMIC database best journals COSMIC database top journals COSMIC database free medical journals COSMIC database famous journals COSMIC database Google Scholar indexed journals cancer genome articles cancer genome Research articles cancer genome review articles cancer genome PubMed articles cancer genome PubMed Central articles cancer genome 2023 articles cancer genome 2024 articles cancer genome Scopus articles cancer genome impact factor journals cancer genome Scopus journals cancer genome PubMed journals cancer genome medical journals cancer genome free journals cancer genome best journals cancer genome top journals cancer genome free medical journals cancer genome famous journals cancer genome Google Scholar indexed journals lung cancer articles lung cancer Research articles lung cancer review articles lung cancer PubMed articles lung cancer PubMed Central articles lung cancer 2023 articles lung cancer 2024 articles lung cancer Scopus articles lung cancer impact factor journals lung cancer Scopus journals lung cancer PubMed journals lung cancer medical journals lung cancer free journals lung cancer best journals lung cancer top journals lung cancer free medical journals lung cancer famous journals lung cancer Google Scholar indexed journals colorectal cancer articles colorectal cancer Research articles colorectal cancer review articles colorectal cancer PubMed articles colorectal cancer PubMed Central articles colorectal cancer 2023 articles colorectal cancer 2024 articles colorectal cancer Scopus articles colorectal cancer impact factor journals colorectal cancer Scopus journals colorectal cancer PubMed journals colorectal cancer medical journals colorectal cancer free journals colorectal cancer best journals colorectal cancer top journals colorectal cancer free medical journals colorectal cancer famous journals colorectal cancer Google Scholar indexed journals
Article Details
Simple Summary
Immune checkpoint inhibitor (ICI) therapy shifted the paradigm for advanced urothelial carcinoma (mUC) treatment, however, majority mUC patients present non-durable clinical benefit (non-DCB). Functional genomic analysis revealed key gene mutations and resistant gene expression in non-DCB group. Transcriptional reprogram reshapes the tumor microenvironment contributing to sensitivity of ICI treatment in mUC. This multi-omics study leverages our understanding of the molecular mechanisms of the intrinsic-or induced immune evasion, helps defining biomarkers for stratifying subgroup of patients for effective treatment.
1. Introduction
Urothelial Bladder Cancer (UBC) is the most common cancer from urinary tract worldwide, causing 150,000 deaths per year, and it is characterized with high rate of relapse, metastasis, and mortality [1]. Median survival for patients with recurrent or metastatic bladder cancer remains 14-15 months with cisplatin-based chemotherapy, and there is no widely recognized second-line therapy [2, 3]. Immunotherapy has played an essential role in UBC with the use of Baccille Calmette Guerin (BCG) in the treatment of non-muscle invasive bladder cancer [4]. Recent approval of a PD-L1 inhibitor, atezolizumab, in patients with locally advanced or metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy [5] or who is ineligible to cisplatin treatment [6] represents a significant milestone in treating urothelial cancer in the past 30 years. In these two clinical trials, patients were selected based on PD-L1 expression on tumor cells or immune cells, the over call objective response rate is about 15 % ~ 23%, tumor mutation burden is associated to the response [5, 6]. The molecular determinants of response and resistance to immune checkpoint inhibitors (ICIs) is a key to improving outcomes and developing new treatment strategies. Genomic and transcriptomic characteristics have been demonstrated to be correlated with response to anti- PD-1, CTLA-4 treatment in metastatic melanoma [7-9], non-small cell lung cancer [10] or colorectal cancer [11, 12] and many other types of cancers [13]. As of metastatic urothelial cancer, high neoantigen or tumor mutational burden, CD8+ T-effector cell phenotype are associated with response to treatment, a signature of transforming growth factor b (TGFb) signalling in fibroblasts is associated with non-response [14], more importantly, the balance of adaptive immunity and protumorigenic inflammation within tumor microenvironments was reported to associate with PD-1/PD-L1 inhibitor resistance [15]. Therefore, it is crucial to continuously identify biomarker(s) to stratify or predict responders to immune checkpoint blockade for better clinical outcome using different cohorts. In this study, we analyzed whole exome sequence and RNA-seq data of urothelial bladder cancer patients who received PD-L1 inhibitor treatment, and the clinical outcomes were categorized into patients with durable clinic benefit (DCB) or without durable clinic benefit (non-DCBs). The Cancer Genome Altas (TCGA), GSE78220 cohort and MSKCC-bladder cancer cohort were also used for cross-validation. We provided genomic and transcriptomic foundation to understand underlying molecular mechanisms of why only a subgroup of bladder cancer patients respond to anti-PD-L1 treatment, and have the long-lasting clinic benefit.
2. Material and Methods
- Somatic mutations shared by bladder cancer patients in COSMIC database. The frequency of the specific point mutation in each gene mutation was estimated as following: (1) genetic mutation data within the top 500 sample numbers of bladder cancer (transition cell carcinoma) were downloaded from COSMIC (GRCh38 v94). COSMIC>>Cancer Browser>> Urinary tract from Tissue selection>>Bladder from Subtissue >> Carcinoma from Histology selection>>Transitional cell carcinoma from subHistology selection.
- TCGA bladder urothelial carcinoma (BLCA) whole exome raw mutation annotation file (n=395) was downloaded from firehose broad institute (firebrowse.org), gene expression RNAseq (polyA+IlluminaHiSeq, n=426, level 3) and phenotype (n=436) was queried from UCSC Xena browser (https://xenabrowser.net/datapages/?cohort=TCGA%20Bladder%20Cancer%20(BLCA)&removeHub=https%3A%2F%2xena.
treehouse.gi.ucsc.edu%3A443). MSKCC-bladder cancer cohort (n=215) with targeted exome sequencing and PD-1/PD-L1 blockade therapy survival data [13] was downloaded from cbioportal website (https://www.cbioportal.org/study/summary?id=tmb_mskcc_2018). Data were processed in R statistic program with maftools, GenVisR packages. - Locally advanced or metastatic bladder cancer patients (n = 29) from Memorial Sloan Kettering Cancer Center were treated with atezolizumab were on protocol NCT02108652 [5]. Their whole exome sequence, bulk RNAseq and clinical information were downloaded from http://doi.org/10.5281/zenodo.546110 and https://github.com/hammerlab/multi-omicurothelialanti-pdl1. Mechanistic-driven analyses were performed on these data. GSE78220 cohort with PD-1 treatment in melanoma [8] was obtained for selected gene expression validation.
- Differentially expressed genes (DEGs). Gene expression matrix from RNAseq data of 26 advanced bladder cancer cases receiving PD-L1 inhibitor treatment were analyzed using EdgeR and limma R package with default parameters. The absolute log2FC >=1.5 and false discovery rate <=0.05 was used as cut-off, thirty-three DEGs were selected for heatmap plot using ComplexHeatmap package.
- Dysfunctional regulations of gene expression program in PD-L1 inhibitor treated urothelial bladder cancer cohort. Gene expression from DCB (n=9) and non-DCB groups (n=17) of advanced bladder cancer cases receiving PD-L1 inhibitor treatment were analysed using DysRegSig algorithm in R packages.
3. Results
3.1 Mutational Burden and Signature in Bladder Cancer
Whole exome sequencing for 391 bladder cancer samples were obtained from The Cancer Genome Atlas (TCGA), non-synonymouse mutation was retained for analysis. The total number of non-synonymous mutations in each patient is heterogeneous with median number of 173.5 (Figure 1A). Somatic point mutations have two types of DNA substitution: transitions (A ↔ G and C ↔ T) and transversions (A ↔ C, G ↔ T, A ↔ T, C ↔ G). Transition mutation is higher than transversion mutation, and C > T transition and C > G transversion are higher than other type of mutations in average (p < 0.05) and in each individual case (Figure 1B, C). We then analyzed the mutational signature against known mutational signatures [16], three predominant signatures were defined that highly correlated with APOBEC signature (signature 2, 13), aging signature (signature 1) and ultra-hypermutators with error-prone polymerase POLE somatic mutation (Signature 10). In addition, microsatellite unstable signature (Signature 6) and defect DNA mismatch repair signature (Signature 20) are also high in bladder cancer (Figure 1D). These broad mutation processes in bladder cancers corresponded with the hyper mutation rate in bladder cancer genome and correlated to the subpopulation with high PD-L1 \/CD8A expression [17]. Recently, Snyder et al [18] systematically analyzed multi-omic data from subset patients who receiving immune checkpoint inhibitor, atezolizumab, in a clinical trial cohort and demonstrated the complex nature of immune response to ICI. We take advantage of whole exome sequencing data from this subset cohort to compare the genomic difference between patients with durable clinic benefit (DCB, response lasting more than 6 months), and patients without durable clinic benefit (non-DCB). We observed that average mutation variants are higher in DCB group than non-DCB group (unpaired two sample t-test, p < 0.05) (Supplementary Figure 1), this is consistent with higher mutation burden in responders relative to non-responders [14]. Interestingly, the distribution of mutation types (T > G, T > A, T > C, C > T, C > G, C > A) in DCB and non-DCB groups are similar. We obtained top 20 frequent mutated cancer genes in bladder cancer from COSMIC (supplementary Figure 2), we did not observe COSMIC cancer mutated genes enriched in either group (Supplementary Figure 2), however, the top 20 mutated gene profiling displayed difference between DCB and non-DCB group, such as DSPP, FAM186A and NBPF10 mutations are only in non-DCB group (Supplementary Figure 3A and 3B). Moreover, pathways analysis showed increased mutated genes in each oncogenic pathways in non-DCB group, such as RTK-RAS, WNT, PI3K, MYC, TGF-beta and NRF2 pathways (Supplementary Figure 3C and 3D). These pathway activations may contribute to the resistance to ICI treatment. We also examined the MSKCC-bladder cancer cohort including clinical and targeted genomic sequencing data [13], the PIK3CA mutation significantly higher in deceased patient group relative to living patient group after ICI treatment (Supplementary Figure 4), suggesting tumors harboring PI3K/Akt pathway activation might have short clinic benefit from ICI therapy.
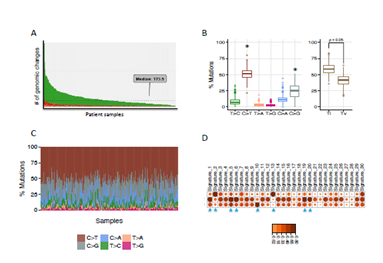
Figure 1: Mutational landscape of urothelial bladder cancer. Whole exom sequence data for TCGA-BLCA were used to investigate the tumor mutational burden (A), mutation type distribution (B, C) and somatic mutational signatures (D). Ti: Transitions, Tv: Transversions.
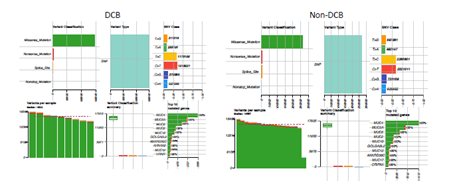
Supplementary Figure 1: Mutational landscape of pre-treatment tumor samples from advanced bladder cancer patients with durable clinical benefit (DCB) or without DCB after receiving immune checkpoint PD-L1 inhibitor (atezolizumab) therapy.
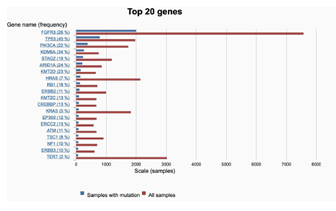
Supplementary Figure 2: The mutational frequency of top 20 cancer genes in bladder cancer derived from COSMIC. Red bar: all bladder cancer samples were tested; blue bar: samples with gene mutations.
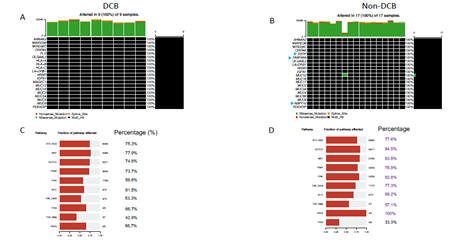
Supplementary Figure 3: Concurrent gene mutations and involved pathways in pre-treatment tumor samples from advanced bladder cancer patients with durable clinical benefit (DCB) or without DCB after receiving immune checkpoint PD-L1 inhibitor (atezolizumab) therapy. (A, B) Oncoprint of gene mutations in DCB, non-DCB group. (C, D) Pathway enrichment from non-synonymous mutated genes in DCB or non-DCB group.
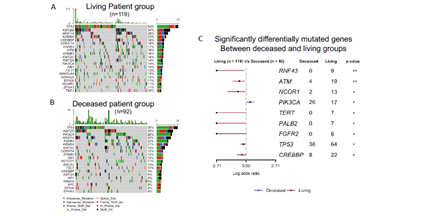
Supplementary Figure 4: Mutational landscape (A, B) and differentially mutated genes (C) between patient living and deceased group after immune checkpoint inhibitor treatment. MSKCC-bladder cancer cohort with clinical and targeted exome sequencing data were obtained from cbioportal website and maftools algorithm was used for genomic analysis.
3.2 Interferon-Receptor Signaling and Antigen Presentation Pathways Defects in Bladder Cancer
Immune checkpoint blockade therapy on melanoma, lung cancer and colorectal cancer provided underlying mechanisms of response, resistance to PD-1 or CTLA-4 inhibitors. From genomic level, JAK1, JAK2 and B2M mutations have been found associated with acquired resistance to anti-PD-1therapy in melanoma [9]. Positive selection for HLA and antigen- processing machinery mutations in tumors with TILs may have implications for potential immunotherapy resistance [12]. Castro et al reported that B2M mutation (occurring relatively early event in tumors) and HLA mutations were highly enriched in patients with microsatellite instability. In addition, these mutations had higher levels of immune infiltration by natural killer and CD8+ T cells and associated with higher levels of cytotoxicity [19]. From TCGA bladder cancer patients (n=396) with whole exome sequence data, we found 66 (~17%) patients harboured mutations associated with either MHC binding molecules (HLA gene A, B, C), antigen-processing machinery (APM) pathway or interferon receptor signalling. Interestingly, these mutations exhibit mutually exclusive mutation pattern, indicating the non-redundant roles of these genes in immune response regulation (Figure 2A). We then explored these gene mutations in the ICI-treated cohort. We found that JAK1, PDIA3, CALR, TAP1, TAPBP have higher mutation frequency in non-DCB group than in DCB group, although there is no significant statistical difference due to a small sample size (Figure 2B). Taken together, genetic alterations tend to increase in non-DCB group from ICI treatment relative to DCB group.
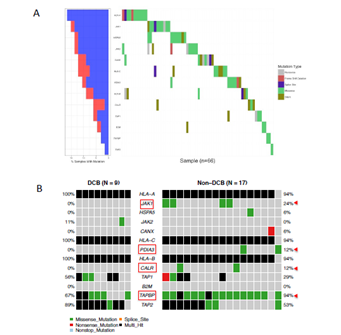
Figure 2: Mutational profiling of genes in antigen-processing machinery and interferon-receptor signalling pathway in urothelial bladder cancer. (A) The oncoprint of 19 genes participating antigen-processing machinery and interferon-receptor signalling pathway from TCGA-BLCA was presented. (B) The immune checkpoint inhibitor-treated bladder cancer cohort was grouped as durable clinical benefit (DCB) and non-durable clinical benefit (non-DCB). Target gene mutational profiling was demonstrated in each group. The color codes represent different type of mutations. Multi_Hit indicates a tumor sample contains two or more different types of mutations.
3.3 Putative Immune Checkpoint Sensitivity Gene Expressions in Bladder Cancer
From transcriptional level, if a subset of genes such as granzyme A (GZMA), perforin (PRF1, CD8 T cell cytolytic score), PDCD1LG2 (PD-L2), and CTLA4 were highly expressed in the pretreatment melanoma tumors, these patients exhibited clinical benefit from CTLA-4 antibodies treatment [20]. On the other hand, immune suppressive surface receptors including PDCD1 (PD-1), LAG3, HAVCR2 (Tim-3), CD160 and CD244 as well as transcription factors such as EOMES, PRDM1 (Blimp-1), and TBX21 (T-BET) were identified from T cell exhaustion, which is reminiscent of non-response to ICI treatment [21]. We were wondering how these gene expressions changed in ICI-treated bladder cancer patients. From bulk RNAseq data, we observed that CD8A, CD8B, PDCD1(PD-1), PRF1, GZMA, EOMES, and TBX21 highly expressed in DCB group than in non-DCB group (p< 0.2, unpaired two sample t-test) (Figure 3A). Hierarchical cluster of the correlation of these genes showed different patterns between two groups (Supplementary Figure 5). We then estimated the relative abundance of different cell types in the tumor microenvironment (TME) using bulk tumor RNAseq data with advanced concensusTME algorithm [22]. In ICI-treated cohort, among 19 different cell types within TME, the abundances of mast cells and fibroblast cells are marginally higher in non-DCB than in DCB group (p = 0.082 and p=0.16, respectively. Supplementary Figure 6). Tumor-infiltrating mast cells have recently reported to associate with resistance to anti-PD-1 therapy in humanized melanoma mouse model [23]. Tumor-associated fibroblasts could activate transforming growth factor β signalling resulting in restricting T-cell infiltration and restrain anti-tumor immunity in urothelial cancer [14]. Because the numbers of tumor-infiltrating CD8+ T cells (TILs) increase overall survival and immune checkpoint inhibitor efficacy. From RNA-seq data, the expression of CD8A (one component of the CD8 dimer) has been used as a surrogate for TIL levels [24]. Within 390 bladder cancer patients with clinical information and RNA-seq data, bladder cancer with high CD8A expression showed significantly logner overall survival time than that with lower CD8A expression (log-rank test, p < 0.05). (Figure 3B). We observed similar trend of higher expression of CD8A and higher CD8+ T cell infiltration in DCB group compared to non-DCB group. These positive findings support the TIL cells and antigen recognition by T cells in DCB group.
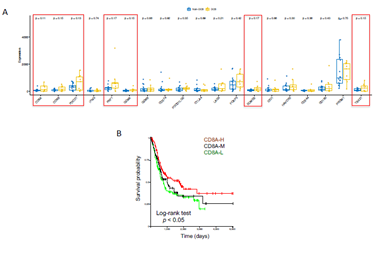
Figure 3: Immune-related gene expressions and clinic association in urothelial bladder cancer. (A) Selected immune checkpoint inhibitor treatment related gene panel of expression in durable clinic benefit (DCB) group and non-DCB group. (B) Kaplan Meier survival plot showed the level of CD8A expression (proxy of tumor infiltrating lymphocytes, TIL) and overall survival time in TCGA-BLCA cohort. Log-rank test was used for survival time difference.
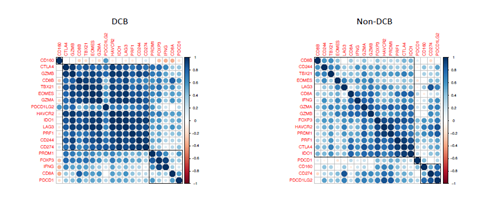
Supplementary Figure 5: The correlation of 19 immune checkpoint blockade-related gene expression in pre-treatment tumor samples from advanced bladder cancer patients with durable clinical benefit (DCB) or without DCB after receiving immune checkpoint PD-L1 inhibitor (atezolizumab) therapy. Pearson correlation coefficient indicated in color bar at right side of each heatmap.
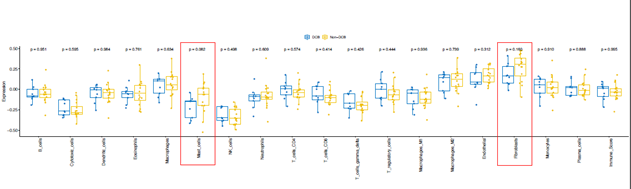
Supplementary Figure 6: The cellular components deconvoluted from RNAseq in pre-treatment tumor samples from advanced bladder cancer patients with durable clinical benefit (DCB) or without DCB after receiving immune checkpoint PD-L1 inhibitor (atezolizumab) therapy. The marginal differentially expressed cell types were highlighted with red rectangle. Blue box: DCB group, yellow box: non-DCB group.
3.4 Differentially Expressed Genes and Transcription Regulators between DCB and non-DCB
Although the ICI-treated advanced bladder cancer cohort has been analyzed by Snyder et al, but the study mainly focused on genomic alteration and T cell receptor functions [18]. We took advantage of RNAseq from pre-treated samples to examinate the differentially expressed genes (DEGs) and master regulators between patients with DCB and non-DCB. Figure 4A showed 33 DEGs (absolute log2FC >1.5 and false discovery rate < 0.1). Among them, the expressions of 9 genes (27.8%) are down-regulated in DCB and the expressions of 24 genes (72.2%) are up-regulated in DCB. These genes involved in multiple GO biological processes such as oxidative stress, metabolism and cell-cell adhesion (Figure 4B). Interestingly, there are several non-coding transcripts in DEGs, their biological functions remain elusive. In order to deeply understand the transcriptional regulation, we used DysRegSig algorithm, a machine learning-based gene dysregulation analysis [25], to explore the gene expression dysregulation between DCB and non-DCB patients, there are 184 significantly dysregulated transcription factor (TF) – targeted genes pairs (Supplementary Table 1). The dysregulated target genes are predominately enriched in immune-related signallings such as TNFs, STAT5-IL2-T cell activation, TNFR2 non-canonical NF-KB pathway, IL-18 signalling and cytokine-cytokine receptor interaction (Supplementary Table 2). Further gene regulatory network analysis revealed 10 top-ranked master transcriptional regulators (Supplementary Table 3). One of them is PATZ1-WNK4 with regulatory intensity 3.107, which is highly correlated to DCB phenotype (Figure 5A). Transcription factor PATZ1 negatively regulates the development of FOXP3 regulatory T cells [26], this may contribute to patients with favourable clinic benefit when receiving ICI treatment. We verified PATZ1-WNK4 high expression pattern in responder group from an independent ICI-treated melanoma cohort [8]. Other TF-target pairs such as TCF7L2-HOXA7, ETV6-ZNF277, and CIC-SGSM2 showed stronger regulatory intensities in DCB group than in non-DCB group (Figure 5B-D). All these TF-targets regulations tend to promote cancer cells or its microenvironments responding to ICI treatment in DCB group, the molecular mechanisms remain to be further investigated.

Figure 4: Differentially expressed genes between bladder cancer patients presenting durable clinic benefit (DCB) and non-DCB after PD-L1 inhibitor treatment. (A) Bulk RNAseq was derived from pretreatment tumor samples, the differentially expressed genes were identified based on |log2 (FC)| >= 1.5, and FDR < 0.05, and visualized in heatmap. (B) The GO terms of differentially expression genes were analyzed with goprofiler with multiple pathway databases. The selected representative GO terms were listed in the table below the plot. P-value for the enrichment < 0.05.
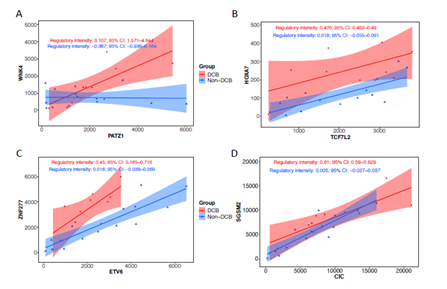
Figure 5: Representative dysregulators and targets in immune PD-L1 inhibitor treated bladder cancer. RNAseq derived from advanced bladder cancer patients with durable clinic benefit (DCB) group and non-DCB group was subjected to systematically identification of dysregulation events associated with treatment benefit using DysRegSig algorithm. The top one ranked transcription factor (PATZ1) and target (MNK4) and other transcription factorstargets (TCF7L2-HOXA7, ETV6-ZNF277, CIC-SGSM2) were highly correlated in DCB group compared to non-DCB group. The regulatory intensity and 95% confidence interval was shown in each group.
4. Discussion
Remarkable clinical efficacy, durable response and low toxicity of immune checkpoint blockade treatment have been observed in various malignancies including UBC [5, 6, 27]. Anti-PD-L1 for advanced bladder cancer could reach 43.3% response rate in early small clinical trial cohort [27], it reached >10% response rate in later large clinical trial cohort [5, 6]. However, a large proportion of patients failed to respond to checkpoint inhibitors, therefore, it is crucial to identify biomarker(s) to stratify or predict responders to achieve better clinical outcome. The molecular determinants of responsiveness to PD-1/PD-L1 and CTLA-4 inhibitors appear to be heterogeneous and complex. We take advantage of bladder cancer cohort with PD-L1 inhibitor treatment to analyze the molecular determinants of immunosensitivity or resistance in bladder cancer. First, urothelial bladder cancer is a genomic disorder with high mutation load, consistent with TCGA-bladder cancer cohort. Three dominant mutation signatures have been identified such as APOBEC, POLE signature and aging signature. The PD-L1 inhibitor (Atezolizumab) treated patients with advanced bladder cancer (n=29) exhibit limited recurrent gene mutations correlated to DCB group or non- DCB group from WES, for example, although relatively higher tumor mutation burden in DCB [14], we found several non-DCB group unique gene mutations (DSPP, FAM186A and NBPF10) are present when compared to DCB group, the functions of these gene mutations are unknown. From the pathway analysis, we observed mutated genes in each pathway increased in non-DCB group compared to DCB group (Supplementary Figure 3), orthogonal MSKCC-bladder cancer cohort showed PIK3CA mutation significantly higher in deceased patient group compared to living patient group after receiving ICI treatment. This suggesting the activation of these pathways (RTK-RAS, PI3K, MYC, TGF-beta, NRF2, Hippo) collectively contribute to unfavorable clinical outcome of ICI-treatment. Second, we found 17% bladder cancer patients from TCGA-BLCA cohort harbouring mutations involving MHC molecules, antigen processing machinery or interferon-receptor signalling pathway. These mutations exhibited mutually exclusive pattern. In the ICI-treated cohort, we observed that JAK1, TAPBP mutation frequency is relative higher in non-DCB group than CDB group (Figure 2). Third, a panel of nineteen immuno-responsive genes found in other types of cancers differentially expressed between DCB group and non-DCB group of bladder cancer patients who received ICI treatment, such as CD8A, CD8B, GZMA (granzyme A), transcription factor EOMES, TBX21 marginally higher in DCB relative to non-DCB, these gene expressions indicate functional activities of immunity (Figure 3A). Moreover, patients with high CD8A expression demonstrated longer survival time in TCGA bladder cancer cohort (Figure 3B). Finally, we identified differentially expressed genes between DCB group and non-DCB group. These transcriptional signature is able to discriminate ICI-treated clinic outcomes (Figure 4). Ten significant transcriptional regulators were further characterized from the RNAseq, a group of transcription factor-target (e.g., PATZ1-WNK4, TCF7L2- HOXA7, ETV6-ZNF277, and CIC-SGSM2) demonstrated higher regulatory intensities in DCB group relative to non-DCB group, which reflects transcriptional reprogramming in tumor cells and or within tumor microenvironment that influence the consequence of immune checkpoint blockade therapy. Transcriptional signatures are potential predictors for responding to antagonists of PD- 1. For instance, up-regulation of mesenchymal transition genes (AXL, ROR2, WNT5A, LOXL2, TWIST2, TAGLN, FAP), immunosuppressive genes (IL10, VEGFA, VEGFC) and monocyte and macrophage chemotactic genes (CCL2, CCL7, CCL8, CCL13) preferentially in non-responding tumors [8], indicating patients with these expression signature most likely do not respond to anti-PD-1 treatment. These findings highlight the complexity of interplay between cancer cells and the immune system which will need further elucidation in urothelial bladder cancer. The limitation of this analysis is that the TCGA-BLCA cohort did not have PD-1 or PD-L1 inhibitor treatment information, and the ICI-treated metastatic bladder cancer cohort contains small samples, particularly patients with durable clinic benefit group. Most the difference between DCB and non-DCB group are marginal. Nevertheless, comprehensive analyses of this valuable cohort with multi-omics and clinic data provide genomic and transcriptional insight into potential molecular mechanisms for ICI-treated durable clinical benefit. The findings remain to be confirmed in a large data in the future.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/
Authors’ Contributions
Conceived by WC, X.F.X, investigated by FX, DF, X.Q.S, JH, data collection and analysis with ZW, Y.H.C. J.P.D, T.L.Z., reviewed and edited by J.P.G.
Funding
Natural Science Foundation of China through grant 81572526 and 81972841, the fifth phase of “333 High-level Talent Cultivation Project” in Jiangsu Province (BRA2018097), Project of Jiangsu Provincial Health Committee (Z2018020), Scientific Research Project of Jiangsu Provincial Health Commission (No. M2022099).
Ethics Approval and Consent to Participate
NA.
Availability of Data and Materials
The datasets analyzed during the current study are public available from weblinks: Somatic mutations shared by bladder cancer patients in COSMIC database (https://cancer.sanger.ac.uk/cosmic/browse/tissue). TCGA-BLCA whole exome mutation annotation file was downloaded from firehose broad institute (http://firebrowse.org/?cohort=BLCA). TCGA-BLCA RNAseq data was downloaded from UCSC Xena browser (https://xenabrowser.net/datapages/?cohort=TCGA%20Bladder%20Cancer%20(BLCA)&removeHub=https%3A%2F%2Fxena.
treehouse.gi.ucsc.edu%3A443). MSKCC-bladdre cancer cohort was downloaded from cbioport website
(https://www.cbioportal.org/study/summary?id=tmb_mskcc_2018). whole exome sequence, bulk RNAseq and clinical information derived from 29 locally advanced or metastatic bladder cancer patients (Memorial Sloan Kettering Cancer Center) were downloaded from http://doi.org/10.5281/zenodo.546110 and https://github.com/hammerlab/multi-omicurothelialanti-pdl1, respectively. GSE78220 RNAseq data was downloaded from GEO website.
Acknowledgement
This research was supported by Natural Science Foundation of China through grant 81572526 and 81972841, the fifth phase of “333 High-level Talent Cultivation Project” in Jiangsu Province (BRA2018097), Project of Jiangsu Provincial Health Committee (Z2018020), Scientific Research Project of Jiangsu Provincial Health Commission(No. M2022099). The early version of the manuscript has been presented in electronic preprint (https://assets.researchsquare.com/files/rs-889171/v1_covered.pdf?c=1631878731).
Conflict of Interests
The authors declare no conflict of interest.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 65 (2015): 87-108.
- von der Maase H, Sengelov L, Roberts JT, et al. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol 23 (2005): 4602-4608.
- Sonpavde G, Sternberg CN, Rosenberg JE, et al. Second-line systemic therapy and emerging drugs for metastatic transitional-cell carcinoma of the urothelium. Lancet Oncol 11 (2010): 861-870.
- Lenis AT, Chamie K. Bladder cancer in 2014: From the genomic frontier to immunotherapeutics. Nat Rev Urol 12 (2015): 74-76.
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a singlearm, multicentre, phase 2 trial. Lancet 387 (2016): 1909-1920.
- Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 389 (2017): 67-76.
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371 (2014): 2189-2199.
- Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 165 (2016): 35-44.
- Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 375 (2016): 819-829.
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscapedetermines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348 (2015): 124-128.
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372 (2015): 2509-2520.
- Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 15 (2016): 857-865.
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51 (2019): 202-206.
- Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554 (2018): 544-548.
- Wang L, Sfakianos JP, Beaumont KG, et al. Myeloid Cell-associated Resistance to PD- 1/PD-L1 Blockade in Urothelial Cancer Revealed Through Bulk and Single-cell RNA Sequencing. Clin Cancer Res (2021).
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 500 (2013): 415-421.
- Chen S, Zhang N, Shao J, et al. Multi-omics Perspective on the Tumor Microenvironment based on PD-L1 and CD8 T-Cell Infiltration in Urothelial Cancer. J Cancer 10 (2019): 697-707.
- Snyder A, Nathanson T, Funt SA, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med 14 (2017):
- Castro A, Ozturk K, Pyke RM, et al. Elevated neoantigen levels in tumors with somatic mutations in the HLA-A, HLA-B, HLA-C and B2M genes. BMC Med Genomics 12 (2019):
- Van Allen EM, Miao D, Schilling B, et al: Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350 (2015): 207-211.
- Wherry EJ. T cell exhaustion. Nat Immunol 12 (2011): 492-499.
- Jimenez-Sanchez A, Cast O, Miller ML. Comprehensive Benchmarking and Integration of Tumor Microenvironment Cell Estimation Methods. Cancer Res 79 (2019): 6238-6246.
- Somasundaram R, Connelly T, Choi R, et al. Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy. Nat Commun 12 (2021):
- Brown SD, Warren RL, Gibb EA, et al. Neoantigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 24 (2014): 743-750.
- Li Q, Dai W, Liu J, et al. DysRegSig: an R package for identifying gene dysregulations and building mechanistic signatures in cancer. Bioinformatics 37 (2021): 429-430.
- Andersen L, Gulich AF, Alteneder M, et al. The Transcription Factor MAZR/PATZ1 Regulates the Development of FOXP3 (+) Regulatory T Cells. Cell Rep 29 (2019): 4447-4459 e4446.
- Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515 (2014): 558-562.
Supplementary Table 1: Dysregulation of Transcription factor-target gene pairs in immune checkpoint inhibitor PD-L1 treated advanced bladder cancer between durable clinic benefit (DCB) group and non-DCB group
Supplementary Table 2: Gene ontology pathway analysis.
Supplementary Table 3: Top 10 ranked master transcriptional regulators between durable clinic benefit (DCB) group and non-DCB group treated with immune checkpoint inhibitor (Atezolizumab) in advanced bladder cancer.
|
Rank |
Gene |
Dysregulation degree |
Main functions in cancer |
PMID |
|
1 |
PATZ1 |
3876.64541 |
Inhibits proliferation and induces apoptosis |
35509848 |
|
2 |
HIC2 |
1772.141003 |
a transcription repressor, |
36650953 |
|
3 |
FOXK1 |
1739.29726 |
promoting the malignant behavior |
32175400 |
|
4 |
ZNF324B |
1685.109337 |
unknown |
|
|
5 |
PAX7 |
1607.067305 |
transcription factor of the forkhead family (FKHR) are associated with alveolar rhabdomyosarcomas. |
9973247 |
|
6 |
ZNF785 |
1593.57106 |
unknown |
|
|
7 |
ELK1 |
1444.439546 |
transcription factor, promotes cancer progression |
34966781 |
|
8 |
ERF |
1254.483741 |
an ETS transcriptional repressor, acts as |
28515055 |
|
9 |
ZNF341 |
1184.604744 |
a transcription factor, low expression is |
34504527 |
|
10 |
PAX2 |
1145.368368 |
induces formation of vascular-like structures, |
35601066 |
